Abstract
The efficient analysis of digital mammograms has an important role in the early detection of breast cancer and can lead to a higher percentage of recovery. The process of mammogram classification can be divided into two steps as follows: first, it has to be established if the tissue contains abnormalities, and, second, the nature of the lesion has to be determined. This second step of a computer-aided diagnosis system is important in order to select the best treatment for the patient and to achieve the highest chance of recovery. In general, digital mammogram analysis consists of preprocessing, feature extraction, feature selection and classification. Feature extraction is crucial in identifying informative characteristics that can differentiate between benign and malignant lesions. The two main types of feature extraction methods are shape features and texture features. In the current paper, we present several experiments in order to compare the performance of different feature extraction methods from the two types mentioned previously. As data, images from the Digital Database for Screening Mammography (DDSM) are used, which has precise ground truth for the cancerous tissue. For classification Decision Trees and Random Forest methods are used to evaluate the performance using the different extracted features. The experiments that were carried out show that shape features perform better than texture features to separate benign and malignant abnormalities. Also, some outliers were found causing a decrease in the accuracy of the system and achieving 66% test accuracy using shape features and Random Forest classifier.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In medicine, X-ray imaging is frequently used to analyze the internal structure and potential abnormalities of the human body/organs. Mammography, a type of X-ray imaging, is commonly used to detect and diagnose breast cancer. It is a non-invasive and low-risk diagnostic tool that allows the early detection of breast cancer, which significantly increases the chances of successful treatment and reduces the mortality rate. The growth in digital mammography has greatly contributed to the early detection and diagnosis of breast cancer, a leading cause of mortality among women worldwide [6]. However, there is still much room for improvement in the development of robust and effective feature extraction methods for lesion type detection, which can further enhance the accuracy and reliability of mammographic analysis.
Mammogram analysis is a complex problem, which involves the following steps: preprocessing, segmentation, feature extraction, feature selection and classification. Feature extraction is a crucial step in the process of mammographic analysis, as it enables the identification and characterization of lesions present in digital mammograms. These features may include but are not limited to texture, shape, intensity, and density of the regions of interest.
The current experiments are addressed to compare the performance of different feature extraction methods, specifically using shape and texture features, to improve lesion type detection in digital mammograms. The combination of these features is also considered in this study, as it may provide a more comprehensive and accurate representation of the mammographic images. The experiments use images from the Digital Database for Screening Mammography (DDSM) [10, 11] that contain precise ground truth information on cancerous tissue. To evaluate the performance of the different feature extraction methods and achieve optimal accuracy for lesion type detection, decision trees and random forests are used as classification techniques. The novelty of the presented approach consists in the use of feature extraction, feature selection and classification method combination on the dataset (DDMS). Furthermore, in our research we increased the number of input images (1424 instead of 323) compared to [13].
The rest of the paper is organized as follows: in Sect. 2 existing studies are discussed from the literature related to lesion classification. Section 3 details the current approach and Sect. 4 analyzes the achieved results. Finally, in Sect. 5 we present the conclusion of our experiments.
2 Related Work
In recent years, there have been numerous studies [1,2,3,4, 7,8,9, 12, 14] presented in the field of computer-aided diagnosis (CAD) for the early detection of cancerous cells. Breast cancer causes the most deaths among women worldwide [6]; hence, developing accurate and reliable CAD systems has been a major focus of research. With the help of CAD systems, the mortality rate due to breast cancer can be significantly reduced. In the literature, there are studies [1,2,3,4, 7,8,9, 12, 14] focusing on the different steps of a CAD system: preprocessing, segmentation, feature extraction, feature selection, and classification. In the following paragraphs, some of the relevant studies related to feature extraction in mammographic analysis will be discussed.
Various feature extraction methods have been proposed for mammographic analysis. Some use texture features such as Gray-Level Co-Occurrence Matrix (GLCM) [1, 4, 5, 7], Gray-Level Run-Length Matrix (GLRLM) [3,4,5], and Local Binary Patterns (LBP) [7, 8]. Others use shape features such as area, perimeter, compactness, roughness and slope [9, 13].
