Abstract
Breast cancer continues to be one of the major health issues across the world and it is mostly observed in females. However, the actual cause of this cancer is still an ongoing research topic. Hence, early detection and diagnosis of breast cancer are considered to be an effective and reliable solution. Mammography is one of the most efficacious medical tools for early detection of breast cancer. The radiologists identify the suspicious regions in the breast by carefully examining the mammograms. However, mammograms are sometimes difficult to analyze when the breast tissues are dense. Therefore, a computer-aided diagnosis (CAD) system is adopted which can improve the decisions of the radiologists. This paper proposes a hybrid CAD framework to classify the suspicious regions into either normal or abnormal, and further, benign or malignant. The proposed framework constitutes four computational modules, namely, ROI generation using cropping operation, texture feature extraction using contourlet transformation, a wrapper-based feature selection algorithm, namely, forest optimization algorithm to select the optimal features, and finally different classifiers like SVM, k-NN, Naïve Bayes, and C4.5 that are employed to classify the inputs into normal or abnormal, and again benign or malignant. The proposed framework is examined on two widely used standard datasets, namely, MIAS and DDSM. The performance measures are computed with respect to normal vs. abnormal, and benign vs. malignant for four different hybrid CAD models, namely, (Contourlet + FOA + SVM), (Contourlet + FOA + k-NN), (Contourlet + FOA + Naïve Bayes), and (Contourlet + FOA + C4.5). The highest classification accuracy of 100% is achieved for normal vs. abnormal classification in case of both MIAS and DDSM. The performance of the proposed hybrid scheme demonstrates its effectiveness with the other state-of-the-art schemes. Experimental results reveal that the proposed hybrid scheme is accurate and robust. Finally, the suggested scheme is considered as a reliable CAD framework to help the physicians for better diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In recent years, cancer has become a decisive significant health problem and is a major cause of the high mortality rate. According to the statistics of GLOBOCAN reported in 2012, 14.1 million new cancer cases and 8.2 million cancer-causing deaths ensued, as compared to 12.7 million and 7.6 million, respectively, in 2008 [36]. In the upcoming years, the death rate due to cancer can be a serious issue in the society. Cancer is the second leading reason for the death rate across the world and was accountable for 8.8 million number of deaths in 2015. Among all types of existing cancers, breast cancer is considered to be one of the major health issues and leading causes of deaths. It is especially a threat to women at the age of 40 or above. Breast cancer is not only seen in females but also affects the males. According to the American Cancer Society (ACS), in 2015, 40,730 death cases were estimated due to breast cancer [32]. Hence, it is of utmost importance to reduce the breast cancer mortality rate through timely detection and effective treatment.
With the invent of modern medical science and information technology, medical imaging techniques are playing a vital role in the early detection and diagnosis of breast cancer. The screen-film mammography is a widely accepted mechanism to identify the breast cancer at its early stage. It examines the breast through imaging technique to provide breast related information such as its morphology, anatomy, and so on. In this process, two types of views are acquired, namely, Medio-lateral oblique (MLO) and Cranio-caudal (CC) for both the right and left breasts. The illustrations of the two views are shown in Fig. 1a and b, respectively. The MLO view of breast projects more breast tissue than that of the CC view because of the slope of the chest wall. These views of the breast enhance the breast tissues as a result of which the probability of detection of suspicious regions is increased. The digital mammograms are always examined by trained and experienced radiologists. The radiologists consider both the views for more accurate identification of the number of patches containing the abnormalities. However, the study of these screen-film mammograms is a repetitive task, and many-a-times, radiologists make errors by missing out the abnormalities present in the mammograms. According to studies, radiologists fail to identify the abnormal lesions 10%-30% of the times as a result of which the severity of breast cancer rises from early stage to last stage [4].
A computer-aided diagnosis (CAD) system merges image processing and pattern recognition theory and uses computers, software, and efficient tools to interpret medical information. Additionally, it provides viable diagnosis suggestions to reduce false detection and false negative rates. So, there is a growing need to design an automated CAD system which can effectively categorize the mammograms into either normal-abnormal or benign-malignant. The prime objective of CAD framework is to automatically take care of the issues related to the abnormal patch identification by collecting and analyzing important features of the mammograms. Moreover, CAD is regarded as a ‘second opinion’ by the medical practitioners and physicians to reach the final diagnosis decision. Therefore, in the present scenario, computer-aided detection (CADe) and computer-aided diagnosis (CADx) are most efficient and widely used techniques among the radiologists for mammograms analysis. The role of CADe system is to automatically identify and segregate the lesions containing abnormalities. On the other hand, CADx method aims at correctly classifying the lesions, and analyzing the mammograms with the help of image processing and pattern recognition approaches [19]. The four prime modules of CADx includes pre-processing, feature extraction, feature selection, and classification. It starts with finding out the region of interest (ROI) of a mammogram that includes the abnormalities. Next, it extracts the essential features from the ROI using effective feature extraction algorithms. Then, it selects the most relevant features with the aid of an appropriate feature selection algorithm. The feature selection algorithm is considered to be one of the most important steps prior to classification as it helps to increase the accuracy as well as to minimize the computational burden of the classifiers.
