Abstract
We propose a new recurrent generative adversarial architecture named RNN-GAN to mitigate imbalance data problem in medical image semantic segmentation where the number of pixels belongs to the desired object are significantly lower than those belonging to the background. A model trained with imbalanced data tends to bias towards healthy data which is not desired in clinical applications and predicted outputs by these networks have high precision and low recall. To mitigate imbalanced training data impact, we train RNN-GAN with proposed complementary segmentation mask, in addition, ordinary segmentation masks. The RNN-GAN consists of two components: a generator and a discriminator. The generator is trained on the sequence of medical images to learn corresponding segmentation label map plus proposed complementary label both at a pixel level, while the discriminator is trained to distinguish a segmentation image coming from the ground truth or from the generator network. Both generator and discriminator substituted with bidirectional LSTM units to enhance temporal consistency and get inter and intra-slice representation of the features. We show evidence that the proposed framework is applicable to different types of medical images of varied sizes. In our experiments on ACDC-2017, HVSMR-2016, and LiTS-2017 benchmarks we find consistently improved results, demonstrating the efficacy of our approach.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Medical imaging plays an important role in disease diagnosis, treatment planning, and clinical monitoring [4, 24]. One of the major challenges in medical image analysis is imbalanced training sample where desired class pixels (lesion or body organ) are often much lower in numbers than non-lesion pixels. A model learned from class imbalanced training data is biased towards the majority class. The predicted results of such networks have low sensitivity, showing the ability of correctly predicting non-healthy classes. In medical applications, the cost of miss-classification of the minority class could be more than the cost of miss-classification of the majority class. For example, the risk of not detecting tumor could be much higher than referring to doctors a healthy subject.
The problem of the imbalanced class has been recently addressed in diseases classification, tumor localization, and tumor segmentation. Two types of approaches being proposed in the literature: data-level approaches and algorithm-level approaches.
At data-level, the objective is to balance the class distribution through re-sampling the data space [35, 52], by including SMOTE (Synthetic Minority Over-sampling Technique) of the positive class [10] or by under-sampling of the negative class [23]. However, these approaches often lead to remove some important samples or add redundant samples to the training set.
Algorithm-level based solutions address class imbalance problem by modifying the learning algorithm to alleviate the bias towards majority class. Examples are cascade training [8, 11], training with cost-sensitive function [47], such as Dice coefficient loss [11, 13, 41], and asymmetric similarity loss [18] that modifying the training data distribution with regards to the miss-classification cost.
In this paper, we mitigate imbalanced training samples: In data-level, we explore the advantage of training network with inverse class frequency segmentation masks, named complementary segmentation masks in addition to ground truth segmentation masks (ordinary masks) which can then be used to improve the overall prediction of the quality of the segmentation. Assume, Y is true segmentation label annotated by expert and \(\bar {Y}\) is synthesized pair of corresponding images with a complementary label. In the complementary masks \(\bar {Y}\), the majority and minority pixels value are changed to skew bias from majority pixels where the negative label for the major class and a positive label for the c − 1 class. Then, our network train with both ordinary segmentation mask Y and complementary segmentation masks \(\bar {Y}\) at the same time but in multiple loss. The final segmentation masks refine by considering ordinary and complementary mask prediction.
In algorithm-level, we study the advantage of mixing adversarial loss with categorical accuracy loss compared to traditional losses such as ℓ1 loss. Hence, image segmentation is an important task in medical imaging that attempts to identify the exact boundaries of objects such as organs or abnormal regions (e.g. tumors). Automating medical image segmentation is a challenging task due to the high diversity in the appearance of tissues among different patients, and in many cases, the similarity between healthy and non-healthy tissues. Numerous automatic approaches have been developed to speed up medical image segmentation [32]. We can roughly divide the current automated algorithms into two categories: those based on generative models and those based on discriminative models.
Generative probabilistic approaches build the model based on prior domain knowledge about the appearance and spatial distribution of the different tissue types. Traditionally, generative probabilistic models have been popular where simple conditionally independent Gaussian models [14] or Bayesian learning [33] are used for tissue appearance. On the contrary, discriminative probabilistic models, directly learn the relationship between the local features of images [3] and segmentation labels without any domain knowledge. Traditional discriminative approaches such as SVMs [2, 9], random forests [27], and guided random walks [12] have been used in medical image segmentation. Deep neural networks (DNNs) are one of the most popular discriminative approaches, where the machine learns the hierarchical representation of features without any handcrafted features [26, 51]. In the field of medical image segmentation, Ronneberger et al. [38] presented a fully convolutional neural network, named UNet, for segmenting neuronal structures in electron microscopic stacks.