Texture features are frequently used on medical images to capture visual characteristics. These features are derived from second-order statistics that examine the gray levels on the image. The use of GLCM was proposed by Ancy and Nair [1] to extract features from data of Mammographic Image Analysis Society (MIAS) [16]. The computed characteristics were fed to a Support Vector Machine (SVM) classifier to detect tumors in the breast tissue. The system resulted in 81% accuracy score. Farhan et al. [8] have used LBP to differentiate benign and malignant breast lesions. LBP descriptors are proficient in efficiently capturing the grayscale contrast along with local spatial patterns in an image. The authors reported 85% accuracy using logistic regression classifier on MIAS [16]. Saeed et al. [7] proposed the classification of LBP features using Random Forest (RF) classifier using images from the same dataset and achieved 75% accuracy. Chaieb and Kalti [5] published a survey comparing different feature extraction methods and concluded that GLRLM features were the most suitable for identifying breast cancer in mammograms. Based on the results presented in [5], in previous research [3, 4] we used GLRLM feature extraction on images from MIAS [16] and DDSM [10, 11]. The computed characteristics were used as input to different feature selection (Principal Component Analysis – PCA and genetic algorithm-based – GA method) and classification (Decision Tree – DT and Random Forest – RF) methods to differentiate between normal, benign and malignant tissues. The reported accuracies were 70% on MIAS and 54.1% on DDSM.
In a study by Li et al. [13], contour features were extracted from the mammograms, by defining a 1-dimensional representation of the lesion boundary. The authors obtained 99.33% accuracy using SVM and 323 images from Digital Database for Screening Mammography (DDSM) dataset [10, 11]. A similar approach is described in [9], where shape features are classified using an Artificial Neural Network (ANN), achieving an accuracy of 97.24% on the Curated Breast Imaging Subset of DDSM (CBIS-DDSM).
Kumari and Jagadesh [12] introduced an advanced GLCM (AGLCM) by combining texture- (GLCM), intensity- (entropy), and shape features. Using SVM to detect lesions on mammograms from MIAS, the authors obtained an accuracy of 92.4% which outperformed the texture feature extraction methods [1]. This experiment shows how the combination of different features can improve the accuracy of the CAD system.
Ansar et al. [2] proposed a deep learning-based approach for breast cancer detection using a model with Convolutional Neural Networks (CNNs). Their proposed model achieved an accuracy of 86.8% on DDSM. Li et al. [14] proposed a two-view system and achieved 94.7% accuracy.
Experiments clearly show the importance of feature extraction in a mammogram classification system. In the current study, we aim to compare the shape feature extraction method presented in [13] (with outstanding results on DDSM) to GLRLM texture feature extraction method in identifying breast abnormality type in mammograms.
3 Proposed Approach
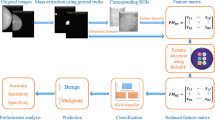
The objective of the current study is to compare the performance of textural- and shape features in distinguishing between benign and malignant lesions in digital mammograms. Specifically, we use gray-level run-length matrix (GLRLM) texture features and shape (geometrical and contour) features to extract relevant information from mammograms, which are then used as inputs for the classification models. Similarly to [4], feature selection is applied to reduce the dimensionality of the feature set and eliminate any redundant or irrelevant features, which can improve the classification accuracy and efficiency.
3.1 Preprocessing
In the preprocessing step, we use the predefined mask of the lesion to isolate the relevant regions of interest in mammograms. In the first step, the bounding box enclosing the lesion is defined. These bounding boxes are increased to have 25 pixels of padding around the lesion. We presume that this padding helps to capture the surrounding tissue and potential microcalcifications that may be indicative of malignancy – especially for textural feature extraction. The image is cropped based on the resulting bounding box. Next, a same-size binary mask is defined – by using the predefined mask – where 1 marks the pixels belonging to the lesion and 0 marks the pixels belonging to the background.
