Abstract
Background
Currently, no recognized evidence is known about the bacterial communities found within seminal vesicles (SV) of men presenting with refractory hematospermia.
Methods and results
Fifteen male patients with refractory hematospermia or anejaculation were enrolled, and 15 SV-Infection (SV-In) samples from SV with hemorrhage and/or stones, 11 SV-Control (SV-C) samples from SV with non-infection, and 14 Urine (Urine) samples from posterior urethra were obtained via transurethral seminal vesiculoscopy. Then the high-throughput 16 S rRNA gene sequencing method was performed to characterize the microbiota profile. Finally, a total of 1535 operational taxonomic units (OTUs) were found, 1295 OTUs were shared across three groups, 7 OTUs, 45 OTUs, and 48 OTUs were unique to SV-C group, SV-In group, and Urine group, respectively. The 5 top bacterial phyla (mean relative abundance) in all samples were Firmicutes (52.08%), Bacteroidetes (21.69%), Proteobacteria (12.72%), Actinobacteria (9.64%), and Fusobacteria (1.62%), the 5 top bacterial genera in all samples were Bacteroides (9.13%), Lactobacillus (5.38%), Bifidobacterium (5.35%), Faecalibacterium (5.10%), and Allobaculum (3.34%), of which Bifidobacterium had the highest level in SV-C samples and had a significant difference (P < 0.05) across all groups. Differential analysis showed genera Leuconostoc and LachnospiraceaeFCS020group were identified as biomarkers in the SV-In microbiota.
Conclusion
Altered microbiota composition in seminal vesicles is related to refractory hematospermia in men, and the distribution of genus Leuconostoc or LachnospiraceaeFCS020group within seminal vesicles may interact with hematospermia. This study provides clues for the diagnosis and treatment of this urologic disorder.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seminal vesicles (SV) are a pair of two tubular glands that secrete fluid to compose around 50–80% of the semen, the lower part of each vesicle joins with the end of the ipsilateral vas deferens to form the ejaculatory duct which enters the prostate gland obliquely and opens into the prostatic urethra around verumontanum [1]. Inflammation, infection, trauma, or congenital malformation within this area of the male genital tract may induce hematospermia, painful ejaculation, or infertility due to obstruction of the ejaculatory duct, and transurethral endoscopic managements are primary treatment options for these conditions [2]. Particularly, transurethral seminal vesiculoscopy (TSV) and related procedures are effective treatments for refractory hematospermia with no obvious postoperative complications [3], and hematospermia was alleviated or disappeared in 80.56–93.2% of patients after surgery [4,5,6,7]. Besides, TSV is an effective technique for diagnosing and treating intractable seminal vesiculitis [8].
The etiology and underlying pathology of hematospermia are often difficult to determine and treatment strategies are not clear and concise. Though it is reported that nonspecific inflammation and/or infection were the most common non-iatrogenic etiology of hematospermia [9], and bacteria were found in more than one-third of patients with hematospermia by routine bacteriological culture [10]. With the fast development of high-throughput sequencing methods, microbial characterization can be quickly and efficiently completed in a culture-independent manner [11]. Microbiomes play important roles in human health and disease, and currently, no recognized evidence is known about the bacterial communities found within seminal vesicles of men presenting with hematospermia. In this study, we use the high-throughput 16 S rRNA gene sequencing method to characterize the microbiota profile present in samples obtained during TSV, we aim to determine if some substantial specific microorganisms or taxa may be associated with hematospermia.
