Abstract
Background
Sporisorium scitamineum is the causative agent of smut disease in sugarcane. The tricky life cycle of S. scitamineum consists of three distinct growth stages: diploid teliospores, haploid sporidia and dikaryotic mycelia. Compatible haploid sporidia representing opposite mating types (MAT-1 and MAT-2) of the fungus fuse to form infective dikaryotic mycelia in the host tissues, leading to the development of a characteristic whip shaped sorus. In this study, the transition of distinct stages of in vitro life cycle and in planta developmental stages of S. scitamineum are presented by generating stable GFP transformants of S. scitamineum.
Methods and results
Haploid sporidia were isolated from the teliospores of Ss97009, and the opposite mating types (MAT-1 and MAT-2) were identified by random mating assay and mating type-specific PCR. Both haploid sporidia were individually transformed with pNIIST plasmid, harboring an enhanced green fluorescent protein (eGFP) gene and hygromycin gene by a modified protoplast-based PEG-mediated transformation method. Thereafter, the distinct in vitro developmental stages including fusion of haploid sporidia and formation of dikaryotic mycelia expressing GFP were demonstrated. To visualize in planta colonization, transformed haploids (MAT-1gfp and MAT-2gfp) were fused and inoculated onto the smut susceptible sugarcane cultivar, Co 97009 and examined microscopically at different stages of colonization. GFP fluorescence-based analysis presented an extensive fungal colonization of the bud surface as well as inter- and intracellular colonization of the transformed S. scitamineum in sugarcane tissues during initial stages of disease development. Noticeably, the GFP-tagged S. scitamineum led to the emergence of smut whips, which established their pathogenicity, and demonstrated initial colonization, active sporogenesis and teliospore maturation stages.
Conclusion
Overall, for the first time, an efficient protoplast-based transformation method was employed to depict clear-cut developmental stages in vitro and in planta using GFP-tagged strains for better understanding of S. scitamineum life cycle development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane smut caused by the basidiomycetous fungus Sporisorium scitamineum (Syd.) is one of the most important diseases of the crop. It causes significant decrease in cane yield and reduction in juice quality leading to a considerable economic loss in sugarcane production worldwide [1, 2]. The most characteristic symptom of smut infected stalk is the development of a whip-like sorus, composed of plant and pathogen cells modified from the apex of the primary meristem. Other associated symptoms include profuse tillering of the affected clumps resulting in poor cane formation [3]. The life cycle of S. scitamineum consists of three distinct growth stages namely: haploid sporidia, dikaryotic mycelia and diploid teliospores [4]. While the non-pathogenic haploid sporidial stage is represented by two distinct and opposite mating type sporidia viz., MAT-1 and MAT-2, the pathogenic dikaryotic mycelial stage is manifested by systemic colonization of the host plant [5].
GFP (Green Fluorescent Protein), a reporter gene, isolated from the jellyfish Aequorea victoria [6] is a useful marker to visualize protein localization and microbial colonization during plant-pathogen interactions [7]. Several researchers have employed GFP-tagged strains to visualize, differentiate and analyze the establishment and development of the fungal colonization within host plants [8]. It is a helpful tool to investigate plant-pathogen interactions, as the chromophore in GFP is inherent to the primary structure of the protein and it does not require any substrate or co-factor. It emits bright green fluorescence, when excited under UV/blue light and is extremely stable under natural conditions [9].
GFP tagging and expression was first reported in Ustilago maydis [10], among the filamentous fungi. Thereafter, host colonization by a wide range of filamentous fungi viz., Leptosphaeria maculans [11], Fusarium graminearum [12], Aspergillus carbonarius [13], Colletotrichum falcatum [14], etc. were investigated using GFP-tagged strains.
Genetic transformation in filamentous fungi has been mainly achieved by Agrobacterium tumefaciens mediated transformation (ATMT) or by Polyethylene glycol (PEG)-mediated protoplast transformation [15]. In S. scitamineum, a reliable gene transformation method using ATMT was first developed by Sun et al. [16] and this strategy was utilized for understanding the life cycle development of S. scitamineum using a GFP-tagged strain [17]. On the other hand, protoplast-based PEG-mediated transformation has been routinely used for visualizing in planta colonization in a wide range of filamentous fungi, but not in S. scitamineum [18]. Though simple and efficient, optimization and implementation of this method requires significant efforts as it is reported to be strain dependent.
