Abstract
A low-frequency variant of sushi, von Willebrand factor type A, EGF, and pentraxin domain-containing protein 1 (SVEP1) is associated with the risk of coronary artery disease, as determined by a genome-wide association study. SVEP1 induces vascular smooth muscle cell proliferation and an inflammatory phenotype to promote atherosclerosis. In the present study, qRT‒PCR demonstrated that the mRNA expression of SVEP1 was significantly increased in atherosclerotic plaques compared to normal tissues. Bioinformatics revealed that EGR1 was a transcription factor for SVEP1. The results of the luciferase reporter assay, siRNA interference or overexpression assay, mutational analysis and ChIP confirmed that EGR1 positively regulated the transcriptional activity of SVEP1 by directly binding to its promoter. EGR1 promoted human coronary artery smooth muscle cell (HCASMC) proliferation and migration via SVEP1 in response to oxidized low-density lipoprotein (ox-LDL) treatment. Moreover, the expression level of EGR1 was increased in atherosclerotic plaques and showed a strong linear correlation with the expression of SVEP1. Our findings indicated that EGR1 binding to the promoter region drive SVEP1 transcription to promote HCASMC proliferation and migration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cardiovascular disease (CVD) remains the leading cause of morbidity and mortality worldwide, most of which is due to myocardial infarction or stroke [1, 2]. Atherosclerosis is an inflammatory disease that induces the development of atherosclerotic plaques and vascular stenosis, which is the main underlying etiology of cardiovascular events [3]. However, the pathogenesis of atherosclerosis has not yet been clarified [4]. Despite improvements in current comprehensive measures, atherosclerosis is still a serious health threat and social burden [5]. Recently, accumulating evidence has revealed that smooth muscle cells play critical roles in atherosclerosis by switching to a proliferative, dedifferentiated, and inflammatory phenotype in response to atherogenic stimuli [6,7,8]. It is important to uncover the underlying mechanism to identify novel therapeutic targets.
SVEP1 encodes sushi (a complement control protein), von Willebrand factor type A, epidermal growth factor (EGF), and pentraxin domain-containing protein 1, an extracellular matrix (ECM) protein that directly interacts with integrin α9β1 [9]. In a large-scale exome-wide association study, a coding missense polymorphism in SVEP1 (p. D2702G) increased the risk of coronary artery disease relative to the wild type genotype (odds ratio, 1.14 per risk allele) [10]. Further study demonstrated that SVEP1 induced vascular smooth muscle cell proliferation and dedifferentiation to aggravate the formation of atherosclerotic plaques [11]. Winkler MJ et al. found that wild-type SVEP1 deficiency increased endothelial CXCL1 expression to enhance the recruitment of proinflammatory leukocytes [12].
SVEP1 is associated with atherosclerosis, but the expression of SVEP1 in coronary artery disease has not been thoroughly examined. In this study, we examined SVEP1 expression and showed that SVEP1 was significantly increased in the atherosclerotic plaques of patients with coronary artery disease compared to normal tissues. Furthermore, we explored the molecular mechanisms involved in the modulation of SVEP1 and found that the transcriptional activity of SVEP1 was dependent on the transcription factor early growth response-1 (EGR1). Finally, we identified that EGR1 promoted human coronary artery smooth muscle cell (HCASMC) proliferation and migration via SVEP1 through its function in the promoter region. This finding may contribute to potential therapeutic strategies to alter SVEP1 expression levels in patients with coronary artery disease.
Materials and methods
Subjects and sample collection
The vascular tissues of five patients undergoing thoracic aorta replacement surgery for aortic pathologies were collected from our hospital between September 2018 and April 2019. This study was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (2016-SR-144). Informed consent was obtained from the patients before participation. The collected arterial tissues were directly placed in cold saline and stored on ice during transport to the laboratory. The arterial samples were then dissected into atherosclerotic regions (plaques) and adjacent normal tissues and placed in tissue-freezing medium for sectioning [13].