In the present work, a hybrid CAD framework is proposed aiming at a better diagnosis of the mammograms. The key contributions of the proposed scheme are as follows:
-
To describe the extracted ROIs of the mammograms, distinct texture features are extracted using the contourlet transform.
-
Next, a wrapper-based feature selection approach, namely, Forest optimization algorithm (FOA) is utilized to select the best optimal features.
-
Different hybrid schemes, namely, (Contourlet + FOA + SVM), (Contourlet + FOA + k-NN), (Contourlet + FOA + Naïve Bayes), and (Contourlet + FOA + C4.5) are formulated to correctly classify the mammograms either as normal or abnormal, and further, into benign or malignant.
The performance of the proposed CAD framework is evaluated in terms of classification accuracy, sensitivity, specificity, Matthew’s correlation coefficient (MCC), area under the ROC curve (AUC).
The remainder of this paper is organized as follows: Section 2 concisely describes the different CAD models. Section 3 provides the details of the proposed framework consisting of the four subsections of ROI generation, feature extraction, feature selection, and classification. Section 4 contains experimental results and a detailed analysis of the results. Finally, Section 5 presents the summary of the proposed scheme and prospects for future work.
2 Related work
For the last couple of years, the medical imaging community is actively involved toward the development of CAD framework. With the advent of CAD, newer issues have surfaced that demand deeper understanding and significant research. Some key modules in CAD like feature extraction, feature selection, and classification are being rigorously researched by the community. So this section presents some of the recent approaches with respect to these three modules to design an effective and automated CAD framework. Verma and Zakos [34] presented a technique for the detection and diagnosis of micro-calcifications in mammograms. In their work, 14 different statistical features are extracted and 3 features are selected. The scheme used a feed forward back propagation neural network (BPNN). El-Naqa et al. [12] proposed a SVM-based classification scheme to classify the abnormalities. After performing the SVM classifier on 76 clinical mammograms, they achieved a sensitivity of 94%. Fu et al. [14] utilized a feature extraction algorithm which extracts 61 features from both the spatial domain and spectral domain. They have used sequential forward search (SFS) feature selection algorithm followed by SVM and general regression neural network (GRNN) classifiers. After selecting the most relevant features, the reported technique achieved an AUC of 0.9800 and 0.9780 with respect to SVM, and GRNN, respectively.
Prathibha and Sadasivam [27] suggested multi-scale wavelet transformation for textural feature extraction. Further, they used k-nearest neighbor (k-NN) to classify the mammogram tissues and obtained an AUC of 0.946. Jona and Nagaveni [21] proposed an approach using gray level co-occurrence matrix (GLCM) for feature extraction. Further, genetical swarm optimization (GSO) is employed to select the relevant features followed by an SVM classifier. They achieved an improved classification accuracy of 94% as compared to its competent schemes. Azar and El-Said [2] used a probabilistic neural network (PNN) for the classification of mammograms and reported an accuracy of 97.66%. Mohamed et al. [24] proposed a CAD system which utilizes GLCM for feature extraction and k-NN, SVM and artificial neural network (ANN) as classifiers. Further, histogram equalization and morphological operation are applied to enhance the mammograms followed by an Otsu’s thresholding technique for ROI segmentation. The accuracy achieved by the above three classifiers are 73%, 83%, and 77%, respectively.
Dheeba et al. [8] suggested an approach for breast cancer diagnosis using particle swarm optimization wavelet neural network (PSOWNN). In their work, a Law’s texture energy measure (LTEM) is used to extract the features. Particle swarm optimization (PSO) was applied to optimize the parameters essential for wavelet neural network (WNN) and achieved an AUC of 0.96853. Eltoukhy et al. [13] presented a hybrid approach using wavelet and curvelet transforms to extract the features from mammograms. For feature selection, they applied t-test method to select the optimal features and then fed to an SVM classifier to classify the mammograms as normal and abnormal, and further, benign and malignant. Beura et al. [6] used GLCM and discrete wavelet transformation (DWT) to extract the texture features from the ROIs. They performed feature selection using the filter methods namely two sample t-test and F-test to get the relevant features. Further, BPNN is used as the classifier. The accuracy obtained for the two standard databases MIAS and DDSM is 98.0%, and 98.8%, respectively. Pawar and Talbar [25] utilized a wrapper method for feature selection. Wavelet co-occurrence feature was used to extract the features from four resolution levels of decomposition. They used a genetic fuzzy system as a feature selection approach and gained an accuracy of 89.47%.
Rouhi et al. [30] classified the mammogram tissues into benign and malignant using two different automated methods. In the first method, segmentation is done using automated region growing whose threshold value is determined by a trained ANN. The second method utilizes a cellular neural network (CNN) for segmentation, whose parameters are obtained by genetic algorithm (GA). Then various classifiers such as MLP, k-NN, SVM, Naïve Bayes, and Random Forest are employed, and a maximum accuracy of 96.47% is attained in case of MLP classifier. Deep neural networks as feature selection are proposed by [31]. The authors used deep neural networks (DNN) to select the most appropriate features in the field of action recognition and achieved an improved classification accuracy of 93.93% with the selected features.