As the rate of success of ATMT was reported to be low in S. scitamineum [17], in the present study, a modified and highly efficient protoplast-based PEG-mediated transformation was developed, and employed for GFP-tagging to enhance the understanding on the transition of fungal developmental stages by direct visualization of in planta colonization in sugarcane on a spatio-temporal pattern.
Materials and methods
Fungal strain and plant material
Teliospores of S. scitamineum Ss97009, one of the standard high virulent isolates collected from the smut susceptible sugarcane cultivar, Co 97009, from the experimental farm of ICAR-SBI, Coimbatore, India was used for isolating compatible haploid sporidia (MAT-1 and MAT-2) for gene transformation experiments. Single budded setts of the sugarcane cultivar, Co 97009 were incubated under high moist conditions for 7 days to induce sprouting and used for studies on in planta colonization.
Plasmid
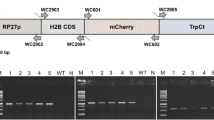
pNIIST plasmid, a shuttle vector containing eGFP and hygromycin phosphotransferase gene under the control of Aspergillus nidulans Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter and Tryptophan synthase (TrpC) terminator was used in this study for gene transformation (Supplementary Fig. S1). The vector was constructed by ligating the eGFP (720 bp) and hygromycin (1020 bp) gene expression cassettes, with GAPDH promoter and TrpC terminator in a pUC19 backbone [19].
Isolation and confirmation of distinct mating types of the S. scitamineum isolate Ss97009
Isolation of compatible haploid sporidia (MAT-1 and MAT-2) was done according to the procedure reported by Barnabas et al. [20] with some minor modifications. Fresh teliospores collected from smut infected meristems of cv. Co 97009 were rinsed three times with sterile distilled water and surface sterilized using streptomycin sulphate (500 µg/mL) for 2 min. The surface sterilized teliospores were plated onto Potato Dextrose agar (PDA) medium (HiMedia, India) amended with streptomycin sulphate (500 µg/mL) and incubated at 28 °C. After 48 h, the culture was serially diluted, plated and incubated as described above to isolate haploid colonies. Single haploid sporidial cultures thus obtained were transferred to Yeast Malt (YM) broth (3 g/L Yeast extract, 3 g/L Malt extract, 10 g/L Dextrose and 5 g/L Peptone) and incubated on a shaker (130 rpm) at 28 °C for 24 h. After incubation, random combinations of single haploid sporidial cultures were subjected to mating on PDA with 1% charcoal and streptomycin sulphate (500 µg/mL) and the plates were incubated at 28 °C under dark condition for 48 h. The haploid sporidia were identified either as identical/same or opposite mating types (+ or −), based on the smooth or fuzzy phenotype, as observed in the random mating assay.
For DNA extraction, the isolated distinct haploid sporidial cultures were mass multiplied in YM broth at 28 °C for 48 h and extracted using CTAB method as described by Abu Almakarem et al. [21]. The identity of MAT-1 and MAT-2 haploid sporidia was further confirmed by PCR using bE mating type gene specific primer sets viz., bE1F1 (FP: 5ʹ-ATGCGTGAATTTGCGC-3ʹ) and bE1-2R1 (RP: 5ʹ-TTGATGAACCAGAGCGTGAG-3ʹ), and bE2F2 (FP: 5ʹ-CTTTCCTACAATCCAAACCAATA-3ʹ) and bE1-2R1 (RP: 5ʹ-TTGATGAACCAGAGCGTGAG-3ʹ) (unpublished data).
Protoplast isolation
Protoplast isolation was optimized from the protocol reported by Yu et al. [22] with modifications in osmotic stabilizer, enzyme composition and digestion time. Wild-type MAT-1 cells cultured in YM broth (O.D - 0.8) were pelleted by centrifugation at 3000 rpm for 5 min at 4 °C, and washed with SCS solution containing D-sorbitol (20 mM trisodium citrate, pH 5.8 and 1 M D-sorbitol) or sucrose (20 mM trisodium citrate, pH 5.8 and 0.4 M sucrose) as an osmotic stabilizer. To optimize the enzyme solution, haploid pellet was mixed with four different enzyme digestion solutions in SCS buffer viz., 20 mg/mL lysing enzymes from Trichoderma harzianum (Sigma–Aldrich, USA); 5 mg/mL β-glucanase (Sigma–Aldrich, USA); 20 mg/mL lysing enzymes from Trichoderma harzianum + 5 mg/mL β-glucanase; 10 mg/mL lysing enzymes from Trichoderma harzianum + 10 mg/mL β-glucanase, and incubated at 80 rpm at 30 °C. SCS buffer without enzyme served as control. To optimize digestion time, the haploid cells were incubated in the enzyme solutions for three different time periods viz., 1, 2 and 3 h. The protoplasts were washed with ice-cold SCS solution twice and then with ice-cold STC solution containing D-sorbitol (10 mM Tris–HCl, pH 7.5; 1 M D-sorbitol and 100 mM CaCl2) or sucrose (10 mM Tris–HCl, pH 7.5; 0.4 M sucrose and 100 mM CaCl2) as an osmotic stabilizer. Finally, the washed protoplasts were resuspended in 1 mL of the same ice-cold STC solution with D-sorbitol or sucrose, as described above.