Materials and methods
Study population
Enrolled in this study were male patients who underwent TSV at the Department of Urology, Beijing Chao-Yang Hospital, Capital Medical University from July 2021 to December 2021. Patients who underwent TSV were included if they had a chief complaint of either refractory hematospermia or anejaculation urging for endoscopic sperm retrieval, men with hematospermia had a hemorrhage in the seminal vesicle were all confirmed by magnetic resonance imaging. At last, 15 men were enrolled, in which 12 men were accompanied by refractory hematospermia and 3 men were accompanied by anejaculation. Patients were excluded if they had urethrostenosis, congenital absence of the vas deferens, prostatitis, acute infection of the urinary tract, a history of sexually transmitted diseases, or positive for HBV, HCV, HIV, or treponema pallidum. All patients included in the study were married men and over 18 years old with serum PSA levels less than 4.0 ng/mL, and written informed consent were signed and all patients were evaluated and followed in compliance with the protocol approved by the local ethics committee of the hospital (No. 2021-KE-482).
Sample collection
A rigid vesiculoscope (4.5/6.5-Fr, Wolf, Germany) was used for performing TSV, patients were placed in the lithotomy position and received general anesthesia or combined spinal and epidural anesthesia. First, urine samples (Urine group), serving as a background control for the intraurethral environment, were obtained directly from the posterior urethra through the working channel using the 5 mL syringe after examination of the urethra and bladder; then, fluid samples inside the seminal vesicle were obtained through the working channel when the vesiculoscope was inserted into the seminal vesicle, either by entering into the natural opening of the ejaculatory duct or by using a transutricle fenestration method. Samples obtained from the seminal vesicle were classified as SV-Infection (SV-In) group if hemorrhage and/or stones existed, whereas samples were allocated to SV-Control (SV-C) group if no aforesaid change was noticed inside the seminal vesicle. To avoid possible sample contamination or potential iatrogenic infection, the working channel of the vesiculoscope was rinsed with sterile normal saline at the interval between collecting specimens from different sites, and the non-hemorrhagic side of the seminal vesicle was explored first if there was merely unilateral hemorrhage observed in preoperative imaging. Samples were stored immediately under freezing at -80 °C for subsequent DNA extraction.
DNA extraction and 16 S rRNA gene V3-V4 sequencing
Total DNA extraction was performed according to the manufacturer’s guidelines for microbial analysis using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The concentration and quality of extracted DNA were tested before sequencing using a NanoDrop instrument (Thermo Fisher, Waltham, MA, USA), and the obtained DNA was stored under freezing at -80°C for subsequent analysis. To characterize the composition and structure of bacterial communities indigenous to the specific location of the male reproductive tract, we sequenced the V3-V4 regions of 16 S rRNA gene amplified by PCR Zone from each sample. DNA was amplified with 1x KAPA HiFi Master Mix (Coyote, Beijing, China) using primers (primer-F: 5’-CCTAYGGGRBGCASCAG-3’; primer-R: 5’-GGACTACNNGGGTATCTAAT-3’). The first PCR conditions were: 95 °C for 3 min; followed by 30 cycles of 98 °C for 15 s, 50 °C for 50 s, and 72 °C for 30 s, and 1 cycle of 72 °C for 10 min. Then the amplification products were purified with VAHTS clean beads and barcoded adapters were attached, the second PCR was performed in only 8 cycles with an annealing temperature of 58 °C for 30 s under similar conditions as above mentioned. Finally, high throughput 16 S rRNA gene sequencing was performed using the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA).
Sequence data analysis
The sequences acquired went through a filtering process to obtain the qualified reads. Total reads were merged, removed low-quality sequences, and removed chimera sequences. The sequences were clustered into operational taxonomic units (OTUs) using the Uclust method in the QIIME software package at 97% similarity, then the taxonomic classification of the representative sequence for each OTU was done based on the Silva Reference database (Release 128). Alpha-diversity indices (chao1 index, shannon index, simpson index, and observed species) based on the OTU level abundance profile were calculated using the Vegan package in R software (version 3.5.1), beta-diversity indices of partial least squares discriminant analysis (PLS-DA) and principal coordinate analysis (PCoA) based on the unifrac distances were also calculated by using the ggplot2 package. Linear discriminant analysis (LDA) of effect size (LEfSe) was applied to determine the most discriminant taxa across groups.