A particle swarm optimization based feature selection algorithm along with SVM classifier is proposed to reduce the false positive rate in mammogram classification [38]. The authors have used multi-scale GLCM (MDGLCM) and second-order statistics of wavelet coefficients (SOSWC) to extract the texture descriptors followed by PSO-based SVM to classify the mammograms as normal and abnormal. The accuracy obtained for GLCM and SOSWC features of MIAS dataset are 89.8% and 95.2%, respectively. Phadke and Rege [26] presented a classification scheme based on the fusion of local and global features. The local features are obtained by using Chebyshev moments and GLCM, and global features are obtained from LTEM, Gabor based texture energy measures and fractal dimension. Further, SVM classifier is used to classify the mammograms as normal and abnormal with an accuracy of 93.17%. A new feature extraction technique using fast finite shearlet transform (FFST) is employed [15]. In this scheme, the features are extracted from the mammograms and ranked using the t-test statistics. Next, SVM classifier is applied to the optimal feature set and accuracy of 98.29% and 98.08% are achieved for MIAS, and DDSM datasets, respectively.
Guo et al. [18] proposed a 3-step micro-calcification (MC) detection approach in mammograms. In the first step, pre-processing is performed for the segmentation of ROI followed by enhancement. ROI is segmented using region-growing and double top-hat transform technique. A grayscale-adjustment function is applied to enhance the quality of the extracted ROI. Secondly, features are extracted using the contourlet transform. Finally, a non-linking simplified pulse-code neural network is employed to detect the MCs with an accuracy of 95.8%. Another mammogram diagnosis method using multi-resolution wavelet and Zernike moments is proposed in [22]. Here, both texture and shape features are extracted using the wavelet and Zernike moments. Then, the features are classified using SVM and Extreme Learning Machine (ELM) in a multi-kernel approach, with an obtained accuracy of 94.11%. Xie et al. [37] presented a CAD model based on Extreme Learning Machine (ELM) to classify the mammograms as benign and malignant. They extracted multidimensional features followed by feature selection using the combination of SVM and ELM. The classification accuracy for MIAS dataset achieved by this ELM-based model is 96.02%.
From the literature study, it is noticed that feature extraction, feature selection, and classification modules are the key modules to improve the overall efficacy of CAD framework. So, there is a growing need to improve these modules. Hence, keeping this in mind, this article proposes an improved hybrid CAD framework to classify the mammograms into either normal or abnormal and benign or malignant. Special emphasis is placed to improve feature extraction and feature selection modules to formulate different hybrid frameworks.
3 Proposed methodology
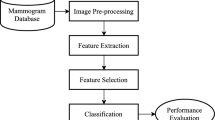
The proposed CAD system consists of three prime modules, namely, feature extraction, feature selection, and classification. The reported CAD model has experimented on two standard mammogram datasets, namely, Mammographic Image Analysis Society (MIAS), and Digital Database for Screening Mammography (DDSM). A multi-resolution and multi-orientation feature extractor, called contourlet transform is employed to extract the texture features along with various directions of the mammograms. Once the feature matrix is generated, the feature selection procedure is performed to select the best optimal features. To do this, an evolutionary algorithm, namely, forest optimization algorithm is utilized. Then, various classifiers such as SVM, Naïve Bayes, C4.5, and k-NN are considered to correctly classify the mammograms into either normal or abnormal, and benign or malignant. Figure 2 represents a generic framework followed by an elaborated discussion of each module for better interpretation. Also, a clear phase-wise visualization of the proposed CAD model is presented in Fig. 3.
3.1 Extraction of ROI
The digital mammograms contain imaging noise, artifacts, and pectoral muscles. The noise and artifacts lead to a low-quality image. Additionally, the pectoral muscles are unwanted regions in the mammograms. Hence, these undesirable regions need to be eliminated to obtain the actual ROI upon which the feature extraction technique needs to be performed. To extract the ROI, a cropping operation is employed on the mammograms taken from the standard dataset. The dataset is given with the radiologist’s truth markings on the region of abnormalities. So, using the information available, such as the center and radius of any abnormal area, the cropping mechanism is performed. In case of normal mammograms, ROI is selected arbitrarily. Finally, the actual ROI is segregated from the original mammogram. For instance, the extracted ROI from MIAS dataset are depicted in Fig. 4a-c for normal, benign, and malignant cases, respectively.
3.2 Feature extraction using contourlet transformation
An image can be examined thoroughly with the help of a multi-resolution based analysis. This analysis allows examining the image at different scales. DWT is one of the most widely accepted multi-resolution approaches. But, the major drawback with DWT is its restricted directional information. As it operates only in two dimensions, it overlooks the smoothness along the contours. This drawback is addressed by contourlet transform (CT), proposed by Do and Vetterli [10]. It has a greater degree of directionality and anisotropy. In addition, contourlet transform is capable of capturing contours and directional information of an image at various scales. Contourlet transform mainly contains two stages: a) the sub-band decomposition, and b) directional filtering. At the first stage, Laplacian pyramid (LP) is employed to find the point discontinuities in an image, followed by a directional filter bank (DFB) to link those point discontinuities into a linear structure in the second phase.