Protoplast enumeration and viability assay
The protoplasts were enumerated under the microscope using a haemocytometer and were checked by staining with two stains; Calcofluor White, a cell wall specific stain and Propidium Iodide, which stains non-viable (dead) protoplasts and cells. Untreated haploid sporidia were taken as a control. Calcofluor White-stained and Propidium Iodide-stained protoplasts/untreated haploids were observed using Leica DM LB2 epifluorescence microscope (Leica Microsystems, Germany) under UV filter (Excitation filter: BP340-380, Dichromatic mirror: 400 and Suppression filter: LP425) and N2.1 filter (Excitation filter: BP515-560, Dichromatic mirror: 580 and Suppression filter: LP590), respectively, and the images were captured with a Leica DMC2900 digital camera. Protoplast viability was calculated according to the equation: protoplast viability (%) = (number of protoplasts not stained/total number of protoplasts) × 100. Regeneration potential of the protoplasts was assessed by inoculating the protoplast suspension onto Regeneration agar (10 g/L Yeast Extract, 4 g/L Peptone, 4 g/L Sucrose, 1 M D-Sorbitol and 20 g/L Agar) cast on sterile glass slides and incubating them at 28 °C. Thereafter, a drop of Calcofluor White stain was added onto the slides at periodic time intervals (2 h, 4 h, 6 h, 8 h and 10 h) to visualize distinct stages of protoplast regeneration under epifluorescence microscope. Statistical analysis of data was performed using the software IBM SPSS Statistics 21.0 (SPSS, Chicago, USA). The data from triplicate observations were analyzed using one-way analysis of variance (ANOVA) and significant differences among treatments were determined at p ≤ 0.05 based on post-hoc Tukey’s test.
PEG-mediated protoplast transformation
Hygromycin B sensitivity of the Ss97009 MAT-1 haploid was tested prior to transformation by plating onto Yeast Malt (YM) agar (3 g/L Yeast extract, 3 g/L Malt extract, 10 g/L Dextrose, 5 g/L Peptone and 20 g/L Agar) containing different concentrations of hygromycin B (Calbiochem, USA) ranging from 25 µg/mL to 300 µg/mL. PEG-mediated transformation using circular plasmid DNA was optimized from the protocol described by Yu et al. [22] with few modifications. A total of 100 µL protoplasts (1 × 107 cells) obtained from MAT-1 and MAT-2 were individually mixed with 5 µg of pNIIST plasmid DNA (circular) and incubated on ice for 10 min. After incubation, 500 µL of PEG4000 dissolved in STC solution was added to the mixture and kept on ice for an additional 10 min. To optimize PEG concentration, different concentrations of PEG4000 viz., 10%, 20%, 30%, 40%, 50% were tested. After that, the protoplasts were regenerated on a 2-layer Regeneration agar, composed of a top layer of YePS soft agar (10 g/L Yeast extract, 20 g/L Peptone, 20 g/L Sugar and 6.5 g/L Agar) and a bottom layer of YePSS agar (10 g/L Yeast extract, 20 g/L Peptone, 20 g/L Sugar, 1 M D-Sorbitol and 20 g/L Agar), with only the bottom YePSS layer containing 50 µg/mL of hygromycin B for primary screening of transformants based on antibiotic resistance. The plates were incubated at 28 °C for 5–6 days. Untransformed Ss97009 haploid sporidia cultured on hygromycin B-amended (50 µg/mL) and hygromycin B-free Regeneration agar served as controls. The mitotic stability of the transformants was analyzed for five successive sub-cultures on YM agar with and without hygromycin B. The selected stable transformants were cryopreserved in 30% glycerol and stored at − 80 °C. The cultures were routinely grown in YM broth amended with hygromycin B (50 µg/mL).