Cell culture
Human embryonic kidney (HEK) 293 T cells and HCASMCs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum, 1% penicillin and streptomycin. The cells were incubated at 37 °C supplied with 5% CO2. Human coronary artery smooth muscle cells were stimulated with 50–100 mg/L ox-LDL (Thermo Fisher Scientific) to establish proliferation and migration models.
Quantitative real-time PCR (qRT‒PCR)
Total RNA was extracted from tissue and cells using TRIzol reagent (Invitrogen) and subsequently reverse-transcribed into cDNA using PrimeScript RT Reagent (Takara).
qRT‒PCR was performed on an Applied Biosystems Step One Plus Real-Time PCR System using SYBR Green technology (Takara). The specificity of amplification was assessed by analyzing the melting curve. Each sample was analyzed in triplicate with GAPDH as a normalization standard. The primers used for qRT‒PCR were as follows:
SVEP1: 5’-CAGCAGTTGCATTCCATGTCC-3’ (sense), 5’-AAGTAACCATTTTCGGGAGGC-3’ (antisense);
EGR1: 5’-GGTCAGTGGCCTAGTGAGC-3’ (sense), 5’-GTGCCGCTGAGTAAATGGGA-3’ (antisense); and.
GAPDH: 5’-ATGACATCAAGAAGGTGGTG-3’ (sense), 5’-CATACCAGGAAATGAGCTTG-3’ (antisense).
Plasmids and small interfering RNA (siRNA)
The SVEP1 genomic DNA fragment (-250 to + 100) was inserted into the pGL3-Basic vector (Promega) and named pGL-250/ + 100. The JASPAR database version 5.0 (jaspar.genereg.net) and hTFtarget software (http://bioinfo.life.hust.edu.cn/hTFtarget) were used to predict potential transcriptional binding sites. According to the site-directed mutagenesis kit (Takara) protocol, the EGR1 binding site mutant promoter (pGL3-mut) was created and purchased from Tsingke Biotech Co. Ltd. The site-specific mutated plasmids were named mut-EGR1-A, mut-EGR1-B, and mut-EGR1-A + B according to the binding sites. The pENTER-EGR1 overexpression plasmids and the corresponding pENTER control plasmid were kept in our laboratory.
For the knockdown assays, double-stranded siRNAs were synthesized and purified by high-performance chromatography (Gene Pharma). Sequences targeting EGR1 and SVEP1, as well as the negative control sequence, were as follows:
EGR1: 5’-GTGACTGTTTGGCTTATAATT-3’ (sense), 5’-TTATAAGCCAAACAGTCACTT-3’ (antisense);
E2F1: 5’-CACTGAATCTGACCACCAATT-3’ (sense), 5’-TTGGTGGTCAGATTCAGTGTT-3’ (antisense);
SP1: 5’-AUCACUCCAUGGAUGAAAUGATT-3’ (sense),
5’-UCSUUUCSUCCSUGGSGUGAUTT-3’ (antisense);
SVEP1: 5’-GCUACUAUCUAUUGGGUGA-3’ (sense),
5’-AGUCUAUAUCGAUGGGAAA-3’ (antisense); and.
Control: 5’-UUCUCCGAACGUGUCACGU-3’ (sense), 5’-ACGUGACACGUUCGGAGAATT-3’ (antisense).
Transfections and dual-luciferase reporter assays
Transfections were carried out in HEK 293 T cells and HCASMCs by using Lipofectamine™ 3000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. HEK 293 T cells in 96-well plates were cotransfected with 100 ng of each of the luciferase-containing plasmids plus 4 ng of a control pRL-TK plasmid as an internal control. After 24 h, the cells were harvested, and a luciferase assay was performed by using the Dual Reporter Assay System (Promega) and a TD-20/20 Turner Designs Iuminometer according to the manufacturer’s instructions. For overexpression or siRNA, the expression plasmid or siRNA was individually cotransfected into HEK 293 T cells and HCASMCs. The results are representative of at least three independent experiments performed in triplicate.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed with the EZ-Magna ChIP™ kit according to the manufacturer’s instructions. A total of 1 × 107 HEK 293 T cells were fixed with 1% formaldehyde for 10 min at room temperature. The cell lysates were sonicated to generate 200–1,000 bp DNA fragments. The soluble chromatin was incubated with anti-EGR1 (Proteintech), negative control anti-IgG (Millipore) or anti-acetyl histone H3 antibodies (Millipore) at 4 °C overnight with rotation. After reverse crosslinking and DNA purification, DNA from input or immunoprecipitated samples was examined by qRT‒PCR with SYBR Green (Takara). The primers are as follows:
5’-CAGACTCCAGGCGCAGAAG-3’ (sense); and 5’-GACTTCGCTGGCGTTTACAG-3’ (antisense).