3.2 Feature Extraction
Feature extraction, as mentioned in Sect. 1, has a key role in computer-aided systems. The classification is highly dependent on its input. Therefore, the process of feature extraction requires a careful selection of appropriate methods and algorithms that can effectively capture the relevant information from the images.
In our proposed approach, we extract texture and shape features from the regions of interest in digital mammograms with the aim of characterizing the lesions. Texture features are extracted using GLRLM [5], which has been shown to effectively capture the texture information in medical – gray-scale – images. These features are quantified by calculating the frequency and distribution of gray-level runs in a given direction. Shape features, on the other hand, are extracted using the mask of the lesion and are categorized as geometrical features and contour features [13]. These characteristics illustrate the size of the lesion and the regularity/irregularity of the border surrounding the lesions. In the following paragraphs, the used feature extraction methods are detailed.
GLRLM [5] is a feature extraction method used to quantify texture features in medical images by calculating the distribution of gray-level runs in a given direction. In case of an image (2-dimensional data) it can be defined in four directions: horizontal, first diagonal, vertical and second diagonal. From the constructed matrices 11 features are extracted – as listed in [3] – describing the relationship between gray levels and their spatial distribution and therefore providing insight into the complex textures present in mammographic images. Taking into account the aforementioned four orientations, a total of 44 characteristics will be derived from an analysis of a mammogram.
Geometrical features are calculated from the mask of a lesion. This set of features includes the perimeter, the area and the compactness (calculated as the perimeter squared divided by the area). These are simple and frequently used features and they are used as a baseline.
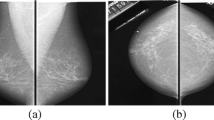
Contour features were introduced in [13]. The image (2-dimensional information) is converted into 1-dimensional data. In order to extract details regarding the circularity of a lesion, we try to fit an ellipse to it. First, the center of the ellipse \(C(x_c, y_c)\) is defined by the average of white pixel coordinates on the mask. This is also presented in Eq. (1), where N is the total number of white pixels. Next, the width and height of the lesion are defined and used as the minor- (b) and major-axis (a) of the ellipse, as shown in Eq. (2). Figures 1a and 2a illustrate the calculated ellipses.
Based on this information we define points on the ellipse and calculate their distance from C. These distances are saved into a list called \(d_e\). For every point on the ellipse, there is a corresponding point on the boundary of the lesion. In \(d_l\), we save the distances of these points from C. Finally, \(\varDelta d\) is defined by the difference between \(d_l\) and \(d_e\) Eq. (3). Figures 1b and 2b show the calculated distances for a benign and malignant lesion respectively.
After defining \(\varDelta d\), the final features are calculated, namely the root mean roughness Eq. (4), the root mean slope Eq. (5) and the circularity Eq. (6). In Eq. (4) to (6) \(\mu \) and \(\sigma \) represent the mean and the standard deviation of the calculated values. In some cases, it is sufficient to look only at a fragment of the border to decide its malignancy. Hence, local features are also extracted by calculating the above-mentioned contour features for smaller sub-regions of the lesion border. These sub-regions are obtained by dividing the border into equal segments and extracting the contour features from each of them.
These contour features provide a measure of irregularity in the shape of the lesion. By analyzing Figs. 1b and 2b we presume that these features are particularly useful for distinguishing between benign and malignant tumors.
3.3 Feature Selection
Feature selection involves selecting a subset of the most relevant features from a larger set of extracted features. This is important because it reduces the dimensionality of data and removes irrelevant features, thus simplifying analysis, improving computational efficiency, and increasing the accuracy of the classification model. Various feature selection techniques have been proposed in the literature (more details in survey [5]). In our experiments, we utilize the principal component analysis (PCA) method [4] and a genetic algorithm-based (GA) method [4] to perform feature selection.