Confirmation of GFP-tagging of haploids by fluorescence microscopy
Overnight grown cultures of the transformants, Ss97009 MAT-1gfp and MAT-2gfp, in YM broth were examined under Leica DM LB2 epifluorescence microscope (Leica Microsystems, Germany) equipped with a GFP filter (Excitation filter: BP470/40, Dichromatic mirror: 500 and Suppression filter: BP525/50), and images were captured with a Leica DMC2900 digital camera.
In vitro mating of compatible haploid transformants
For confirmation of the mating compatibility, equal proportion of overnight grown cultures of both Ss97009 MAT-1gfp and MAT-2gfp transformants were mixed and grown on YM agar supplemented with hygromycin B (50 µg/mL) at 28 °C for 2 days. Distinct stages of mating viz., emergence of conjugation hyphae, haploid fusion and formation of dikaryotic mycelia were observed with Calcofluor staining at different time intervals (4 h, 6 h, 8 h and 12 h) under epifluorescence microscope (Leica Microsystems, Germany).
Validation of GFP transformants by PCR
For PCR confirmation, GFP (GFP-F: 5ʹ-GACGTAAACGGCCACAAGTTC-3ʹ and GFP-R: 5ʹ-GGGGTGTTCTGCTGGTAGTG-3ʹ) and hygromycin (HYG-F: 5ʹ-ATTTGTGTACGCCCGACAGT-3ʹ and HYG-R: 5ʹ-AATCTCGTGCTTTCAGCTTCG-3ʹ) specific primers yielding amplicons of 500 bp and 829 bp, respectively, were used. The GFP-tagged MAT-1gfp and MAT-2gfp were grown overnight separately in YM broth amended with hygromycin B (50 µg/mL) at 28 °C. After incubation, the cultures were pelleted, frozen with liquid nitrogen and ground to a fine powder. For establishing dikaryotic mycelia, equal proportions of the haploid cultures were mixed, plated onto YM agar and incubated at 28 °C for 3–5 days. DNA was extracted from Ss97009 MAT-1gfp, MAT-2gfp and DM by CTAB method as described earlier, and PCR was performed in a final reaction volume of 20 µl containing 1X buffer, 200 µM dNTPs, 0.25 µM each of forward and reverse primers, 1 U Taq polymerase and 100 ng template DNA. The thermocycler conditions consisted of an initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 1 min, annealing temperature (60 °C and 58 °C for GFP and HYG primers, respectively) for 30 s, 72 °C for 1 min followed by a final extension at 72 °C for 5 min.
Histopathological analysis of GFP transformants in sugarcane
GFP-tagged haploid sporidial cultures viz., Ss97009 MAT-1gfp and SS97009 MAT-2gfp were mixed in equal proportion and inoculated onto the pre-germinated buds of cv. Co 97009. Similarly, the control plants were inoculated with a mixture of wild type Ss97009 MAT-1 and Ss97009 MAT-2 haploids. At least, 25 plants were inoculated per treatment and maintained under ideal glasshouse conditions. After inoculation, three bud/meristem samples each from both challenge inoculated and control plant tissues were drawn at different time points viz., 2 dpi, 5 dpi and 10 dpi, and examined under GFP filter as described earlier. The remaining plants were monitored regularly for the emergence of smut whips over a period of 110 days.
Results
Identification of distinct mating types of S. scitamineum haploids
Haploid sporidia were distinctly identified either as same or opposite mating types (MAT-1 or MAT-2), based on the appearance of smooth or fuzzy phenotype, respectively, using random mating assay method (Supplementary Fig. S2). Further, the identity of distinct mating types was confirmed using two specific primer sets, viz., bE1F1/bE1-2R1 MAT-1 specific) and bE2F2/bE1-2R1 (MAT-2 specific). PCR amplification using the primers bE1F1/bE1-2R1 yielded a specific amplicon of 668 bp with MAT-1 haploid sporidia, and bE2F2/bE1-2R1 resulted in an amplicon of 458 bp with MAT-2 haploid sporidia (Supplementary Fig. S3). Dikaryotic mycelia, which served as a positive control yielded specific amplicons with both the primers. Mating type-specific PCR confirmed the identity of the distinct haploid sporidia obtained from the random mating assay and one colony representing individual mating type (MAT-1 and MAT-2) was randomly selected for further experiments.