Statistical analysis
The metric variables were shown as the mean ± standard deviation, the categorical variables were shown as the median and interquartile range and statistical significances were assessed using the Kruskal-Wallis test. All calculations were performed using SPSS statistical software (version 23, IBM SPSS, Chicago, IL, USA). Probability values of less than 0.05 were considered significant.
Results
Population characteristics
Fifteen male patients who underwent TSV were enrolled in this study, the patients had a mean age of 38.9 ± 10.3 years and a mean disease duration time of 28.8 ± 23.3 months. Finally, 15 SV-In samples, 11 SV-C samples, and 14 Urine samples were obtained for further microbiota analysis, and the mean operation time for TSV was 79.0 ± 27.3 min. Of the 12 men with refractory hematospermia, the mean age, disease duration time, and operation time were 39.6 ± 11.5 years, 24.0 ± 19.6 months, and 86.3 ± 25.7 min, respectively. Treatments of irrigation with sterile normal saline in the seminal vesicle during TSV, perioperative antibiotic prophylaxis, and postoperative regular semen discharge were used in men presenting with hematospermia, and no recurrent hematospermia was informed with a mean follow-up of 9.3 ± 1.4 months. All patient clinical manifestations and sample types retrieved during TSV were shown in Table 1, an illustration including magnetic resonance imaging and three groups of samples retrieved during TSV from Patient 11 was shown in Fig. 1.
Magnetic resonance imaging, endoscopic findings, and samples retrieved during TSV in men presenting with refractory hematospermia. (A&B) Relatively high-intensity signals on T1WI and low-intensity signals on T2WI were shown in magnetic resonance imaging, indicating a hemorrhage in the LSV from Patient 11; (C) chronic hemorrhagic foci and brown sediments were found in LSV during TSV; (D) no hemorrhage was found in RSV during TSV; (E) an illustration showing three groups of samples retrieved from Patient 11. TSV, transurethral seminal vesiculoscopy; T1WI, T1 weighted image; T2WI, T2 weighted image; RSV, right seminal vesicle; LSV, left seminal vesicle; SV, seminal vesicle; SV-C, the SV-Control group; SV-In, the SV-Infection group; Urine, the Urine group
Taxonomic diversity and microbial community
A total of 6,620,373 paired-end (PE) reads were obtained from all 40 samples, and after sequence assembly, quality filtering, and chimera removal, a total of 2,418,978 high-quality classifiable sequences were obtained, with a mean of 60,474 ± 26,374 sequences per sample. Clustering analysis showed an average of 730 ± 154 OTUs, 731 ± 145 OTUs, and 682 ± 201 OTUs were identified in each sample in the SV-C group, the SV-In group, and the Urine group respectively, though no significant difference was observed across groups. The total number of OTUs obtained from all samples was 1535, of these, 1295 OTUs were shared across three groups, 7 OTUs were unique to the SV-C group, 45 OTUs were unique to the SV-In group, and 48 OTUs were unique to the Urine group (Fig. 2A).
Taxonomic classification and microbial diversity of samples retrieved during TSV. (A) Venn diagram demonstrating the overall overlap of OTUs according to sample source, OTUs are defined at 97% sequence similarity level; (B) alpha-diversity indices of chao1 index, shannon index, simpson index, and observed species plots of microbiota based on the OTUs level abundance profile among three groups; (C) beta-diversity indice of PLS-DA plots of microbiota based on sampling unit distribution among three groups; (D) beta-diversity indice of PCoA plots of microbiota based on weighted unifrac distances among three groups. TSV, transurethral seminal vesiculoscopy; OTUs, operational taxonomic units; PLS-DA, partial least squares discriminant analysis; PCoA, principal coordinate analysis; SV, seminal vesicle; SV-C, the SV-Control group; SV-In, the SV-Infection group; Urine, the Urine group
The diversity index is used to measure the heterogeneity of microbial communities. Although there were no statistically significant differences in richness and evenness (chao1, shannon, and simpson indexes) or observed species of microbiota between either two groups (Fig. 2B), we observed SV-C samples had lower observed richness than other samples, which was consistent with Venn diagram. Distinct clusters of microbiota based on the unifrac distances were observed among three groups in PLS-DA plots but not in PCoA plots (Fig. 2 C&D).