Recently, GANs [15] have gained a lot of momentum in the research fraternities. Mirza et al. [28] extended the GANs framework to the conditional setting by making both the generator and the discriminator network class conditional. Conditional GANs (cGANs) have the advantage of being able to provide better representations for multi-modal data generation since there is a control over the modes of the data being generated. This makes cGANs suitable for image semantic segmentation task, where we condition on an observed image and generate a corresponding output image.
Unlike previous works on cGANs [22, 29, 48], we investigate the 2D sequence of medical images into 2D sequence of semantic segmentation. In our method, 3D bio-medical images are represented as a sequence of 2D slices (i.e. as z-stacks). We use bidirectional LSTM units [16] which are an extension of classical LSTMs and are able to improve model performance on sequence processing by enhancing temporal consistency. We use time distribution between convolutional layers and bidirectional LSTM units on bottleneck of the generator and the discriminator to get inter and intra-slice representation of features.
Summarizing, the main contributions of this paper are:
We introduce RNN-GAN, a new adversarial framework that improves semantic segmentation accuracy. The proposed architecture shows promising results for small lesions segmentation as well as anatomical regions.
Our proposed method mitigates imbalanced training data with biased complementary masks in task of semantic segmentation.
We study the effect of different losses and architectural choices that improve semantic segmentation.
The rest of the paper is organized as follows: in the next section, we review recent methods for handling imbalanced training data and semantic segmentation tasks. Section 3 explains the proposed approach for semantic segmentation, while the detailed experimental results are presented in Section 4. We conclude the paper and give an outlook on future research in Section 5.
2 Related work
This section briefs the previous studies carried out in the area of learning from imbalanced datasets, generative adversarial networks, and medical image semantic segmentation mostly in recent years.
- Handling imbalanced training dataset.:
Cascade architecture [8] and ensemble approaches [43] provided best performance on highly imbalanced medical dataset like LiTS-2017 for segmentation of very small lesion(s). Some have focused on balancing recall and precision with asymmetric loss [18], others used accuracy loss [41] and weighted the imbalanced class according to its frequency in the dataset [8, 36]. Similar to some recent work [39, 41], we mitigate the negative impact of the class imbalanced, by mixing adversarial loss and categorical accuracy loss and training deep model with complementary masks.
- Learning with complementary labels.:
Recently, the complementary labels in context of machine learning [21] has been used by assuming the transition probabilities are identical with modifying traditional one-versus-all and pairwise-comparison losses for multi-class classification. Ishida et al. [21] theoretically prove that unbiased estimator to the classification risk can be obtained by complementary labels. Yu et al. [50] study learning from both complementary labels and ordinary labels can provide a useful application for multi-class classification task. Inspired by recent success [21, 50], we train the proposed RNN-GAN with both complementary labels and ordinary labels for the task of semantic segmentation to skew the bias from majority pixels.
- Generative Adversarial Network.:
Previous works [22, 54] show the success of conditional GANs as a general setting for image-to-image translation. Some recent works applied GANs unconditionally for image-to-image translation by forcing generator to predict desired output under ℓ1 [48] or ℓ2 [31, 53] regression. Here, we study the mixing of adversarial loss in conditional setting with traditional loss and accuracy loss motivated to attenuate imbalanced training dataset. Our method also differs from the prior works [22, 25, 29, 55] by the architectural setting of the generator and the discriminator, we use bidirectional LSTM units on top of the generator and discriminator architecture to capture temporal consistency between 2D slices.
- Medical image semantic segmentation.:
The UNet has achieved promising results in medical image segmentation [38] since it allows low-level features concatenated with high-level features which provided better learning representation. Later, UNet with combination of residual network [6], in cascade of 2D and 3D [20] were used for cardiac image segmentation or heterogeneous liver segmentation [8]. The generator network in RNN-GAN, is modified UNet where high resolution features are concatenated with up-sampled of global low-resolution features to help the network learn both local and global information.
3 Method
In this section we present the recurrent generative adversarial network for medical image semantic segmentation. To tackle with miss-classification cost and mitigate imbalanced pixel labels, we mixed adversarial loss with categorical accuracy loss Section 3.1. Moreover, we explain our intuition for skewing the biased from majority pixels with proposed complementary labels Section 3.2.
3.1 Recurrent generative adversarial network
In a conventional generative adversarial network, generative model G tries to learn a mapping from random noise vector z to output image y; G : z → y. Meanwhile, a discriminative model D estimates the probability of a sample coming from the training data xreal rather than the generator xfake. The GAN objective function is two-player mini-max game with value function V (G, D):
In a conditional GAN, a generative model learns the mapping from the observed image x and a random vector z to the output image y; G : x, z → y. On the other hand the D attempts to discriminate between generator output image and the training set images. According to the (2), in the cGANs training procedure both G and D are conditioned on desired output y.