Optimization of protoplast isolation
To develop an efficient protocol for protoplast isolation, different factors such as choice of osmotic stabilizer, enzyme composition and digestion time were optimized. The use of SCS/STC buffer with D-sorbitol yielded protoplasts, whereas those with sucrose were found to be ineffective for protoplast isolation. Hence, protoplast yield and viability were checked with all the four enzyme treatments at three different time periods (1, 2 and 3 h). Among the enzyme treatments, the use of lysing enzymes with β-glucanase enzyme resulted in higher yield (~ 9 × 106 protoplasts/mL) of clearly visible and spherical protoplasts (Supplementary Table S1). Low protoplast yield was obtained with lysing enzymes alone, whereas β-glucanase alone was found to be inefficient for protoplast isolation. Significant differences in viability were observed among different combinations, and higher yield of viable protoplasts was obtained after 1 h of digestion followed by a gradual decrease after 2 and 3 h of incubation (Supplementary Table S1). Viability of the protoplasts was comparatively higher with those isolated using 20 mg/mL lysing enzymes from Trichoderma harzianum + 5 mg/mL β-glucanase than that obtained by using 10 mg/mL lysing enzymes from Trichoderma harzianum + 10 mg/mL β-glucanase for all the digestion times. After 1 h of incubation, Propidium Iodide staining of the protoplasts obtained using 20 mg/mL lysing enzymes from Trichoderma harzianum + 5 mg/mL β-glucanase, showed that > 95% of the protoplasts did not take up the stain, indicating the presence of intact cell membrane, and thus confirming their viability (Fig. 1a and b). Meanwhile, Calcofluor White staining of the protoplasts revealed that > 95% of the protoplasts did not take up the stain, indicating the absence of cell wall (Fig. 1c). Untreated haploid sporidia which served as a control did not take up the Propidium Iodide, but were stained with Calcofluor White (Fig. 1d–f). Further monitoring of protoplasts in regeneration medium revealed that the protoplasts remained to be in spherical shape, until 4 h and get elongated with the emergence of septate hyphae by 8 h (Supplementary Fig. S4). Hence, the protoplast isolation method employing 20 mg/mL lysing enzymes from Trichoderma harzianum + 5 mg/mL β-glucanase in SCS buffer with D-sorbitol that rendered highest yield of pure and viable protoplasts after 1 h incubation was optimized for isolating protoplasts from MAT-1 and MAT-2 haploid sporidia for further experiments.
Representative images of protoplasts isolated from S. scitamineum Ss97009 MAT-1. a Protoplasts isolated using 20 mg/mL lysing enzymes from Trichoderma harzianum + 5 mg/mL β-glucanase observed under bright field, b stained with Propidium Iodide and observed under N2.1 filter, c stained with Calcofluor White and observed under UV filter; d Ss97009 MAT-1 control (untreated) under bright field, e stained with Propidium Iodide and observed under N2.1 filter, f stained with Calcofluor White and observed under UV filter. Arrows in a indicate viable protoplasts, and in d and f indicate untreated haploid sporidia
GFP-tagging of Ss97009 MAT-1 and MAT-2 haploid sporidia by protoplast-based PEG-mediated transformation
A preliminary sensitivity assay to hygromycin B was performed prior to transformation and it was observed that the growth of the wild type Ss97009 MAT-1 haploid sporidia was completely inhibited at a minimum concentration of 50 µg/mL of hygromycin B, thus indicating a higher sensitivity of the fungus to the antibiotic. The circular pNIIST plasmid DNA was transformed into the MAT-1 protoplasts by PEG-mediated transformation and screened on Regeneration agar containing 50 µg/mL of hygromycin B. The effects of different concentrations of PEG4000 were assessed, and the transformation efficiency was found to increase along with an increasing concentration of PEG4000 from 10 to 40%. Meanwhile, slight decline in transformation efficiency was observed at 50% PEG4000 (data not shown). Maximum transformation efficiency yielding about 80 transformants/µg of DNA was obtained at a concentration of 40% PEG4000, and thus used for subsequent transformation experiments. The ability of hygromycin resistance of about 20% of transformants was lost upon successive sub-culturing owing to the transient expression. Hence, the transformants which remained stable even after 10 successive sub-cultures on hygromycin B-added agar were selected for further experiments. Notably, no difference in the colony morphology of the transformants was observed, when compared to that of the wild type strain.