Microbiota composition
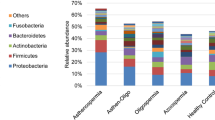
The 5 top bacterial phyla (mean relative abundance) in all samples were Firmicutes (52.08%), Bacteroidetes (21.69%), Proteobacteria (12.72%), Actinobacteria (9.64%), and Fusobacteria (1.62%) (Fig. 3 A), of which Proteobacteria and Fusobacteria had significant differences (P < 0.05) across three groups (Fig. 3 C&D). The 15 top bacterial genera (mean relative abundance) in all samples included Bacteroides (9.13%), Lactobacillus (5.38%), Bifidobacterium (5.35%), Faecalibacterium (5.10%), Allobaculum (3.34%), Blautia (2.90%), Escherichia-Shigella (2.83%), Streptococcus (2.39%), Prevotella9 (2.35%), Subdoligranulum (1.61%), Lachnospira (1.56%), Megamonas (1.44%), Bacillus (1.41%), Catenibacterium (1.40%), and Anaerobacillus (1.29%) (Fig. 3B), of which Bifidobacterium had the highest level in SV-C samples and had significant difference (P < 0.05) across three groups (Fig. 3E).
Frequencies of the most common bacterial phyla and genera found in samples. (A) The mean relative abundances of the 5 top bacterial phyla in total samples and in samples of each group; (B) the mean relative abundances of the 15 top bacterial genera in total samples and in samples of each group; the relative abundances of the phyla of Proteobacteria (C) and Fusobacteria (D) and the genera of Bifidobacterium (E) in samples of each group. #P < 0.05 across three groups, *P < 0.05 between SV-C and Urine groups. SV, seminal vesicle; SV-C, the SV-Control group; SV-In, the SV-Infection group; Urine, the Urine group
Differential analysis
LEfSe analysis was used to explore bacterial biomarkers across groups, and discriminative taxa with LDA score > 2 were selected. The SV-In microbiota was characterized by the dominance of the genera of Leuconostoc and LachnospiraceaeFCS020group, the family of Spirochaetaceae, and the order of Aeromonadales. And the Urine microbiota was characterized by the dominance of the genera of Neisseria, Eubacterium ruminantiumgroup, Porphyromonas, Aeromonas, Vulcaniibacterium, and Actinomyces, the families of Enterobacteriaceae, Fusobacteriaceae, Neisseriaceae, Aeromonadaceae, and Actinomycetaceae, the orders of Enterobacteriales, Fusobacteriales, Neisseriales, and Actinomycetales, the class of Fusobacteriia, and the phyla of Proteobacteria and Fusobacteria (Fig. 4 A&B). The relative abundances of biomarkers in the genera level of the SV-In microbiota and the Urine microbiota in all samples were shown in Fig. 4 C&D, while the SV-C microbiota had no significant biomarker identified.