More specifically, in our proposed RNN-GAN network, a generative model learns the mapping from a given sequence of 2D medical images xi to the semantic segmentation of corresponding labels \(y_{i_{seg}}\); \(G : {x_{i},z} \rightarrow \{y_{i_{seg}}\}\) (where i refers to 2D slices index between 1 and 20 from a total 20 slices acquired from ACDC-2017). The training procedure for the semantic segmentation task is similar to two-player mini-max game (3). While the generator predicted segmentation in pixel level, the discriminator takes the ground truth and the generator’s output to determine whether predicted label is real or fake.
We mixed the adversarial loss with ℓ1 distance (4) to minimize the absolute difference between the predicted value and the existing largest value. Hence the ℓ1 objective function takes into account CNN features and differences between the predicted segmentation and the ground truth, resulting in less noise and smoother boundaries.
where j and i indicate the number of semantic classes and the number of 2D slices for each patients respectively.
Moreover, we mixed categorical accuracy loss ℓacc, (5), in order to mitigate imbalanced training data by assigning a higher cost to the less represented set of pixels, boosting its importance during the learning process. Categorical accuracy loss checks whether the maximal true value is equal to the maximal predicted value regarding each category of the segmentation.
Then, the final adversarial loss for semantic segmentation task by RNN-GAN is calculated through (6).
In this work, similar to the work of Isola et al. [22], we used Gaussian noise z in the generator alongside the input data x. As discussed by Isola et al. [22], in training procedure of conditional generative model from conditional distribution P(y|x), that would be better, a trained model produces more than one sample y, from each input x. When the generator G, takes plus input image x, random vector z, then G(x, z) can generate as many different values for each x as there are values of z. Specially for medical image segmentation, the diversity of image acquisition methods (e.g., MRI, fMRI, CT, ultrasound), regarding their settings (e.g., echo time, repetition time), geometry (2D vs. 3D), and differences in hardware (e.g., field strength, gradient performance) can result in variations in the appearance of body organs and tumour shapes [19], thus learning random vector z with input image x makes network robust against noise and act better in the output samples. This has been confirmed by our experimental results using datasets having a large range of variation.
3.2 Complementary label
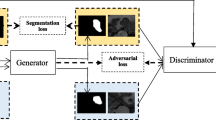
In order to mitigate the impact of imbalanced pixels labels on medical images, the proposed RNN-GAN as described in Fig. 1, is trained with complementary mask (Fig. 2, third column) in addition of the ordinary masks (Fig. 2, columns 4–6). Similar to Yu et al. [50], we assumed transition probabilities are identical then the adversarial loss (i.e. categorical cross entropy loss) provides an unbiased estimator for minimizing the risk. Since we have the same assumption we skip the proof of theoretical side and here we experimentally show that complementary labels in addition of ordinary losses are able to provide more accurate results for a task of semantic segmentation.
The architecture of RNN-GAN consists of two deep networks: a generative network G and a discriminative network D. G takes sequence of 2D images as a condition and generates the sequence of 2D semantic segmentation outputs, D determines whether those outputs are real or fake. RNN-GAN captures inter and intra-slice feature representation with bidirectional LSTM units on bottleneck of both G and D network. Here, G is modified UNet architecture and D is fully convolutional encoder
The chest MR image, from ACDC-2017 after pre-processing. The first column is semantic segmentation mask correspond to MR images in second column. Columns 3-6 present complementary labels mask, right ventricle, myocardium vessel, and left ventricle where we map 2D images from second column into four segmentation masks presented in columns 3-6
3.3 Network architecture
The proposed architecture is shown in Fig. 1, where the generator network G in the left followed by the discriminator network D in the right side of the figure. We design bidirectional LSTM units on circumvent bottleneck of both G and D, to capture the non-linear relationship between previous, current, and next 2D slices which is important key to process sequential data.
3.3.1 Recurrent generator
The recurrent generator takes a random vector z plus sequence of 2D medical images. Similar to the UNet architecture, we added skip connections between each layer r and the corresponding layer t − 1 − r, where t represents the total number of layers. Each skip connection simply concatenates all channels at layer r with those at layer t − 1 − r. Feature maps from the convolution part in the down-sampling step are fed into the up-convolution part in the up-sampling step. The generator is trained on a sequence input images from same patient and same acquisition plane. We use the convolutional layer with kernel size 5 × 5 and stride 2 for down-sampling, and perform up-sampling by the image resize layer with a factor of 2 and convolutional layer with kernel size 3 × 3 and stride 1.