cHCASMCs were lysed in ice-cold lysis buffer containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF; KeyGENE BioTECH). The samples were subjected to 10% SDS‒PAGE and transferred to nitrocellulose membranes. To block nonspecific sites, the membranes were incubated in 5% dry milk in TBS-T (0.25 M Tris–HCl; pH 7.6, 0.19 M NaCl, 0.1% Tween 20) for 2 h, and then the blots were incubated overnight at 4 °C with primary antibodies against GAPDH (Proteintech), EGR1 (Proteintech) and SVEP1 (R&D Systems). The membranes were washed three times with TBS-T and treated with goat anti-rabbit IgG or anti-mouse IgG (Proteintech). The signals were measured by a chemiluminescence (ECL) system, and the membranes were scanned and analyzed by Image Lab Software (Bio-Rad).
Enzyme-linked immunosorbent assay (ELISA)
Cell culture supernatants were harvested by centrifugation at 1000 × g for 10 min. ELISA kits (Invitrogen, #BMS223INST, #BMS213HS, and #KAC1211) were used to analyze the levels of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interleukin-1β (IL-1β) according to the manufacturer’s instructions. The concentrations of TNF-α, IL-6, and IL-1β were determined based on standard curves.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 (Dojindo) was used to measure cell viability according to the manufacturer’s instructions. HCASMCs were seeded in 96-well plates at a density of 2.5 × 104 per well. After cell transfection and ox-LDL treatment, the medium was replaced with 100 µL of complete medium, and then 10 µL of CCK-8 reagent was added per well. The cells were incubated at 37 °C for 2 h. The optical density (OD) was measured at 450 nm by a microplate reader (Thermo Fisher).
Cell scratch wound assay
HCASMCs were seeded in a six-well plate, transfected and treated. After 24 h, a straight line was drawn through the cells with a pipette tip, and medium containing 2% FBS was added. Images of the scratched cells were taken at 0 h and 24 h using a microscope. HCASMC migration was determined by analyzing the healed area of the scratch.
Flow cytometry assay
2.11. Apoptosis of HCASMCs was assessed by Annexin V-FITC Apoptosis Detection Kits (BD Pharmingen, Franklin Lakes, NJ, USA). After incubation for 24 h, transfected HCASMCs were harvested as a single cell suspension (1 × 106/mL) by trypsin digestion. The staining buffer containing Annexin V-FITC and propidium iodide was added to incubate HCASMCs at 4℃ for 30 min. The Flow Cytometer (Beckman Coulter, Miami, FL, USA) was used for apoptosis assay.
Statistical analysis
Experiments were performed at least in triplicate, and the data wwere expressed as mean ± standard deviation (SD). Statistical analysis was performed using GraphPad Prism 7. Differences among groups were analyzed using unpaired Student’s t test. P < 0.05 was considered statistically significant.
Results
SVEP1 expression in atherosclerotic plaques and HCASMCs treated with ox-LDL
The mRNA expression of SVEP1 was investigated by qRT‒PCR in 5 pairs of atherosclerotic plaques and matched normal tissues from patients with coronary artery disease. Our results demonstrated a significant increase in SVEP1 expression in atherosclerotic plaques compared to normal tissues (Fig. 1a). Then, we developed a HCASMC cell model treated with ox-LDL. Consistently, the mRNA and protein levels of SVEP1 were markedly increased in HCASMCs treated with ox-LDL (Fig. 1b, c and d).