3.2.1 Laplacian pyramid (LP)
Laplacian pyramid (LP) is one of the acceptable mechanisms to decompose the image at different scales [7]. In this mechanism, each level of decomposition induces one down-sampled low-pass variant of the original image and one band-pass image which is the resultant image of the difference between the original and the predicted images. This mechanism can be performed iteratively on the coarse image (down-sampled low-pass images) to produce a series of decomposed images. The generated images are so arranged (one above the other) that a pyramid-like shape is formed and is referred to as the Laplacian pyramid.
3.2.2 Directional filter bank (DFB)
The directional filter bank is designed by Bamberger and Smith [3]. It can be effectively constructed through j-level binary tree decomposition and generates 2j sub-bands having wedge-structured frequency division. To attain the required frequency division in the original architecture of DFB, a complex tree expanding principle needs to be followed to get the enhanced directional sub-bands. However, an improved DFB [9] has been proposed that excludes modulating the image. It follows a simple approach in order to expand the decomposition tree. This advanced DFB stems upon two building blocks, wherein the former block is a two-channel quincunx filter bank with a fan filter [35]. It segments a 2D-spectrum into two directions: parallel and perpendicular. The second block holds a shearing operator that represents the restructuring of the image. By including both a shearing operator and its inverse before and after the two-channel quincunx filter bank, several directional frequency segments are attained while retaining the perfect transformation. Hence, the fundamental principle of DFB is to utilize the combination of shearing operator and two-channel quincunx filter bank effectively at every level in a binary tree-shaped filter bank so as to achieve the desirable 2D-spectrum partitions. Figure 5 illustrates the wedge-structured frequency division where j = 3 and hence, there are 23 = 8 real wedge-structured frequency segments.
Figure 6 depicts the decomposition in multi-scale and directions by combining the LP and DFB. The band-pass images produced by the LP are given as inputs to the DFB to capture the directional facts. This schema can be performed iteratively on the coarse image. The output of the scheme is a two-fold iterated filter bank framework, known as contourlet filter bank and it decomposes the input image into sub-bands in multiple directions and scales.
Let Io[n] be an input image upon which a LP stage is applied. This results in M number of band-pass images, bm[n], m = 1,2,...M (in the fine-to-coarse order) and one low-pass image IM[n]). In other words, a mth level of LP decomposes the image Im− 1[n] into a coarser image Im[n] and a band-pass image bm[n]. Each of the band-pass images bm[n] is again decomposed by a jm-level DFB into \(2^{j_{m}}\) band-pass directional images \(x_{m,z}^{j_{m}}[n], z = 0,1,...,2^{j_{m}}-1\). Further, some of the important characteristics of the discrete contourlet transform is presented in [10].
As the scheme is an iterative low-pass filtering, the texture information is present in the directional sub-bands at each of the resolution levels. Therefore, only the band-pass images are taken into consideration while determining the feature vector. Once the transformation operation is over, the generated sub-band images contain all the directional information which is needed for classifying the images. To acquire more textural information from the sub-band images in the contourlet domain, a set of several statistical properties such as energy, mean, absolute mean, standard deviation, skewness, and kurtosis are calculated in this scheme instead of taking the contourlet coefficients. Let an image Imn be one of the sub-images in mth level and nth direction with size P × Q, then the aforementioned statistical measures can be denoted mathematically as follows.
3.3 Feature selection using forest optimization algorithm
Feature extraction process plays a vital role in classifying the ROIs of the mammograms either as normal or abnormal. Further, if the ROI is found to be abnormal, it is again classified as benign or malignant. Moreover, it may not be necessary that all the extracted features contribute toward the classification process. Therefore, along with the feature extraction, it becomes necessary to select the most optimal features that actually contribute to improving the classification accuracy. So, in the proposed CAD scheme, a wrapper-based feature selection algorithm, namely, Forest Optimization Algorithm (FOA) is employed to obtain the optimal features [17]. FOA is an evolutionary-based approach, which is influenced by the growing process of the trees in a forest. The algorithm is composed of three fundamental steps: 1) Local Seeding Operation, 2) Limiting the population of the forest, and 3) Global Seeding Operation.
Before the steps are performed, the forest needs to be initialized with a predefined number of trees. Every element of a tree is initialized with a binary value, 0/1, arbitrarily. The size of each tree is equal to T + 1, where T is the dimension of the feature vector and the extra 1 is added to indicate the ‘age’ of the tree. Initially, the age of each of the newly created trees is set to zero. These newly created trees are referred to as parent trees (0-aged trees). Next, a local seeding operation is performed only on the parent trees with the help of a parameter referred to as the local seeding changes (LSC). Here, depending on the value of the LSC parameter, each of the parent trees generates new trees (referred to as child trees) with age 0. In each of the newly generated trees, any one variable is arbitrarily chosen (except the age variable) and the value of that variable is flipped from 0 to 1 or 1 to 0. Once the local seeding operation is performed on every parent tree (0-aged trees), the age of each tree is incremented by 1. Then, a population limitation operation is performed with the help of two parameters, namely, ‘area limit’ (AL), and ‘lifetime’ (LT). The trees having ‘age’ larger than the predetermined value of LT are eradicated from the forest to create the candidate population. Then, the remaining trees are individually passed through a classification stage and are arranged in descending order of the obtained classification accuracy (fitness value). Further, the trees which fall beyond the preset value of AL are transferred from the forest to the candidate population. Next step is to perform the global seeding operation on a specified percentage of the candidate population. This percentage is determined by a parameter, known as the ‘transfer rate’ (TR). The global seeding is accomplished by a parameter, namely, ‘global seeding changes’ (GSC), wherein gsc number of elements are randomly chosen from each selected tree. Further, the values of the randomly chosen elements are flipped (0 → 1/1 → 0) at a time. Then, the modified trees are again passed through the classifiers and the tree with the highest classification accuracy is selected as the optimal tree. This age of the selected tree is updated to zero and will act as the parent tree in the next iteration. The flowchart of the detail process is depicted in Fig. 7.