3.3.2 Recurrent discriminator
The discriminator network is a classifier and has similar structure as an encoder of the generator network. Hierarchical features are extracted from fully convolutional encoder of discriminator and used to classify between the generator segmentation output and ground truth. More specifically, the discriminator is trained to minimize the average negative cross-entropy between predicted and the true labels.
Then, two models are trained through back propagation corresponding to a two-player mini-max game (see (3)). We use categorical cross entropy [30] as an adversarial loss. In this work, the recurrent architecture selected for both discriminator and generator is a bidirectional LSTM [16].
4 Experiments
We validated the performance of RNN-GAN on three recent public medical imaging challenges: real patient data obtained from the MICCAI 2017, automated cardiac MRI segmentation challenge (ACDC-2017) [5], CT liver tumour segmentation challenge (LiTS-2017), and the 2016 whole-heart and great vessel segmentation challenge (HVSMR).
4.1 Datasets and pre-processing
Our experiments are based on three independent datasets consisting of two cardiac MR images, and an abdomen CT dataset that all segmented manually by radiologists at pixel level.
- ACDC.:
The ACDC datasetFootnote 1 comprised of 150 patients with 3D cine-MR images acquired in a clinical routine. The training database was composed of 100 patients. For all these data, the corresponding manual references were given by a clinical expert. The testing database consisted of 50 patients without manual references. Figure 3 shows a cardiac MR images from the ACDC dataset.
- HVSMR.:
Thirty training cine MRI scans from 10 patients were provided by the organizers of the HVSMR challenge.Footnote 2 Three images were provided for each patient: a complete axial cine MRI, the same image cropped around the heart and the thoracic aorta, and a cropped short-axis reconstruction.
- LiTS.:
In third experiment, we applied the LiTS-2017 benchmarkFootnote 3 that comprised of 130 CT training and 70 test subjects. The examined patients were suffering from different liver cancers. The challenging part is segmentation of very small lesion target on a high unbalanced dataset. Here, pre-processing is carried out in a slice-wise fashion. We applied Hounsfield unit (HU) values, which were windowed in the range of [100, 400] to exclude irrelevant organs and objects as shown in Fig. 4. Furthermore, we applied histogram equalization to increase the contrast for better differentiation of abnormal liver tissue.
- Pre-processing of MR images.:
The gray-scale distribution of MR images is dependent on the acquisition protocol and the hardware. This makes learning difficult since we expect to have the same data distribution from one subject to another. Therefore, pre-processing is an important step toward bringing all subjects under similar distributions. We applied a bias field correction on the MR images from HVSMR and ACDC datasets to correct the intensity non-uniformity using N4ITK [42]. Lastly, we applied histogram matching normalization on the all 2D slices from sagittal, coronal, and axial planes.
The cardiac MR image, from ACDC 2017 after pre-processing left side image shows end of systolic sample and right side is end of diastolic phase. We extracted complementary mask from inverse of ground truth file annotated by medical expert, presented in the second and seventh column. Other binary masks extracted from ground truth file in columns 3-5 and 8-10 respectively are right ventricles, myocardium vessel, and left ventricles which they are used by the discriminator. The first and sixth columns are an example input of the generator
4.2 Implementation and configuration
The RNN-GAN architecture is implemented based on Keras [7] and TensorFlow [1] library. The implemented code is available on the author GitHub.Footnote 4 All training was conducted on a workstation equipped with NVIDIA TITAN X GPU.
The model was trained for up to 120 epochs with batch size 10, iteration 450 and initial learning rate 0.001 on ACDC dataset. Similarly, in HVSMR, we had initial learning rate 0.001, batch size 10, iteration 2750, and 100 epochs where we used all 2D slices from coronal, sagittal, and axial planes with size 256 × 256. The generator and discriminator for all layers use the tanh activation function except the output layer which uses softmax. We use categorical cross-entropy as an adversarial loss mixed with categorical accuracy and ℓ1. The RMSprop optimizer was used in both the generator and the discriminator. The RMSprop divides the learning rate by an exponentially decaying average of squared gradients.
The training took eight hours on ACDC for a total of 120 epochs on parallel NVIDIA TITAN X GPUs and with same configuration, it was 12 hours on HVSMR dataset. With this implementation, we are able to produce a cardiac segmentation mask between 500-700 ms per patient on same cardiac phase from ACDC dataset on an axial plane.
The proposed approach is trained on 75% training data released by the HVSMR-2016 and LiTS-2017 benchmarks. We used all provided images from three axes of sagittal, coronal, and axial for training, validation and testing. We trained our system on 75 exams from axial, coronal, and sagittal plane and validated it on the remaining 25 exams for the ACDC dataset.