SVEP1 is increased in atherosclerotic plaques and HCASMCs treated with ox-LDL. (a) Total RNA was extracted from atherosclerotic plaques and matched normal tissues. The mRNA level of SVEP1 was analyzed by qRT‒PCR. (b) The mRNA level of SVEP1 in HCASMCs with or without ox-LDL treatment was analyzed by qRT‒PCR. The difference was determined using an unpaired t test (***P < 0.001). (c, d) The protein expression of SVEP1 in HCASMCs with or without ox-LDL treatment was analyzed by Western blotting. The difference was determined using an unpaired t test. The data were representative of three independent experiments (***P < 0.001, *P < 0.05)
SVEP1 silencing alleviates the effect of ox-LDL on inflammation, proliferation and migration in HCASMCs
Two siRNAs (siRNA-SVEP1#1 and siRNA-SVEP1#2) were used to silence SVEP1. SVEP1 expression in HCASMCs was successfully decreased by siRNA-SVEP1#1 (Supplementary Fig. 1). To determine the effect of SVEP1 on HCASMC inflammation, proliferation, apoptosis and migration, ELISA, CCK-8 assays, flow cytometry assays and cell scratch wound assays were performed. The results showed that knockdown of SVEP1 inhibited inflammation in HCASMCs treated with ox-LDL (Fig. a, b and c). Similarly, silencing SVEP1 significantly attenuated HCASMC proliferation in response to ox-LDL treatment (Fig. 2d). As shown in Fig. 2e, wound healing in HCASMCs subjected to ox-LDL treatment was significantly reduced after transfection with siRNA-SVEP1. There was no significant difference in HCASMC apoptosis by SVEP1 downregulation (Fig. 2).
SVEP1 silencing alleviates the effect of ox-LDL on inflammation, proliferation and migration in HCASMCs. (a, b, c) The levels of TNF-α, IL-6, and IL-1β were measured by ELISA. (d) HCASMC proliferation was assessed using a CCK-8 assay. (e) HCASMC migration was measured by scratch wound assays. (f) The apoptosis of HCASMCs was assessed by flow cytometry assays ***P < 0.001, **P < 0.005 vs. NC; #P < 0.05, ##P < 0.005 vs. control
EGR1 positively regulates the basal transcriptional activity of SVEP1 by directly binding to its promoter
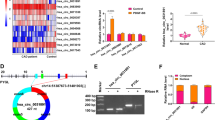
To determine the regulatory mechanism of the SVEP1 gene, we cloned the 350 bp fragment (-250/ + 100 relative to the transcriptional start site) into the pGL3-Basic luciferase reporter plasmid and named it pGL-250/ + 100. As shown in Fig. 3a, HEK 293 T cells transfected with pGL-250/ + 100 showed a high level of luciferase activity (13.1-fold more than pGL3-basic), indicating that the fragment we cloned (-250/ + 100) contained the human SVEP1 promoter region.
EGR1 positively regulates the basal transcriptional activity of SVEP1 (a) The human SVEP1 promoter sequence (from -250 to + 100) was inserted into the pGL3-basic plasmid. Numbering was relative to the TSS. The constructed vectors were transiently transfected into HEK 293 T cells, and luciferase activities were measured. The data are representative of three independent experiments and are expressed as the mean ± SD of normalized luciferase activity. (b) The specific binding site bases of each transcription factor. (c) HEK 293 T cells were cotransfected with the pGL-250/ + 100 plasmid and SP1 siRNA, E2F1 siRNA or EGR1 siRNA. (d) HEK 293 T cells were cotransfected with the pGL-250/ + 100 plasmid and the EGR1 overexpression plasmid
To identify potential transcription factor binding sites in -250/ + 100 bp, we conducted a series of analyses using Jaspar software (http://jaspar.genereg.net/) and hTFtarget software (http://bioinfo.life.hust.edu.cn/hTFtarget). The results showed that the SVEP1 promoter region contained binding sites for several transcription factors, such as SP1, E2F1 and EGR1 (Fig. 3b). The region had two EGR1 transcription factor binding sites.