The aforementioned steps are repeated until one of the following stopping criteria is met: a) a fixed count of iterations, b) negligible difference in empirical fitness value of subsequent iterations, or c) a stipulated degree of accuracy. In the above-mentioned approach, a predetermined threshold for the classification accuracy within the fixed count of iterations is considered to be the stopping criterion [16]. Algorithm 1 represents the steps involved in FOA for feature selection.

3.4 Classification
As most of the schemes operate on real datasets, sometimes the most relevant features are not inferable. Hence, it is required to perform a classification task using the selected features in order to compute the performance of the feature selection algorithms. Additionally, the class prediction relies on the classifiers applied for the classification purpose. So, it is a standard approach to utilize various classifiers so as to make the obtained performance classifier-independent. Therefore, in this work, four well-known classifiers such as SVM, k-NN, Naïve-Bayes, and Decision tree are employed.
3.4.1 Support vector machine (SVM)
SVM is a supervised learning approach widely applicable for classification and regression problems [12]. It linearly classifies the input dataset by constructing a hyperplane in the high dimensional space. The constructed hyper-plane is an optimal one if it has the maximum distance from the proximate data element of any class. The SVM aims at producing a classification model which can correctly assign a new data element to the class it belongs to.
3.4.2 k-Nearest neighbor (k-NN)
k-NN classifier [1] is considered to be a ‘lazy learner’. A new test data is assigned to the class which is most recurrent among its k-nearest neighbors, where k is a small odd integer. The nearest neighbor can be determined by the smallest distance measure well-known as the Euclidean distance. This classifier is suitable for both numeric and discrete values.
3.4.3 Decision tree (C4.5)
The decision tree is a widespread classification technique. It consists of a sequence of well-crafted questions and conditions regarding the attributes in a tree-like structure. In a decision tree, the internal nodes indicate the test conditions of attributes and the leaf node indicate the class labels. Each of the internal nodes contains a threshold connecting to one or more attributes to partition the data into its successors. The process is said to be terminated when it reaches a leaf or terminal node, and the class label of the terminal node is assigned to the new data. A widely used algorithm for generating the decision tree is C4.5 developed by Quinlan [28]. The advantage of C4.5 over ID3 is that it classifies both numeric and symbolic data as well.
3.4.4 Naïve-Bayes
Naïve-Bayes classifier [29] is quite a simple statistical classifier. It employs the principle of Bayes’ theorem and can predict the class membership probability. This classifier assumes that the impact of one feature on a given class is unassociated with the other feature values. Hence, the basic principle of this classifier is the class conditional independence. As the computational overhead of the classifier is simple, so it is termed as Naive. Bayesian classifier performs very well in real and discrete data, regardless of their simple design and assumptions.
The pseudo-code of the proposed Contourlet+FOA-based CAD model is presented in Algorithm 2.

4 Experimental results and discussions
The proposed Contourlet+FOA-based CAD framework with the different combination of classifiers, namely, SVM, k-NN, Naïve-Bayes, and C4.5 is validated using MATLAB 2017a, on a personal computer with a Core-i7 processor and 32 GB RAM, running under Windows 10 operating system. The proposed scheme is analyzed with mammographic images taken from the two standard datasets, namely, MIAS and DDSM [20, 33]. The MIAS dataset has a sample size of 314 mammograms containing normal, benign, and malignant cases. Out of 314 images, 207 cases are normal and the rest 107 are abnormal. In the abnormal set of mammograms, 59 mammograms are benign and the remaining 48 cases are malignant. The mammogram images taken from MIAS are 8-bit gray images with a size of 1024 × 1024 pixels. Similarly, 1500 mammogram images are taken from DDSM. Out of 1500 samples, 519 are normal, 479 are benign, and the rest 502 are malignant images. The efficacy of the proposed scheme is compared with other benchmark schemes in terms of accuracy, sensitivity, specificity, MCC, ROC, and AUC. These performance parameters are computed using the confusion matrix. A confusion matrix (see Fig. 8) contains the counts of the actual and predicted class attained by a classifier. Here, the class of normal and abnormal mammograms is considered to be negative, and positive, respectively. Similarly, the classes of benign and malignant are taken as negative, and positive, respectively.