In both the training and testing phase, the mini-batch consists of 2D images from the same patient, the same acquisition plane and same cardiac phase. We initially normalize the inputs where the mean and variance are computed on a specific patient from the same acquisition plane and from all available images in the same cardiac phase (ED, ES). This normalization helps to restrict the effect of outliers. With batch norm, we normalized the inputs (activations coming from the previous layer) going into each layer using the mean and variance of the activations for the entire mini-batch.
Let us mention that Wolterink’s method (using an ensemble of six trained CNNs) took 4 seconds to compute predictions mask per patient with a system equipped NVIDIA TITAN X GPU in ACDC benchmark as reported in [46], while the RNN-GAN took 500 ms in average per patient with a system equipped single of NVIDIA TITAN X GPUs.
4.3 Evaluation criteria
The evaluation and comparison performed using the quality metrics introduced by each challenge organizer. Semantic segmentation masks were evaluated in a five-fold cross-validation. For each patient, a corresponding images for the End Diastolic (ED) instant and for the End Systolic (ES) instant has provided. As described by ACDC-2017, cardiac regions are defined as right-ventricle region labeled 1, 2 and 3 representing respectively myocardium and left ventricles. In order to optimize the computation of the different error measures, the Dice coefficient (7) and Hausdorff distance (8) python script code were obtained from the ACDC for all participants.
The average distance boundary (ADB) in addition Dice and Hausdorff considered for evaluating the blood pool and myocardium in HVSMR-2016 and similarly, for validating of liver lesions segmentation on LiTS-2017. Besides these parameters, we calculated sensitivity and specificity since they are a good indicator for miss-classified rate (false positives and false negatives) (see Tables 5 and 6).
where P and T indicates predicted output by our proposed method and ground truth annotated by medical expert respectively.
4.4 Comparison with related methods and discussion
As shown in Table 1, our method outperforms other top-ranked approaches from the ACDC benchmark. Based on Table 1, in Dice coefficient, our method achieved slightly better than the Wolterink et al. [46] on ACDC challenge in left ventricle and myocardium segmentation. However, Rohe et al. [37] achieved outstanding performance for right ventricle segmentation since they applied the multi-atlas registration and segmentation at the same time. Poudel et al. [34] achieved competitive results on left ventricle segmentation with overall Dice 0.93, based on recurrent fully convolutional networks.
Based on Tables 1 and 2, the right ventricle is a difficult organ for all the participants mainly because of its complicated shape, the partial volume effect close to the free wall, and intensity of homogeneity. Our achieved accuracy in term of Hausdorff distance, in average is 1.2 ± 0.2mm lower than other participants. This is a strong indicator for precision of boundary that RNN-GAN architecture substituted with bidirectional LSTM units is suitable solution for capturing the temporal consistency between slices. Compared to cGAN (Tables 1 and 2) RNN-GAN provides better results when the network is trained with complementary segmentation mask and even sensitivity and precision.
Compared to the expert annotated file on the original ED phase instants, individual Dice scores of 0.968 for the left ventricle (LV), 0.933 for the myocardium (MYO), and 0.940 for the right ventricle (RV) (see Table 1) were achieved in test time on 25 patients. Qualitatively, the RNN-GAN segmentation results are promising (see Fig. 5 and 7) where we can see robust and smooth boundaries for all substructures.
The cardiac segmentation results at test time by RNN-GAN from ACDC 2017 benchmark on Patient084. The red, green, and blue contour present respectively right ventricle, myocardium, and left ventricle region. The top two rows show the diastolic phase from different slices from t = 0 till t = 9 circle. Respectively the third and fourth rows present systolic cardiac phase from t = 0 till t = 9 circle
We report the effect of different losses for RNN-GAN in Table 3. As we expected, the best performance obtained when the network was trained with mixing of categorical cross-entropy (as adversarial loss) with ℓ1 and categorically accuracy. Using an ℓ1 loss encourages the output respect the input, since the ℓ1 loss penalizes the distance between ground truth outputs, which match the input and synthesized outputs. Using categorical accuracy force the network to assign a higher cost to less represented set of objects, by boosting its importance during the learning process.
As depicted on Fig. 5 and Table 1 right ventricle is complex organ to segment. The most failure happened in systolic phase. Based on Fig. 5 the achieved accuracy in the test time on ACDC benchmark, we observed that the average results in diastolic phase (first and second rows) are better than the average results on systolic phase (third and fourth rows). We evaluated quantitatively the results using Hausdorff distance and Dice as shown in Fig. 6. As expected, the achieved Dice score on left ventricle (median of 6.82/8.02 for the ED/ES frames) tend to be lower than for the two other regions of interest with myocardium at 8.08/8.69 and right ventricle at 8.95/12.07 for ED/ES.