Confirmation of GFP-tagged Ss97009 haploid sporidia and their mating ability by epifluorescence microscopy
Both MAT-1 and MAT-2 transformants expressed enhanced green fluorescence under fluorescence microscope confirming the expression of the eGFP gene (Fig. 2). However, there was a notable difference in the intensity of GFP expression among the transformants. Overall, GFP fluorescence was observed in the whole cell, except vacuoles, which appeared to be dark. The MAT-1 and MAT-2 transformants exhibiting bright fluorescence were designated as, MAT-1gfp and MAT-2gfp, respectively, and were chosen for further experiments. No green fluorescence was observed with the Ss97009 wild type, which served as the control.
The mating compatibility of the transformants was confirmed in vitro by fusion of MAT-1gfp and MAT-2gfp, which exhibited growth of white fuzzy colonies. Under fluorescence microscopy, formation of conjugation hyphae was observed from 4 h of incubation and fusion of opposite mating types was observed by 6 h of incubation (Fig. 3a–d). Dikaryotic mycelial formation was initiated by 8 h and the opposite mating types successfully fused to form dikaryotic mycelia by 12 h, thus confirming their mating compatibility (Fig. 3e–g). The hyphal fusion stages as well as the dikaryotic mycelia exhibited green fluorescence, when observed by fluorescence microscopy.
Expression of GFP in S. scitamineum during hyphal fusion and dikaryotic mycelia formation in vitro. a Formation of conjugation hyphae after 4 h incubation, b–d fusion of compatible mating types (Ss97009 MAT-1gfp and Ss97009 MAT-2gfp) by 6 h incubation, e–f initiation of dikaryotic mycelia by 8 h, g mycelial clumps formed by 12 h. Arrows in a–d indicate site of fusion between compatible haploid sporidia, and e–f indicate dikaryotic mycelia
Validation of GFP-tagging of Ss97009 haploid sporidia and dikaryotic mycelia by PCR
Transformants of Ss97009 MAT-1gfp, MAT-2gfp and the dikaryotic mycelia were validated using PCR with GFP-specific and hygromycin B-specific primers. Results showed that a GFP-specific amplicon of 500 bp and a hygromycin-specific amplicon of 829 bp were amplified in the haploids and mycelial transformants, confirming the integration of the GFP gene and hygromycin resistance cassette, respectively (Supplementary Fig. S5). The pNIIST plasmid, which served as a positive control yielded both GFP- and HYG-specific amplicons, whereas these amplicons were absent in the wild types- MAT-1 and MAT-2 control samples.
Visualization of GFP-expressing Ss97009 during interaction with sugarcane
GFP-expressing haploid sporidial mixture (Ss97009 MAT-1gfp and MAT-2gfp) were inoculated onto sugarcane cv. Co 97009 to examine the in planta developmental stages and the extent of in planta colonization of S. scitamineum over a period of 80 days (until smut whip emergence). Inoculated plantlets showed intensive colonization of external bud surface with GFP-expressing haploid sporidia. The haploid sporidia were found to be randomly attached to the outer surface of the bud layer and the opposite mating types fused to form infective dikaryotic mycelia at 2–5 dpi (Fig. 4a–b). The pathogen was also able to colonize the internal tissues after penetration of the infective mycelia. Intercellular colonization of parenchyma cells and, intracellular colonization of xylem and surrounding tissues at 10 dpi were observed, indicating successful establishment of fungal colonization in the host tissues (Fig. 4c–d). Nevertheless, plants treated with mixture of Ss97009 MAT-1 wild type and MAT-2 wild type did not show any green fluorescence, except for the slight autofluorescence of plant tissues (Fig. 4e–f).