To assess the effects of SP1, E2F1 and EGR1 on the regulation of SVEP1 promoter activity, we cotransfected the pGL-250/ + 100 plasmid with SP1, E2F1 or EGR1 siRNA. The results showed that only siRNA-EGR1 significantly reduced luciferase activity, while siRNA-SP1 and siRNA-E2F1 had no effect (Fig. 3c). In addition, overexpression of EGR1 markedly increased the activity of the SVEP1 promoter (Fig. 3d). These results indicated that the EGR1 binding site was essential for the basal activity of the SVEP1 promoter.
To confirm the critical role of the EGR1 binding site in SVEP1 transcription, a series of plasmids with 2–3 bp point mutations in EGR1 binding sites were constructed and transiently transfected into HEK 293 T cells (Fig. 3e). As shown in Fig. 3f, mutations in EGR1-A and EGR1-B reduced SVEP1 promoter activity by 60% and 80%, respectively, compared with wild-type promoter activity. Furthermore, double site mutations in EGR1 decreased promoter activity by 86%. Next, we examined the effect of EGR1 on SVEP1 promoter activity in response to mutations in its binding sites. The results showed that mutations of the EGR1 binding sites failed to response to silence or overexpression of EGR1 (Fig. 3g and h), further confirming that EGR1 positively regulated EGR1 gene transcription via its consensus binding site.
To investigate whether EGR1 binds to the promoter region of SVEP1 in vivo, a ChIP assay was performed on HEK 293 T cells. HEK 293 T cells were lysed, and chromatin was sonicated. EGR1-associated DNA fragments were immunoprecipitated with anti-EGR1 or anti-IgG antibodies. The DNA fragments were then analyzed by qRT‒PCR with primers targeting the potential binding site. The results showed that the anti-EGR1 antibody precipitated proteins bound to the tested sequence, but nonspecific IgG failed to precipitate proteins bound to this sequence (Fig. 4a and b).
EGR1 directly binds to the SVEP1 promoter region. (a) ChIP analysis of the binding of EGR1 to the SVEP1 promoter in vivo. The protein precipitated by the anti-EGR1 antibody could bind to the amplified sequence of the SVEP1 promoter, whereas the anti-IgG antibody did not precipitate the bound protein in the sequence. (b) The immunoprecipitated chromatin fragments were analyzed by quantitative PCR using primer pairs spanning the putative EGR1 binding site. Data were expressed as relative fold increase of specific antibody over normal IgG control
Overall, EGR1 positively regulates the basal transcriptional activity of SVEP1 by directly binding to its promoter region.
(e) A schematic map showing the potential binding of EGR1 in the SVEP1 gene promoter. The mutant bases are listed in the corresponding regions. (f) Site-directed mutagenesis was performed on the pGL-250/ + 100 construct. Various constructs were transfected into HEK 293 T cells, and luciferase activities were measured (***P < 0.001). (g) HEK 293 T cells were cotransfected with each mutant promoter plasmid and EGR1 siRNA. (h) HEK 293 T cells were cotransfected with each mutant promoter plasmid and the EGR1 overexpression plasmid. These results were representative of three independent experiments and were expressed as mean ± SD of the normalized luciferase activity (*P < 0.05, ***P < 0.001).
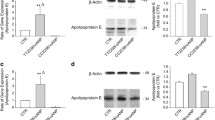
EGR1 directly drives the expression of SVEP1 in HCASMCs treated with ox-LDL
To assess the effect of EGR1 on the expression of SVEP1, the expression level of SVEP1 was measured in HCASMCs treated with ox-LDL after transient transfection with the EGR1 overexpression plasmid or siRNA. The results showed that SVEP1 mRNA levels were reduced by 60% in the presence of siRNA against EGR1 (Fig. 5a). In addition, the mRNA level of SVEP1 increased by 2.17-fold when EGR1 was overexpressed (Fig. 5b). These results were consistent with the results at the protein level (Fig. 5c and d). These data demonstrated that EGR1 positively regulated the expression of SVEP1 in HCASMCs treated with ox-LDL.