The aforementioned performance measures are defined below and are represented as:
-
Sensitivity (True Positive Rate) deals only with the positive cases; it exhibits the ratio of the classified positive cases to the actual positive cases. The higher the sensitivity, less is the false negative rate.
$$\begin{array}{@{}rcl@{}} Sensitivity = \frac{TP}{TP+FN} \end{array} $$(7) -
Specificity (True Negative Rate) takes on the negative cases; it shows the ratio of the classified negative cases to the actual negative cases, the greater the specificity, less is the false positive rate.
$$\begin{array}{@{}rcl@{}} Specificity = \frac{TN}{TN+FP} \end{array} $$(8) -
Accuracy signifies the precision of classification outcomes. The more the accuracy, more efficient the system is.
$$\begin{array}{@{}rcl@{}} Accuracy = \frac{TP+TN}{TP+FP+TN+FN} \end{array} $$(9) -
MCC is one more performance metric used for determining the quality of a classifier proposed in [23]. It produces an effective evaluation of the classifier when the cases of different classes in a sample are highly imbalanced. The more the MCC, higher is the efficiency of the classifiers.
$$\begin{array}{@{}rcl@{}} MCC = \frac {TP\times TN-FP\times FN}{{\sqrt {(TP+FP)(TP+FN)(TN+FP)(TN+FN)}}} \end{array} $$(10)
The ROC plot is computed between the true positive rate and false positive rate. The true positive rate is taken in the vertical axis and the false positive rate is taken in the horizontal axis. The AUC is the descriptor of the results obtained from testing the samples. The AUC close to 1.0 signifies the certainty of the diagnostic experiment; on the contrary, AUC near to 0.5 implies an inaccurate result.
During experimentation for the proposed CAD model, at first, the ROIs need to be extracted from the mammogram images. A simple cropping mechanism is employed on the mammograms to get the desired ROIs and the extracted ROIs are of size 256 × 256 pixels. Once the ROIs are obtained, the texture attributes are explored using the contourlet transformation. Contourlet is used to obtain the textural properties of an image in different scales and directions. In this proposed scheme, a 4-level contourlet transform is employed on the ROIs. The ROIs are decomposed into 4-pyramidal levels, impending 4, 8, and 16 directional sub-images. The texture properties are extracted from the sub-band images of the 4th-level due to the fact that this level generates 16 multiple directions. Further, at each of the sub-band images of the 4th-level decomposition, a basic set of six statistical textural features are generated. The statistical features, namely, energy, mean, absolute mean, standard deviation, skewness, and kurtosis are evaluated using (1)-(6). So, a total of 96 features (6 × 16) are extracted from each of the ROIs.
To perform the experimental analysis, the input dataset is separated into non-overlapping training and testing groups. In this article, the training samples are created by considering 70% of the original dataset and the rest 30% as the testing group. Further, the performance analysis is divided into two groups, namely, without feature selection, and with feature selection.
4.1 Without feature selection
In this case, the total of 96 features is used to measure the performance of the classifiers with respect to the aforementioned metrics. Tables 1 and 2 depict the results obtained with different classifiers for normal vs. abnormal, and benign vs. malignant classification, respectively. Additionally, the ROC curves for normal vs. abnormal, and benign vs. malignant classification are represented in Figs. 9 and 10, respectively.
4.2 With feature selection
If the size of the generated feature matrix is R × F, then R indicates the number of the ROIs (or samples) and F represents the count of the features which is 96. Further, among the F features, it may be realized that some features are not relevant and do not make a proper contribution toward the classification accuracy. So, to find out the most relevant features, FOA is employed to select the best set of features. Hence, the size of the new feature matrix after feature selection is R × f, where f represents the set of a reduced number of features, and it is a subset of F containing the best relevant features.
FOA is an iterative wrapper-based feature selection approach in which the best features are selected based on a fitness value. In this case, the fitness value is considered to be the classification accuracy. As described in Section 3.3, the value of LSC and GSC parameters are set to be \(\frac {1}{5}^{th}\) and \(\frac {1}{4}^{th}\) of the size of the feature vector, respectively. As FOA is an iterative process, the termination condition of the algorithm is a specified classification accuracy with a pre-defined count of iterations.
Table 3 depicts the qualitative measures of the proposed CAD system to classify the mammogram ROIs as normal and abnormal for MIAS dataset. For each of the suggested classifiers, five independent runs are made to show the consistency of the proposed scheme. Additionally, the average of each of the measures is obtained and shown in Table 3. In the proposed work, for k-NN, the results are determined for different k values (k = 3,5,and 7). From Table 3, it is noticed that the accuracy (in %) obtained for all the classifiers come out to be 100 except for Naïve-Bayes classifier (Accuracy = 97.86%). Without applying the feature selection technique, the classifiers have to deal with all the 96 extracted features for each of the ROIs in the sample. However, post feature selection scheme reduces the size of the feature vector by half. From this table, it can be seen that SVM produces the least number of features, 38 with the maximum accuracy of 100%. Figure 11 represents the bar plot of all the performance measures for different classifiers for classifying normal and abnormal.
Similarly, the performance measures for classifying the ROIs as benign and malignant for MIAS are listed in Table 4. For benign vs. malignant classification also, the results for five independent runs are generated, and then the average value of each of the measures is evaluated. Here, the highest accuracy of 98.74% is obtained by C4.5 classifier with 44 number of relevant features. The MCC for C4.5 is also very prominent with a value of 0.9753. Figure 12 exhibits the bar plot for the performance measures obtained by various classifiers for benign vs. malignant classification.