PCA is a mathematical technique that involves transforming data into a new coordinate system, where the features are represented as a set of linearly uncorrelated variables. This method maximizes the variance of data and minimizes the number of variables required to represent it.
GA is a meta-heuristic optimization technique that is inspired by the natural selection process in biology. It involves simulating a population of potential feature subsets. In case of feature selection, the goal is to find the subset of features that maximizes a selected evaluation metric.
3.4 Classification
Classification refers to the process of assigning input data to one of several predefined classes based on a set of rules or models. In the current approach, we utilize two different supervised classification algorithms, namely the Decision Tree (DT) and Random Forest (RF) algorithms because of their interpretability. To construct the models the CART (Classification and Regression Tree) algorithmFootnote 1 is used.
The DT algorithm involves recursively splitting the training data into binary partitions based on a set of if-then rules inferred from the input features. The partitioning is performed in a way that the resulting subsets are as homogeneous as possible with respect to the class labels of their members.
On the other hand, the RF algorithm constructs an ensemble of decision trees and aggregates their results to make a final prediction. This approach has been shown to yield higher classification accuracy compared to using a single decision tree, as it reduces the risk of overfitting. In our research, we perform experiments with every possible combination of feature selection and classification, in order to define the best solution.
4 Experiments and Results
The scope of our experiment is to distinguish the character of lesions based on different extracted features. The constructed system was presented in the previous section (Sect. 3). In the following sections the used data, parameters and the achieved results are presented.
4.1 Dataset
In order to extract shape features from the contour of the abnormality it is necessary to know its exact boundary. Therefore, DDSM [10, 11] is used, because for each mammogram from breast tissue with cancerous cells it contains the boundary mask of the lesion. DDSM contains in total 7809 images: half of them taken from the side and the other half from the top. In the current experiment the mammograms taken from the side are used. Among these samples there are 2465 normal, 712 benign and 728 malignant examples. To build a classifier to distinguish the type of the lesion, the normal images are excluded. Further, to achieve a better result the same number of mammograms are used from each class. Therefore, 1424 images are used in the current experiments (712 examples from each class).
The next step is to split these images into train and test sets. Stratified sampling is used to ensure that the proportion of each set remains the same in both sets. Moreover, if there are more samples from the same patient they will be placed into the same set to prevent bias. 75% of the data (1067) are used to train the model, while 25% (357) to validate to the correctness of the built model.
4.2 Experimental Setup
In Sect. 3 the used methods and the proposed approach are presented. In the following paragraphs, we will further detail the parameter setting for the proposed algorithms.
As mentioned in Sect. 3.2, besides GLRLM features, local contour features are also extracted, by splitting the boundary into smaller segments and applying one of functions presented in Eqs. (4) to (6). As proposed by Li et al. [13], we consider the number of segments (S) to be from \(\{2, 4, 6, 8, 10, 12, 14, 16, 18, 20\}\). The value of S also defines the number of extracted features from the 1-dimensional data (\(\varDelta d\)).
The parameters of the feature selection methods are selected by running preliminary experiment by following the methodology presented in our previous work [3]. According to these results, the minimum explained variance of the PCA is set to 0.99 and for GA’s fitness function a DT is used and the classification accuracy is maximized.
4.3 Results
In the current experiment, we built different systems – detailed in Sect. 3 – for the binary classification problem of determining the type of a lesion. The performance of the proposed method is evaluated using measures such as accuracy, sensitivity, specificity, and f1-score which are commonly used in medical classification problems. The geometrical features are implemented as baseline, and their results are compared to more advanced feature extraction methods.
In our experiments, first, we defined the best shape feature and the optimal number of segments S. The results are presented in Table 1. As we can see Rq (as defined by Eq. (4)) has usually the best performance indifferent from S. We also conducted experiments where all three were used as input to the classifier. We refer to these features as ‘combo’ in Table 1. This combined feature has slightly better performance than \(R\varDelta q\) (as defined by Eq. (5)) or circularity (as defined by Eq. (6)) on its own but the results are still behind Rq. Furthermore, based on the information in the same table we decided to use \(S = 16\) in further experiments.