Differential analysis of bacterial biomarkers found in seminal vesicles relating to refractory hematospermia. (A) Histogram of the LDA scores computed for differentially abundant bacterial taxa in SV-In and Urine groups by LEfSe analysis, results with LDA score > 2 were selected; (B) cladogram of differentially abundant bacterial taxa in SV-In and Urine groups, differences are represented in the color of the most abundant classes (red indicating SV-In, green indicating Urine, and yellow indicating non-significant), each circle’s diameter is proportional to the taxon’s abundance; (C) the abundances of the genera of Leuconostoc and LachnospiraceaeFCS020group in samples of each group; (D) the abundances of the genera of Neisseria, Eubacterium ruminantiumgroup, Porphyromonas, Aeromonas, Vulcaniibacterium, and Actinomyces in samples of each group. LDA, linear discriminant analysis; LefSe, linear discriminant analysis effect size; SV, seminal vesicle; SV-C, the SV-Control group; SV-In, the SV-Infection group; Urine, the Urine group
Discussion
Our study provides some evidence that the altered bacterial communities in the male genital tract may be related to refractory hematospermia, and specific pathogenic microorganisms may be important factors for inflammation, infection, and hemorrhage within seminal vesicles. It is reported that hematospermia occurs due to multiple reasons including infectious, inflammatory, cystic, lithiasis, vascular, and iatrogenic issues, and it originated anatomically from seminal vesicles, prostate, bladder, spermatic cord, or epididymis [12]. In a prospective study to investigate the natural history of hematospermia, Furuya et al. found that hematospermia resolved spontaneously in 88.9% of the patients with a median disease duration of 1.5 months, and hemorrhage from seminal vesicle was important for the prediction of hematospermia duration [13]. Besides, nonspecific inflammation and/or infection were reviewed as the most common non-iatrogenic etiology of hematospermia [9], and bacteria were found in 36% of patients who suffered from hematospermia [10].
Predominant bacterial genera presenting in the male genital tract share an overlapping profile with that in the gastrointestinal tract. In this study, we found that the most abundant bacterial genera in the seminal vesicle include Bacteroides, Lactobacillus, Bifidobacterium, Streptococcus, and Prevotella, which was consistent with the literature report. Evidence showed that the common bacterial genera in the urine of a healthy individual include Lactobacillus, Streptococcus, Staphylococcus, and Gardnerella [14], the core microbes residing in a healthy human colon include Lactobacillus, Streptococcus, Prevotella, Bacteroides, and Clostridium [15], the most abundant genera in the penile microbiota of black South African men are Corynebacterium and Prevotella [16], and the most abundant genera in the semen are Lactobacillus, Pseudomonas, Prevotella and Gardnerella [17].
Besides, our results showed that Lactobacillus and Bifidobacterium were among the three most abundant bacterial genera distributed in urine and seminal vesicle, though Lactobacillus had no significant difference across groups, and samples from the non-hemorrhage seminal vesicle had significantly higher levels of Bifidobacterium, suggesting the genus Bifidobacterium may play as potential probiotics in the male genital tract. Strains of Bifidobacterium and Lactobacillus have been safely and effectively used as probiotics for a long history to modulate the gut microbiota in intestinal health and disease [18], and it is reported that Lactobacillus-predominant semen was associated with good-quality sperm [19].
In addition, it is recognized that the Leuconostoc genus was classified as an opportunistic pathogen that causes a few clinically human infections [20], gut LachnospiraceaeFCS020group was found to be associated with very-low-density lipoprotein particles subclasses and metabolic disease etiology [21]. In this study, differential analysis of LEfSe results showed that genera Leuconostoc and LachnospiraceaeFCS020group were enriched in SV-In microbiota and were identified as biomarkers in the hemorrhage seminal vesicle, so the distribution of genus Leuconostoc or LachnospiraceaeFCS020group within seminal vesicles may be pathogenic and interplay with hematospermia. The impacts of microbiota in the male genital tract on the health of men still need additional studies.
Considering ethical issues, it is hardly possible to retrieve samples invasively from normal healthy volunteers, so direct comparing microbiota within seminal vesicles between patients with hematospermia and healthy men is infeasible. So, in this study, samples obtained from seminal vesicles with no obvious pathological change or from seminal vesicles of men with anejaculation were assigned to SV-C group against SV-In group and/or Urine group, and we designed Urine samples serving as a background control for intraurethral environment. Moreover, samples from the non-hemorrhagic side of the seminal vesicle were retrieved first if there was only unilateral hemorrhage suspected, to avoid potential sample contamination or iatrogenic infection.