The ACDC 2017 challenge results using RNN-GAN and cGAN architecture. The left figure shows Dice coefficient in two cardiac phase as follows the right sub figure presents Hausdorff distance. The y-axis shows the Dice metrics and x-axis shows segmentation performance based on cGAN and RNN-GAN in ED and ES cardiac phase. In each sub figure, the mean is presented in red. The ACDC 2017 challenge results using RNN-GAN and cGAN architecture. The sub figure (b) y-axis codes the Hausdorff distance in mm and x-axis presents segmentation performance based on cGAN and RNN-GAN in ED and ES cardiac phase
Based on Tables 4, 5 and Fig. 7, the results show good relation to the ground truth for the blood pool. The average value of the Dice index is around 0.94. The main source of error here is the inability of the method to completely segment all the great vessels where the average Dice score is 0.86. Regarding the results on Tables 4 and 5, by comparing the first and second row the achieved accuracy is better when we conditional GAN substituted with bidirectional LSTM units. These architecture provide a better representation of features by capturing spatial-temporal information in forward and backward dependency. In this context, Poudel et al [34] designed unidirectional LSTMs on top of UNet architecture to capture inter-intra slice features and achieved competitive results for segmentation of left ventricle.
The cardiac segmentation results in test time by RNN-GAN from HVSMR 2016 benchmark. The top row shows the predicted output by RNN-GAN and the second row presents the corresponding ground truth annotated by medical expert. The contour with cyan colour describes blood pool and dark blue shows the myocardium region
The qualitative results of liver tumour segmentation are presented in Fig. 8. Based on Fig. 8 and Table 6, RNN-GAN is able to detect complex and heterogeneous structure of all lesions. The RNN-GAN architecture trained with complementary masks yielded better results and trade off between Dice and sensitivity. Dice score is a good measure for class imbalance where indicate the true positive rate by considering false negative and false positive pixels. The effect of class balancing can be seen with comparison of first and second row of Table 6. As we expected the RNN-GAN trained with complementary segmentation labels and binary segmentation masks computed more accurate result with average 3% and 6% improvement respectively in Dice and sensitivity.
We compared predicted results by RNN-GAN at test time with other top-ranked and related approaches on LiTS-2017 in terms of volume overlap error (VOE), relative volume difference (RVD), average symmetric surface distance (ASD), and maximum surface distance or Hausdorff distance (HD), as introduced by challenge organizer. As depicted results in Table 6 cascade UNet [8] or ensemble network [6, 17] architectures has achieved better performance compared to trained only with fully convolutional neural network (FCN) [44]. In contrast to prior work such as [6, 8, 17], our proposed method could be generalized to segment the very small lesion and also multiple organs in medical data in different modalities.
5 Conclusion
In this paper, we introduced a new deep architecture to mitigate the issue of imbalanced pixel labels in the task of medical image segmentation task. To this end, we developed a recurrent generative adversarial architecture named RNN-GAN, consists of two architecture: a recurrent generator and a recurrent discriminator. To mitigate imbalanced pixel labels, we mixed adversarial loss with categorical accuracy loss and train the RNN-GAN with ordinary and complementary masks. Moreover, we analyzed the effects of different losses and architectural choices that help to improve semantic segmentation results. Our proposed method shows outstanding results for segmentation of anatomical regions (i.e. cardiac image semantic segmentation). Based on the segmentation results on two cardiac benchmarks, the RNN-GAN is robust against slice misalignment and different CMRI protocols. Experimental results reveal that our method produces an average Dice score of 0.95. Regarding the high accuracy and fast processing speed, we think it has the potential to use for the routine clinic task. We validated also the RNN-GAN on tumor segmentation based on abdomen CT images and achieved competitive results on LiTS benchmark.
The impact of learning from complementary labels from different imbalanced ratio may also be useful in the context of semantic segmentation. We will investigate this issue in the future. In term of application, we plan to investigate the potential of RNN-GAN network for learning multiple clinical tasks such as diseases classification and semantic segmentation.