The scope of the current paper was to compare the performance of the textural features against shape features to distinguish benign and malignant lesions of the breast tissue. In the first row of Table 2, the performance of the baseline (geometrical features) is presented for the test set. All of the proposed feature extraction methods outperformed the baseline. Table 2 lists the results for the test set produced by the different feature extraction methods. Hence, we can conclude that the shape features are more suitable for the current classification of cancerous tissue. We conducted an experiment, using the combination of texture- and shape (contour) features and the achieved results are presented in the last row of Table 2.
Separate experiments were conducted to decide which classification and feature selection method combination performs the best. Figure 3 shows the performance achieved using Rq and indicates that with the increase in the number of segments the performance of the system also increases. Another observation is that the DT has lower results than RF (with 100 DTs in the ensemble). This can be explained by the “majority rule”, that the decision is made based on the opinion of the majority, hence reducing overfitting. Finally, although RF with GA has better accuracy using \(S = 16\), by looking at the total picture the classification results using PCA are more stable (the differences are smaller between the segments).
4.4 Discussion
In this subsection, we aim to compare the results reported by the proposed approach (as given in Sect. 4.3) with those of related relevant methods from the literature. In our experiments we are facing the problem of overfitting: on the train set the model can predict with 100% accuracy, but on the test set it has a performance of 65.55%.
Li et al. [13] presented a 99.66% accuracy using the same shape features (more precisely \(R\varDelta q\)) as input to an SVM classifier. The difference in the results can be explained by a different classification method, but also by a different experiment setup as Li et al. used 323 images from the total of 1440 samples with abnormalities from DDSM. Considering this difference we looked in more detail into the dataset and found that there are outliers regarding the regularity/irregularity of the lesions. An example is shown in Fig. 4. Therefore, we will further investigate if the removal of the outliers increase the precision of our system. On the other hand, we will examine other feature extraction methods on these outliers.
Muramatsu et al. [15] presented a CNN for this binary classification problem and achieved 72.5% by using images from DDSM to train the network. Our results approximate the result of the presented system.
In our previous study [3] we presented 92.23% accuracy using GLRLM features, PCA feature selection and RF classification, but on another dataset (Mammographic Image Analysis Society – MIAS). Another difference, compared to the current study is that the features are extracted from the full preprocessed image, not only from the lesion and its surrounding.
5 Conclusions and Further Work
In the current study we present a computer aided system to define the type of an abnormality in the breast tissue. Our aim was to compare the performance of textural- and shape features for the binary classification of breast cancer. Based on the results of our experiments, we can conclude that using shape features over textural features is more successful in distinguishing benign and malignant lesions. Also, by the combination of textural- and shape features the result increases and slightly outperforms the shape features. In the experiment basic geometrical features were also included, but based on their result we propose to further investigate and combine them with other features. The best results were achieved using a combination of features (Rq of 18 segments and GLRLM), GA feature selection and RF classification. The high train accuracy (100%) and fair test accuracy (65.55%) shows the problem of overfitting. Therefore, in future studies we will look into pruning methods and creating other splits to remove the outliers.