EGR1 positively regulated SVEP1 expression. (a) Silencing EGR1 reduced mRNA expression in HCASMCs treated with ox-LDL. (b) Overexpression of EGR1 caused an increase in mRNA expression in HCASMCs treated with ox-LDL. (c-d) Silencing or overexpressing EGR1 caused a reduction or an increase in protein expression, respectively, in HCASMCs treated with ox-LDL Each experiment was performed in triplicate, and significant differences were determined with Student’s t test (*P < 0.05, **P < 0.005)
EGR1 promotes HCASMC proliferation and migration via SVEP1
As shown in Fig. 6a and b, siRNA-EGR1 inhibited HCASMC proliferation and migration in response to ox-LDL treatment. Moreover, pEGR1 enhanced HCASMC proliferation and migration (Fig. 6c and d). Furthermore, siRNA-SVEP1 rescued the effect of pEGR1 on promoting the proliferation and migration of ox-LDL-treated HCASMCs (Fig. 6c and d).
The transcription of EGR1 correlates with the level of SVEP1 in atherosclerotic plaques and matched normal tissues
To examine the expression level of EGR1 and determine whether the expression level of EGR1 correlates with the expression of SVEP1, we extracted total RNA from atherosclerotic plaques and matched normal tissues from patients with coronary artery disease and analyzed their expression by qRT‒PCR. As shown in Fig. 7a, the expression level of EGR1 was increased in atherosclerotic plaques compared with matched normal tissues and showed a strong linear correlation with the expression of SVEP1 (Fig. 7b).
The transcription level of EGR1 correlates with the level of SVEP1 in atherosclerotic plaques and matched normal tissues. (a) The mRNA levels of EGR1 in atherosclerotic plaques and matched normal tissues were analyzed by qRT‒PCR. The differences in the expression levels of EGR1 were analyzed by using an unpaired t test (***P < 0.001). (b) The linear correlation between EGR1 and SVEP1 was tested with the Pearson correlation test (r2 = 0.9471, ***P < 0.001)
Discussion
In this study, we examined SVEP1 expression and showed that SVEP1 expression was significantly increased in atherosclerotic plaques compared to matched normal tissues from patients with coronary artery disease. However, little is known about the mechanism underlying the high expression of SVEP1. Thus, we found that the expression of SVEP1 was regulated by the transcription factor EGR1 through its promoter. Furthermore, EGR1 promoted HCASMC proliferation and migration in response to ox-LDL treatment via SVEP1. The expression level of EGR1 was increased in atherosclerotic plaques compared to normal tissues and showed a strong linear correlation with the expression of SVEP1.
Ayari H et al. found that SVEP1 mRNA levels were higher in human carotid atherosclerotic plaques than in paired adjacent normal tissues [14]. This result is consistent with our findings. Under atherogenic conditions, vascular smooth muscle cells adopt alternative phenotypes, including macrophage-like, foam cell-like, myofibroblast-like, and mesenchymal stem cell-like phenotypes [15, 16]. Vascular smooth muscle cell-derived fibroblasts synthesize matrix proteins in atherosclerotic plaques [17]. SVEP1, which is an extracellular matrix protein, might also be synthesized by vascular smooth muscle cell-derived cells.
Vascular smooth muscle cells play a particularly complex and crucial role in the development of atherosclerosis. Recent studies have revealed that vascular smooth muscle cells switch to form a large proportion of cells within plaques in response to exposure to pathologic stimuli [15, 16, 18]. Furthermore, a large percentage of coronary artery disease loci can regulate gene expression in vascular smooth muscle cells and influence behavior to promote atherosclerosis [19,20,21]. SVEP1 may affect vascular smooth muscle cells through pathways including integrin, Notch, and FGF receptor signaling [11], each of which is related to atherosclerosis [22,23,24,25].
EGR1 is an immediate-early gene that encodes a Cys2-His2 type zinc finger transcription factor [26]. EGR1 is activated by various stimuli, including cytokines, growth factors, hormones, mechanical injury, and hypoxia [27]. EGR1 regulates the transcriptional activity of proliferative, migratory and proinflammatory genes, including platelet-derived growth factor (PDGF)-A, PDGF-B, PDGF-C, collagen, suppressor of cytokine signaling-1, and colony-stimulating factor 1 receptor [26, 28]. The SVEP1 gene can be added to this group of genes because it also has two EGR1-binding motifs close to the TSS.