References
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viégas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, Zheng X (2015) TensorFlow: Large-scale machine learning on heterogeneous systems. https://www.tensorflow.org/. Software available from tensorflow.org
Afshin M, Ayed IB, Punithakumar K, Law M, Islam A, Goela A, Peters T, Li S (2014) Regional assessment of cardiac left ventricular myocardial function via mri statistical features. IEEE Trans Med Imaging 33(2):481–494
Avola D, Cinque L (2008) Encephalic nmr image analysis by textural interpretation. In: Proceedings of the 2008 ACM symposium on applied computing, pp 1338–1342. ACM
Avola D, Cinque L, Di Girolamo M (2011) A novel t-cad framework to support medical image analysis and reconstruction. In: International conference on image analysis and processing, pp 414–423. Springer
Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng PA, Cetin I, Lekadir K, Camara O, Ballester MAG et al (2018) Deep learning techniques for automatic mri cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Transactions on Medical Imaging
Bi L, Kim J, Kumar A, Feng D (2017) Automatic liver lesion detection using cascaded deep residual networks. arXiv:1704.02703
Chollet F et al (2015) Keras
Christ PF, Ettlinger F, Grun F, Elshaer MEA, Lipkova J, Schlecht S, Ahmaddy F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Hofmann F, D’Anastasi M, Ahmadi S, Kaissis G, Holch J, Sommer WH, Braren R, Heinemann V, Menze BH (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv:1702.05970
Ciecholewski M (2011) Support vector machine approach to cardiac spect diagnosis. In: International workshop on combinatorial image analysis, pp 432–443. Springer
Douzas G, Bacao F (2018) Effective data generation for imbalanced learning using conditional generative adversarial networks. Expert Syst Appl 91:464–471
Drozdzal M, Chartrand G, Vorontsov E, Shakeri M, Di Jorio L, Tang A, Romero A, Bengio Y, Pal C, Kadoury S (2018) Learning normalized inputs for iterative estimation in medical image segmentation. Med Image Anal 44:1–13
Eslami A, Karamalis A, Katouzian A, Navab N (2013) Segmentation by retrieval with guided random walks: application to left ventricle segmentation in mri. Med Image Anal 17(2):236– 253
Fidon L, Li W, Garcia-Peraza-Herrera LC, Ekanayake J, Kitchen N, Ourselin S, Vercauteren T (2017) Generalised wasserstein dice score for imbalanced multi-class segmentation using holistic convolutional networks. In: International MICCAI Brainlesion workshop, pp 64–76. Springer
Fischl B, Salat DH, Van Der Kouwe AJ, Makris N, Ségonne F, Quinn BT, Dale AM (2004) Sequence-independent segmentation of magnetic resonance images. Neuroimage 23:S69–S84
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative Adversarial Networks ArXiv e-prints
Graves A, Schmidhuber J (2005) Framewise phoneme classification with bidirectional lstm and other neural network architectures. Neural Netw 18(5-6):602–610
Han X (2017) Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv:1704.07239
Hashemi SR, Salehi SSM, Erdogmus D, Prabhu SP, Warfield SK, Gholipour A (2018) Tversky as a loss function for highly unbalanced image segmentation using 3d fully convolutional deep networks. arXiv:1803.11078
Inda Maria-del-Mar RB, Seoane J (2014) Glioblastoma multiforme: A look inside its heterogeneous nature. In: Cancer archive 226-239
Isensee F, Jaeger PF, Full PM, Wolf I, Engelhardt S, Maier-Hein K H (2017) Automatic cardiac disease assessment on cine-mri via time-series segmentation and domain specific features. In: International workshop on statistical atlases and computational models of the heart, pp 120–129. Springer
Ishida T, Niu G, Hu W, Sugiyama M (2017) Learning from complementary labels. In: Advances in neural information processing systems, pp 5639–5649
Isola P, Zhu JY, Zhou T, Efros AA (2017) Image-to-image translation with conditional adversarial networks. In: The IEEE conference on computer vision and pattern recognition (CVPR)
Jang J, Eo T, Kim M, Choi N, Han D, Kim D, Hwang D (2014) Medical image matching using variable randomized undersampling probability pattern in data acquisition. In: 2014 international conference on electronics, information and communications (ICEIC), pp 1–2. https://doi.org/10.1109/ELINFOCOM.2014.6914453
Kaur R, Juneja M, Mandal A (2018) A comprehensive review of denoising techniques for abdominal ct images. Multimedia Tools and Applications pp 1–36
Kohl S, Bonekamp D, Schlemmer H, Yaqubi K, Hohenfellner M, Hadaschik B, Radtke J, Maier-Hein KH (2017) Adversarial networks for the detection of aggressive prostate cancer. arXiv:1702.08014
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521 (7553):436–444
Mahapatra D (2014) Automatic cardiac segmentation using semantic information from random forests. J Digit Imaging 27(6):794–804
Mirza M, Osindero S (2014) Conditional generative adversarial nets. arXiv:1411.1784
Moeskops P, Veta M, Lafarge MW, Eppenhof KAJ, Pluim JPW (2017) Adversarial training and dilated convolutions for brain MRI segmentation. arXiv:1707.03195