Notes
References
Ancy, C.A., Nair, L.S.: An efficient cad for detection of tumour in mammograms using SVM. In: 2017 International Conference on Communication and Signal Processing (ICCSP), pp. 1431–1435 (2017). https://doi.org/10.1109/ICCSP.2017.8286621
Ansar, W., Shahid, A.R., Raza, B., Dar, A.H.: Breast cancer detection and localization using MobileNet based transfer learning for mammograms. In: Brito-Loeza, C., Espinosa-Romero, A., Martin-Gonzalez, A., Safi, A. (eds.) ISICS 2020. CCIS, vol. 1187, pp. 11–21. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-43364-2_2
Bajcsi, A., Andreica, A., Chira, C.: Towards feature selection for digital mammogram classification. Procedia Comput. Sci. 192, 632–641 (2021). https://doi.org/10.1016/j.procs.2021.08.065
Bajcsi, A., Chira, C., Andreica, A.: Extended mammogram classification from textural features. Stud. Univ. Babes-Bolyai Inf. 67, 5–20 (2023). https://doi.org/10.24193/subbi.2022.2.01
Chaieb, R., Kalti, K.: Feature subset selection for classification of malignant and benign breast masses in digital mammography. Pattern Anal. Appl. 22(3), 803–829 (2019). https://doi.org/10.1007/s10044-018-0760-x
Chhikara, B.S., Parang, K.: Global cancer statistics 2022: the trends projection analysis. Chem. Biol. Lett. 10(1), 451 (2022)
Darweesh, M.S., et al.: Early breast cancer diagnostics based on hierarchical machine learning classification for mammography images. Cogent Eng. 8(1), 1968324 (2021). https://doi.org/10.1080/23311916.2021.1968324
Farhan, A.H., Kamil, M.Y.: Texture analysis of mammogram using local binary pattern method. J. Phys: Conf. Ser. 1530(1), 012091 (2020). https://doi.org/10.1088/1742-6596/1530/1/012091
Gurudas, V.R., Shaila, S.G., Vadivel, A.: Breast cancer detection and classification from mammogram images using multi-model shape features. SN Comput. Sci. 3(5), 404 (2022). https://doi.org/10.1007/s42979-022-01290-y
Heath, M., et al.: Current status of the digital database for screening mammography. In: Karssemeijer, N., Thijssen, M., Hendriks, J., van Erning, L. (eds.) Digital Mammography. Computational Imaging and Vision, vol. 13, pp. 457–460. Springer, Netherlands (1998). https://doi.org/10.1007/978-94-011-5318-8_75
Heath, M., Bowyer, K., Kopans, D., Moore, R., Kegelmeyer, P.: The digital database for screening mammography. In: Yaffe, M. (ed.) Proceedings of the Fifth International Workshop on Digital Mammography, pp. 212–218. Medical Physics Publishing (2001)
Kumari, L.K., Jagadesh, B.N.: A robust feature extraction technique for breast cancer detection using digital mammograms based on advanced GLCM approach. EAI Endorsed Trans. Pervasive Health Technol. 8(30), e3 (2022). https://doi.org/10.4108/eai.11-1-2022.172813
Li, H., Meng, X., Wang, T., Tang, Y., Yin, Y.: Breast masses in mammography classification with local contour features. Biomed. Eng. Online 16(1), 44 (2017). https://doi.org/10.1186/s12938-017-0332-0
Li, H., Niu, J., Li, D., Zhang, C.: Classification of breast mass in two-view mammograms via deep learning. IET Image Process. 15(2), 454–467 (2021). https://doi.org/10.1049/ipr2.12035
Muramatsu, C.: Improving breast mass classification by shared data with domain transformation using a generative adversarial network. Comput. Biol. Med. 119, 103698 (2020). https://doi.org/10.1016/j.compbiomed.2020.103698
Suckling, J., Parker, J., Dance, D.: The mammographic image analysis society digital mammogram database. In: International Congress Series, vol. 1069, pp. 375–378 (1994)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Bajcsi, A., Chira, C. (2023). Textural and Shape Features for Lesion Classification in Mammogram Analysis. In: García Bringas, P., et al. Hybrid Artificial Intelligent Systems. HAIS 2023. Lecture Notes in Computer Science(), vol 14001. Springer, Cham. https://doi.org/10.1007/978-3-031-40725-3_64
Download citation
DOI: https://doi.org/10.1007/978-3-031-40725-3_64
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-40724-6
Online ISBN: 978-3-031-40725-3
eBook Packages: Computer ScienceComputer Science (R0)