Fasolo F et al. showed that the mRNA and protein expression levels of EGR1 were upregulated in human and mouse carotid plaques compared to adjacent normal tissues [29]. Previous studies also revealed that the protein kinase C β/EGR1 axis plays a central role in animal models of atherosclerosis [30, 31]. Our study showed a similar.
result. EGR1 may be activated in response to atherogenic stimuli and then promotes atherosclerosis progression.
EGR1 has been implicated in the proliferation, differentiation, and death of vascular cells [32]. Yue Li et al. found that miR-191 suppressed the expression of EGR1 to reduce intimal thickening after carotid injury [33]. Zhang J et al. showed that an EGR1-specific DNAzyme regulated EGR1 to inhibit rat vascular smooth muscle cell proliferation [34]. EGR1 decoy ODNs inhibit vascular smooth muscle cell proliferation and neointimal hyperplasia in the arteries of rats with balloon injury [35]. These results demonstrate that EGR1 can promote HCASMC proliferation and migration. The effect of EGR1 on HCASMCs may be partly dependent on SVEP1.
We didn’t perform plaque SVEP1 IHC due to scarce tissue samples. Previous studies have showed that smooth muscle cells represent a major cellular source of SVEP1 under pathological conditions [11, 12]. And our study showed that there was a signifcant increase in SVEP1 expression level in HCASMCs treated with ox-LDL. So we chose HCASMCs for these in vitro studies.
In summary, our study revealed that the mRNA expression of SVEP1 was significantly increased in atherosclerotic plaques compared to normal tissues from patients with coronary artery disease and that the expression of the SVEP1 gene was regulated by the transcription factor EGR1. EGR1 promotes the proliferation and migration of HCASMCs via SVEP1 in response to oxidized low-density lipoprotein (ox-LDL) treatment. Thus, understanding this regulatory mechanism is critical for understanding the dysregulation of SVEP1 gene expression in atherosclerosis. Targeting EGR1 may provide novel therapeutic strategies for the treatment of atherosclerosis and other vascular diseases.
Data availability
The data in the current study are available. The WB images in Supporting Fig. 5 were uploaded.
References
Roth GA et al (2020) Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol 76(25):2982–3021
Benjamin EJ et al (2019) Heart disease and stroke statistics-2019 update: a report from the American Heart Association. Circulation 139(10):e56–e528
Hansson GK (2005) Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 352(16):1685–1695
Libby P (2021) The changing landscape of atherosclerosis. Nature 592(7855):524–533
Arnett DK et al (2019) 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines. Circulation 140(11):e596–e646
Pan H et al (2020) Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation 142(21):2060–2075
Miano JM et al (2021) Fate and state of vascular smooth muscle cells in atherosclerosis. Circulation 143(21):2110–2116
Cao G et al (2022) How vascular smooth muscle cell phenotype switching contributes to vascular disease. Cell Commun Signal 20(1):180
Sato-Nishiuchi R et al (2012) Polydom/SVEP1 is a ligand for integrin α9β1. J Biol Chem 287(30):25615–25630
Myocardial infarction genetics and CARDIoGRAM exome consortia investigators, Stitziel NO et al (2016) Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med 374(12):1134–1144
Jung IH et al (2021) SVEP1 is a human coronary artery disease locus that promotes atherosclerosis. Sci Transl Med. 13(586):eabe0357
Winkler MJ et al (2020) Functional investigation of the coronary artery disease gene SVEP1. Basic Res Cardiol 115(6):67
Zhang H et al (2021) Augmenting ATG14 alleviates atherosclerosis and inhibits inflammation via promotion of autophagosome-lysosome fusion in macrophages. Autophagy 17(12):4218–4230
Ayari H et al (2013) Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J Biosci 38(2):311–315