Nasr GE, Badr E, Joun C (2002) Cross entropy error function in neural networks: Forecasting gasoline demand. In: FLAIRS conference, pp 381–384
Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA (2016) Context encoders: Feature learning by inpainting. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2536–2544
Peng P, Lekadir K, Gooya A, Shao L, Petersen SE, Frangi AF (2016) A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging. Magn Reson Mater Phys, Biol Med 29(2):155–195
Pohl KM, Fisher J, Grimson WEL, Kikinis R, Wells WM (2006) A bayesian model for joint segmentation and registration. Neuroimage 31(1):228–239
Poudel RP, Lamata P, Montana G (2016) Recurrent fully convolutional neural networks for multi-slice mri cardiac segmentation. In: Reconstruction, segmentation, and analysis of medical images, pp 83–94. Springer
Prabhu V, Kuppusamy P, Karthikeyan A, Varatharajan R (2018) Evaluation and analysis of data driven in expectation maximization segmentation through various initialization techniques in medical images. Multimed Tools Appl 77(8):10375–10390
Qiu Q, Song Z (2018) A nonuniform weighted loss function for imbalanced image classification. In: Proceedings of the 2018 international conference on image and graphics processing, pp 78–82. ACM
Rohé MM, Sermesant M, Pennec X (2017) Automatic multi-atlas segmentation of myocardium with svf-net. In: Statistical atlases and computational modeling of the heart (STACOM) workshop
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 234–241. Springer International Publishing
Rota Bulo S, Neuhold G, Kontschieder P (2017) Loss max-pooling for semantic image segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2126– 2135
Shahzad R, Gao S, Tao Q, Dzyubachyk O, van der Geest R (2016) Automated cardiovascular segmentation in patients with congenital heart disease from 3d cmr scans: combining multi-atlases and level-sets. In: Reconstruction, segmentation, and analysis of medical images, pp 147–155
Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso M J (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, pp 240–248. Springer
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4itk: improved n3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Vorontsov E, Tang A, Pal C, Kadoury S (2018) Liver lesion segmentation informed by joint liver segmentation. In: 15th IEEE international symposium on biomedical imaging (ISBI 2018), pp 1332– 1335
Vorontsov E, Tang A, Pal C, Kadoury S (2018) Liver lesion segmentation informed by joint liver segmentation. In: 15th IEEE international symposium on biomedical imaging (ISBI 2018), pp 1332–1335
Wolterink JM, Leiner T, Viergever MA, Išgum I (2016) Dilated convolutional neural networks for cardiovascular mr segmentation in congenital heart disease. In: Reconstruction, segmentation, and analysis of medical images, pp 95–102. Springer
Wolterink JM, Leiner T, Viergever MA, Isgum I (2017) Automatic segmentation and disease classification using cardiac cine mr images. arXiv:1708.01141
Xu J, Schwing AG, Urtasun R (2014) Tell me what you see and i will show you where it is. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3190–3197
Xue Y, Xu T, Zhang H, Long LR, Huang X (2017) Segan: Adversarial network with multi-scalel1 loss for medical image segmentation. arXiv:1706.01805
Yu L, Yang X, Qin J, Heng PA (2016) 3d fractalnet: dense volumetric segmentation for cardiovascular mri volumes. In: Reconstruction, segmentation, and analysis of medical images, pp 103–110. Springer
Yu X, Liu T, Gong M, Tao D (2018) Learning with biased complementary labels. In: The european conference on computer vision (ECCV)
Zhang YD, Muhammad K, Tang C (2018) Twelve-layer deep convolutional neural network with stochastic pooling for tea category classification on gpu platform. Multimedia Tools and Applications pp 1–19
Zhang YD, Zhao G, Sun J, Wu X, Wang ZH, Liu HM, Govindaraj VV, Zhan T, Li J (2017) Smart pathological brain detection by synthetic minority oversampling technique, extreme learning machine, and jaya algorithm. Multimedia Tools and Applications pp 1–20
Zhou Y, Berg TL (2016) Learning temporal transformations from time-lapse videos. In: European conference on computer vision, pp 262–277
Zhu J Y, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: The IEEE international conference on computer vision (ICCV)
Zhu W, Xie X (2016) Adversarial deep structural networks for mammographic mass segmentation. arXiv:1612.05970
Zotti C, Luo Z, Humbert O, Lalande A, Jodoin PM (2017) Gridnet with automatic shape prior registration for automatic mri cardiac segmentation. arXiv:1705.08943
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rezaei, M., Yang, H. & Meinel, C. Recurrent generative adversarial network for learning imbalanced medical image semantic segmentation. Multimed Tools Appl 79, 15329–15348 (2020). https://doi.org/10.1007/s11042-019-7305-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-7305-1