Comparing Tables 1 and 3, it is seen that feature selection does not have a significant effect on the performance of the classifiers for normal vs. abnormal case. However, while comparing Tables 2 and 4, it is observed that feature selection play a vital role in enhancing the performance of the classifiers (with feature selection) for benign vs. malignant classification. Further, similar findings are noticed from the ROC plots (refer Figs. 9 and 13, and Figs. 10 and 14).
To validate the qualitative measures obtained by the proposed model, corresponding ROC curves are also generated. The ROC curves for MIAS dataset obtained by various classifiers for normal vs. abnormal classification are shown in Fig. 13. The value of AUC for SVM, k-NN, and C4.5 is 1, whereas for Naïve-Bayes, it is 0.9995. Similarly, the ROC curves for benign vs. malignant classification for MIAS are shown in Fig. 14.
The performance measures of the proposed model on DDSM dataset are listed in Table 5. The highest accuracy for normal vs. abnormal classification is obtained by SVM classifier with a value of 100% from 39 features. The ROC curves for DDSM dataset with different classifiers are shown Fig. 15. From the figure, it is clear that the highest value of AUC is 1 for SVM classifier. Moreover, similar results are observed for benign vs. malignant classification.
In order to show the complexity of the proposed model, the execution time taken for both MIAS and DDSM are listed in Table 6. The training time and testing time taken by various classifiers are calculated in terms of seconds. Also, the execution time of the proposed model is compared with some of the other techniques. The overall execution time of the proposed model is found out to be 9.805 secs.
To further justify the proposed CAD system, the performance measures obtained are compared with that of some recent state-of-the-art schemes. Table 7 depicts a comprehensive comparison of the proposed scheme with respect to its counterparts in terms of accuracy and AUC. From Table 7, it is noted that the presented scheme prevails over the other compared schemes. The results of the proposed work surpass in both types of classification, i.e. normal vs. abnormal, and benign vs. malignant. The highest accuracy for normal vs. abnormal classification is 100% and for that of benign vs. malignant, it is 98.74% for MIAS dataset. Similarly, for DDSM dataset, the highest accuracy achieved for normal vs. abnormal, and benign vs. malignant are 100%, and 98.72%, respectively. Moreover, these high values of classification accuracy are achieved from a remarkably less number of features.
5 Conclusion
This paper deals with a hybrid CAD framework to correctly classify the mammograms into normal or abnormal, and then, benign or malignant. The proposed approach first applies a pre-processing method in terms of ROI extraction using cropping. Then, it employs the contourlet transform to extract the texture features from the mammogram images. Moreover, the forest optimization technique is performed to select the optimal features which result in a more efficient and accurate classifier. As FOA is a wrapper-based approach, the optimal features are selected as per the classification accuracy. Finally, four different classifiers, namely, SVM, k-NN, Naïve-Bayes, and C4.5 are employed to correctly classify the mammograms as normal or abnormal, and further benign or malignant. In the case of normal-abnormal classification, highest accuracy of 100% is achieved with all the classifiers considered except for Naïve-Bayes. Further, for benign vs. malignant, maximum accuracy of 98.74% is obtained for the C4.5 classifier.
Designing an automated CAD system for breast cancer detection and diagnosis remains an open problem. There exist several future directions which might further improve the CAD framework for mammogram images: (1) The acquisition of large databases from other standard dataset and from different medical institutions with different image qualities for correct clinical evaluation, and to improve the overall efficiency of CAD system, (2) To further improve the classification accuracy, alternate multi-resolution transformation techniques should be investigated to obtain more robust features, (3) There exists enormous scopes for researchers to utilize advanced machine learning techniques, namely, deep learning, and extreme learning for classification, (4) Further, the suggested hybrid framework applicable for correct classification of other types of cancer could be thought of another area of extension.