Of course, there are some limitations to this study. The potential contamination of samples among different groups, the possibility of bacteria translocation from surrounding areas, the lack of further functional validation, the sequencing of v3-v4 region instead of the full length 16 S rRNA gene, or the small sample size enrolled in the study may compromise the main conclusions, though this study provides clues for the diagnosis and treatment of this urologic condition. Undoubtedly, the altered microbiota composition within seminal vesicles is linked to men’s health and disease, including refractory hematospermia, but the extent of this relationship is still unclear. Concurrently, species-level resolution of microbial communities and further exploration of probiotics and microbiota targeted treatment strategies for potentially infection-related issues are warranted.
Conclusion
In summary, we explored the microbiota profile within seminal vesicles by using the high-throughput 16 S rRNA gene sequencing method and identified some specific microorganisms that may interplay with hematospermia. The study concluded that altered microbiota composition in seminal vesicles is related to refractory hematospermia in men, and the distribution of genus Leuconostoc or LachnospiraceaeFCS020group within seminal vesicles may interact with hematospermia. This study provides clues for the diagnosis and treatment of this urologic disorder. Future studies investigating the potential diagnostic and therapeutic opportunities for the microbiota in the male genital tract are needed.
Data availability
16 S rRNA gene sequence data for each sample have been deposited at NCBI’s Sequence Read Archive (SRA; BioProject ID: PRJNA892056).
References
Modgil V, Rai S, Ralph DJ, Muneer A (2016) An update on the diagnosis and management of ejaculatory duct obstruction. Nat Rev Urol 13(1):13–20. https://doi.org/10.1038/nrurol.2015.276
Avellino GJ, Lipshultz LI, Sigman M, Hwang K (2019) Transurethral resection of the ejaculatory ducts: etiology of obstruction and surgical treatment options. Fertil Steril 111(3):427–443. https://doi.org/10.1016/j.fertnstert.2019.01.001
Liao LG, Li YF, Zhang Y, Li K, Zhu T, Li BJ, Wang Q, Liu XD, Luo Y, Zhou B, Jiang J (2019) Etiology of 305 cases of refractory hematospermia and therapeutic options by emerging endoscopic technology. Sci Rep 9(1):5018. https://doi.org/10.1038/s41598-019-41123-2
Chen WK, Yu DD, Chen ZX, Li PF, Cai J, Liu YP, Wu ZG (2021) Transurethral seminal vesiculoscopy for intractable hematospermia: experience from 144 patients. BMC Urol 21(1):48. https://doi.org/10.1186/s12894-021-00817-4
Wang XS, Li M, Shao GF, Sun WD, Zhang XL, Xiao ZY, Ma Z, Yuan MZ, Guo LQ (2020) Real-time transrectal ultrasound-guided seminal vesiculoscopy for the treatment of patients with persistent hematospermia: a single-center, prospective, observational study. Asian J Androl 22(5):507–512. https://doi.org/10.4103/aja.aja_134_19
Ren ZJ, Yang B, Lu DL, Liu SZ, Yang LC, Wang LC, Peng ZF, Liu LR, Dong Q (2020) Transurethral resection of ejaculatory duct combined with seminal vesiculoscopy for management of persistent or recurrent hemospermia in men with ejaculatory duct obstruction. BMC Urol 20(1):34. https://doi.org/10.1186/s12894-020-00589-3
Chen R, Wang L, Sheng X, Piao SG, Nian XW, Cheng X, Zhou T, Li HZ, Liu YW, Chen GH, Zhang CL, Kong DP, Xiao GA, Lu X, Jia ZY, Liu ZY, Sun YH (2018) Transurethral seminal vesiculoscopy for recurrent hemospermia: experience from 419 cases. Asian J Androl 20(5):438–441. https://doi.org/10.4103/aja.aja_76_17