Grootaert MOJ et al (2021) Vascular smooth muscle cells in atherosclerosis: time for a reassessment. Cardiovasc Res 117(11):2326–2339
Tang HY et al (2022) Vascular smooth muscle cells phenotypic switching in cardiovascular diseases. Cells 11(24):4060
Wirka RC et al (2019) Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med 25(8):1280–1289
Kansakar U et al (2021) Targeting the phenotypic switch of vascular smooth muscle cells to tackle atherosclerosis. Atherosclerosis 324:117–120
Aherrahrou R et al (2020) Genetic regulation of atherosclerosis-relevant phenotypes in human vascular smooth muscle cells. Circ Res 127(12):1552–1565
Tcheandjieu C et al (2022) Large-scale genome-wide association study of coronary artery disease in genetically diverse populations. Nat Med 28(8):1679–1692
Solomon CU et al (2022) Effects of coronary artery disease-associated variants on vascular smooth muscle cells. Circulation 146(12):917–929
Finney AC et al (2017) Integrin signaling in atherosclerosis. Cell Mol Life Sci 74(12):2263–2282
Chen PY et al (2016) Smooth muscle FGF/TGFβ cross talk regulates atherosclerosis progression. EMBO Mol Med 8(7):712–728
Martos-Rodriguez CJ et al (2021) Fibrous caps in atherosclerosis form by notch-dependent mechanisms common to arterial media development. Arterioscler Thromb Vasc Biol 41(9):e427–e439
Peng X et al (2023) Role of the notch1 signaling pathway in ischemic heart disease (review). Int J Mol Med 51(3):27
Khachigian LM (2021) Early growth response-1, an integrative sensor in cardiovascular and inflammatory disease. J Am Heart Assoc 10(22):e023539
Xie Y et al (2023) Early growth response-1: key mediators of cell death and novel targets for cardiovascular disease therapy. Front Cardiovasc Med 10:1162662
Wang B et al (2010) Phosphorylation and acetylation of histone H3 and autoregulation by early growth response 1 mediate interleukin 1beta induction of early growth response 1 transcription. Arterioscler Thromb Vasc Biol 30(3):536–545
Fasolo F et al (2021) Long noncoding RNA MIAT controls advanced atherosclerotic lesion formation and plaque destabilization. Circulation 144(19):1567–1583
Yan SF et al (2006) Protein kinase C beta/early growth response-1 pathway: a key player in ischemia, atherosclerosis, and restenosis. J Am Coll Cardiol 48(9):A47–55
Andrassy M et al (2005) Centrol role of PKC beta in neointimal expansion triggered by acute arterial injury. Circ Res 96(4):476–483
Khachigian LM et al (1996) Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science 271(5254):1427–1431
Li Y et al (2016) MicroRNA miR-191 targets the zinc finger transcription factor Egr-1 and suppresses intimal thickening after carotid injury. Int J Cardiol 212:299–302
Zhang J et al (2013) An Egr-1-specific DNAzyme regulates Egr-1 and proliferating cell nuclear antigen expression in rat vascular smooth muscle cells. Exp Ther Med 5(5):1371–1374
Han W et al (2010) EGR-1 decoy ODNs inhibit vascular smooth muscle cell proliferation and neointimal hperplasia of balloon-injured arteries in rat. Life Sci 86(7–8):234–243
Funding
This work was supported by grants from the National Natural Science Foundation of China (81970579 to GZP, 82101816 to QC, 82270068 to RJ) and the Natural Science Foundation of Jiangsu Province (BK20210965 to QC).
Author information
Authors and Affiliations
Contributions
Qiang Tian, Jia-He Chen, and Yi Ding wrote the main manuscript. Xin-Yu Wang, Jia-Yun Qiu and Li–Li Zhuang assisted in the methodology. Qian Cao, Rui Jin and Guo-Ping Zhou received funding for the study and led the study. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (2016-SR-144). Informed consent was obtained from patients before participation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tian, Q., Chen, JH., Ding, Y. et al. EGR1 transcriptionally regulates SVEP1 to promote proliferation and migration in human coronary artery smooth muscle cells. Mol Biol Rep 51, 365 (2024). https://doi.org/10.1007/s11033-024-09322-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09322-x