References
Aha DW, Kibler D, Albert MK (1991) Instance-based learning algorithms. Mach Learn 6(1):37–66
Azar AT, El-Said SA (2013) Probabilistic neural network for breast cancer classification. Neural Comput Appl 23(6):1737–1751
Bamberger RH, Smith MJ (1992) A filter bank for the directional decomposition of images: Theory and design. IEEE Trans Signal Process 40(4):882–893
Berlin L (2014) Radiologic errors, past, present and future. Diagnosis 1(1):79–84
Berraho S, El Margae S, Kerroum MA, Fakhri Y (2017) Texture classification based on curvelet transform and extreme learning machine with reduced feature set. Multimed Tools Appl 76(18):18,425–18,448
Beura S, Majhi B, Dash R (2015) Mammogram classification using two dimensional discrete wavelet transform and gray-level co-occurrence matrix for detection of breast cancer. Neurocomputing 154:1–14
Burt P, Adelson E (1983) The laplacian pyramid as a compact image code. IEEE Trans Commun 31(4):532–540
Dheeba J, Singh NA, Selvi ST (2014) Computer-aided detection of breast cancer on mammograms: A swarm intelligence optimized wavelet neural network approach. J Biomed Inform 49:45–52
Do MN (2002) Directional multiresolution image representations
Do MN, Vetterli M (2005) The contourlet transform: an efficient directional multiresolution image representation. IEEE Trans Image Process 14(12):2091–2106
Do Nascimento MZ, Martins AS, Neves LA, Ramos RP, Flores EL, Carrijo GA (2013) Classification of masses in mammographic image using wavelet domain features and polynomial classifier. Expert Syst Appl 40(15):6213–6221
El-Naqa I, Yang Y, Wernick MN, Galatsanos NP, Nishikawa RM (2002) A support vector machine approach for detection of microcalcifications. IEEE Trans Med Imaging 21(12):1552–1563
Eltoukhy MM, Faye I, Samir BB (2012) A statistical based feature extraction method for breast cancer diagnosis in digital mammogram using multiresolution representation. Comput Biol Med 42(1):123–128
Fu J, Lee S, Wong S, Yeh J, Wang A, Wu H (2005) Image segmentation feature selection and pattern classification for mammographic microcalcifications. Comput Med Imaging Graph 29(6):419–429
Gedik N (2016) A new feature extraction method based on multi-resolution representations of mammograms. Appl Soft Comput 44:128–133
Ghaemi M, Feizi-Derakhshi MR (2014) Forest optimization algorithm. Expert Syst Appl 41(15):6676–6687
Ghaemi M, Feizi-Derakhshi MR (2016) Feature selection using forest optimization algorithm. Pattern Recogn 60:121–129
Guo Y, Dong M, Yang Z, Gao X, Wang K, Luo C, Ma Y, Zhang J (2016) A new method of detecting micro-calcification clusters in mammograms using contourlet transform and non-linking simplified pcnn. Comput Methods Prog Biomed 130:31–45
Gupta S, Chyn PF, Markey MK (2006) Breast cancer cadx based on bi-radsdescriptors from two mammographic views. Med Phys 33(6):1810–1817
Heath M, Bowyer K, Kopans D, Moore R, Kegelmeyer WP (2000) The digital database for screening mammography. In: Proceedings of the 5th international workshop on digital mammography, Medical Physics Publishing, pp 212–218
Jona J, Nagaveni N (2012) A hybrid swarm optimization approach for feature set reduction in digital mammograms. WSEAS Trans Inf Sci Appl 9:340–349
de Lima SM, da Silva-Filho AG, dos Santos WP (2016) Detection and classification of masses in mammographic images in a multi-kernel approach. Comput Methods Prog Biomed 134:11–29
Matthews BW (1975) Comparison of the predicted and observed secondary structure of t4 phage lysozyme. Biochim Biophys Acta (BBA)-Protein Struct 405(2):442–451
Mohamed H, Mabrouk MS, Sharawy A (2014) Computer aided detection system for micro calcifications in digital mammograms. Comput Methods Prog Biomed 116(3):226–235
Pawar MM, Talbar SN (2016) Genetic fuzzy system (gfs) based wavelet co-occurrence feature selection in mammogram classification for breast cancer diagnosis. Perspect Sci 8:247–250
Phadke AC, Rege PP (2016) Fusion of local and global features for classification of abnormality in mammograms. Sādhanā 41(4):385–395
Prathibha B, Sadasivam V (2010) Breast tissue characterization using variants of nearest neighbour classifier in multi texture domain. IE (I) J 91:7–13
Quinlan JR (2014) C4. 5: programs for machine learning. Elsevier, Amsterdam
Rish I (2001) An empirical study of the naive bayes classifier. In: IJCAI 2001 workshop on empirical methods in artificial intelligence, vol 3. IBM, pp 41–46
Rouhi R, Jafari M, Kasaei S, Keshavarzian P (2015) Benign and malignant breast tumors classification based on region growing and cnn segmentation. Expert Syst Appl 42(3):990–1002
Roy D, Murty KSR, Mohan CK (2015) Feature selection using deep neural networks. In: 2015 International Joint Conference on Neural Networks (IJCNN). IEEE, pp 1–6
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA: a Cancer J Clin 65(1):5–29
Suckling J, Parker J, Dance D, Astley S, Hutt I, Boggis C, Ricketts I, Stamatakis E, Cerneaz N, Kok S et al (1994) The mammographic image analysis society digital mammogram database. In: Exerpta Medica. International Congress Series, vol 1069, pp 375–378
Verma B, Zakos J (2001) A computer-aided diagnosis system for digital mammograms based on fuzzy-neural and feature extraction techniques. IEEE Trans Inf Technol Biomed 5(1):46–54
Vetterli M (1984) Multi-dimensional sub-band coding: Some theory and algorithms. Signal Process 6(2):97–112
WHO (2013) Latest world cancer statistics global cancer burden rises to 14.1 million new cases in 2012: Marked increase in breast cancers must be addressed. international agency for research on cancer and others. World Health Organization, pp 12
Xie W, Li Y, Ma Y (2016) Breast mass classification in digital mammography based on extreme learning machine. Neurocomputing 173:930–941
Zyout I, Czajkowska J, Grzegorzek M (2015) Multi-scale textural feature extraction and particle swarm optimization based model selection for false positive reduction in mammography. Comput Med Imaging Graph 46:95–107
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mohanty, F., Rup, S., Dash, B. et al. Mammogram classification using contourlet features with forest optimization-based feature selection approach. Multimed Tools Appl 78, 12805–12834 (2019). https://doi.org/10.1007/s11042-018-5804-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-018-5804-0