Liu B, Li J, Li P, Zhang J, Song N, Wang Z, Yin C (2014) Transurethral seminal vesiculoscopy in the diagnosis and treatment of intractable seminal vesiculitis. J Int Med Res 42(1):236–242. https://doi.org/10.1177/0300060513509472
Drury RH, King B, Herzog B, Hellstrom WJG (2021) Hematospermia Etiology, Diagnosis, Treatment, and Sexual Ramifications: A Narrative Review. Sexual medicine reviews https://doi.org/10.1016/j.sxmr.2021.07.004
Saracoglu M, Ozturk H, Duran A, Atalay S (2015) Effect of microorganisms on etiology of hematospermia. Arch Ital Urol Androl 87(1):80–82. https://doi.org/10.4081/aiua.2015.1.80
Davidson RM, Epperson LE (2018) Microbiome sequencing methods for studying Human Diseases. Methods in molecular biology. (Clifton NJ) 1706:77–90. https://doi.org/10.1007/978-1-4939-7471-9_5
Suh Y, Gandhi J, Joshi G, Lee MY, Weissbart SJ, Smith NL, Joshi G, Khan SA (2017) Etiologic classification, evaluation, and management of hematospermia. Transl Androl Urol 6(5):959–972. https://doi.org/10.21037/tau.2017.06.01
Furuya S, Masumori N, Takayanagi A (2016) Natural history of hematospermia in 189 japanese men. Int J urology: official J Japanese Urol Association 23(11):934–940. https://doi.org/10.1111/iju.13176
Shoemaker R, Kim J (2021) Urobiome: an outlook on the metagenome of urological diseases. Investig Clin Urol 62(6):611–622. https://doi.org/10.4111/icu.20210312
Ruan W, Engevik MA, Spinler JK, Versalovic J (2020) Healthy human gastrointestinal microbiome: composition and function after a Decade of Exploration. Dig Dis Sci 65(3):695–705. https://doi.org/10.1007/s10620-020-06118-4
Onywera H, Williamson AL, Cozzuto L, Bonnin S, Mbulawa ZZA, Coetzee D, Ponomarenko J, Meiring TL (2020) The penile microbiota of Black South african men: relationship with human papillomavirus and HIV infection. BMC Microbiol 20(1):78. https://doi.org/10.1186/s12866-020-01759-x
Weng SL, Chiu CM, Lin FM, Huang WC, Liang C, Yang T, Yang TL, Liu CY, Wu WY, Chang YA, Chang TH, Huang HD (2014) Bacterial communities in semen from men of infertile couples: metagenomic sequencing reveals relationships of seminal microbiota to semen quality. PLoS ONE 9(10):e110152. https://doi.org/10.1371/journal.pone.0110152
Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA (2019) Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol 16(10):605–616. https://doi.org/10.1038/s41575-019-0173-3
Tomaiuolo R, Veneruso I, Cariati F, D’Argenio V (2020) Microbiota and Human Reproduction: the case of male infertility. High Throughput 9(2). https://doi.org/10.3390/ht9020010
Ogier JC, Casalta E, Farrokh C, Saihi A (2008) Safety assessment of dairy microorganisms: the Leuconostoc genus. Int J Food Microbiol 126(3):286–290. https://doi.org/10.1016/j.ijfoodmicro.2007.08.012
Vojinovic D, Radjabzadeh D, Kurilshikov A, Amin N, Wijmenga C, Franke L, Ikram MA, Uitterlinden AG, Zhernakova A, Fu J, Kraaij R, van Duijn CM (2019) Relationship between gut microbiota and circulating metabolites in population-based cohorts. Nat Commun 10(1):5813. https://doi.org/10.1038/s41467-019-13721-1
Acknowledgements
None.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization, H.L. and L.T.; Methodology, H.L., H.H., and Y.F.; Formal Analysis, H.L., H.H., and Y.F.; Investigation, H.L.; Writing - Original Draft, H.L.; Writing - Review & Editing, X.Z., Z.X., and L.T.; Visualization, X.Z. and Z.X.; Supervision, L.T.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University (No. 2021-KE-482).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lei, H., Han, H., Feng, Y. et al. Altered microbiota profile in seminal vesicles of men presenting with refractory hematospermia. Mol Biol Rep 50, 2381–2389 (2023). https://doi.org/10.1007/s11033-022-08139-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08139-w