Visualization of GFP tagged S. scitamineum Ss97009 during initial stages of in planta colonization of sugarcane. a GFP expressing haploid sporidia and b dikaryotic mycelia on the external surface of buds at 2–5 dpi; c intercellular colonization of parenchyma cells and d intracellular colonization of vascular bundles and surrounding tissues at 10 dpi; e external surface and f transverse section of sugarcane buds inoculated with wild type Ss97009 showing no GFP expression (control). Arrows in a–d indicate GFP expressing fungal structures
The plants inoculated with the GFP-expressing Ss97009 haploid mixture exhibited first whip emergence at 80 dpi. A total of nine whips emerged from both GFP-tagged (4 whips) and wild type haploid sporidia (5 whips) inoculated plants between 80 and 110 dpi. Microscopic examination portrayed a comparable degree of colonization in GFP-tagged Ss97009 inoculated plants, when compared to that of the Ss97009 wild type. The whips emerged with the GFP-expressing haploid sporidia were collected, and the portion of the sorus hidden by leaf sheaths was dissected into three distinct regions: apical region, basal region and young stem portion beneath the whip. At the apical region of the whip with mature teliospores, no green fluorescence was observed under epifluorescence microscopy (Fig. 5a). But in some areas, development of sporulating loci with mature teliospores in the central portion was observed (Fig. 5b). The basal region of the whip with active sporogenesis showed sporulating loci with green fluorescing hyphal fragmentation and non-melanized immature teliospores in the peripheral part (Fig. 5c–d). In addition, both apical and basal regions of the whip exhibited inter- and intracellular colonization of parenchyma cells and vascular tissues in the central part. In the young stem beneath the whip, intracellular and intercellular hyphal colonization were distributed systemically throughout the parenchyma cells and vascular tissues (Fig. 5e–f). Comparatively, a higher hyphal colonization was observed in the nodal tissues, with an overall indication that the use of GFP-tagged strains enabled visualization of in planta colonization in sugarcane on a spatio-temporal pattern.
Fluorescence microscopic analyses of GFP tagged S. scitamineum in three distinct portions of smut whip developed at 80 dpi (i) apical region (ii) basal region and (iii) young stem beneath whip. a apical region of the whip with mature teliospores showing no green fluorescence, b sporulation pockets with non-fluorescing mature teliospores in the center; c–d basal region of the whip with active sporogenesis in the peripheral part showing green fluorescing hyphal fragmentation and non-melanized immature teliospores; e young stem beneath sporogenesis displaying inter- and intracellular colonization of parenchyma cells and f vascular tissues. Arrows in a–f indicate sites of green fluorescing fungal colonization
Discussion
GFP-tagging of a fungal strain greatly depends on the implementation of an efficient transformation protocol and stable expression of the GFP gene under natural conditions. In S. scitamineum, a genetic transformation method based on ATMT has been used for expressing GFP to investigate in vitro and in planta developmental stages of S. scitamineum [17, 23]. Alternatively, protoplast-based transformation was reported to facilitate the visualization of many plant-fungal interactions [24, 25]. PEG-mediated transformation method developed by Yu et al. [22] for Ustilago esculenta remained to be a most plausible method to be followed for S. scitamineum, and attempted earlier by Deng et al. [26] and Sun et al. [27]. However, the method resulted in very less protoplast yield and poor viability (data not shown), as the success of such methods depends on cell wall composition of individual fungus.
Hence, in this study, we have optimized an efficient protoplast isolation method for S. scitamineum, and MAT-1 and MAT-2 haploid sporidia of Ss97009 isolate were successfully GFP-tagged using an improved PEG-mediated transformation method. The yield and quality of the protoplasts were often influenced by several parameters including osmotic pressure stabilizer, enzyme and digestion time [18]. In this study, among the osmotic stabilizers, D-sorbitol was found to be the most effective, whereas sucrose was inefficient for protoplast isolation. This is in contrary to the report of protoplast isolation from U. esculenta [22], wherein sucrose had the best effect, highlighting the need for developing strain-specific protocols for each fungus. Though, the use of two different concentrations of lysing enzymes in combination with β-glucanase resulted in higher yield of protoplasts, maximum percent of protoplast viability was achieved when 20 mg/ml lysing enzyme was combined with 5 mg/ml β-glucanase and incubated for 1 h. With this combination, a high yield of protoplasts (~ 9 × 106 protoplasts/mL) with 95% viability was obtained, which is one of the most influencing factors that determine the transformation efficiency.
Despite the fact that the transformation efficiency could be enhanced by using linearized plasmid [28], we could still develop an efficient transformation system for circular plasmid by employing high quality protoplasts. And this has aided in overcoming the requirement for time consuming restriction digestion of the plasmid. Through this optimized PEG-mediated transformation method, about 80 transformants/µg of DNA were obtained, indicating higher efficiency compared to the ATMT method by other workers [17, 23]. For GFP-tagging, pNIIST plasmid harboring eGFP gene as a visual marker and hygromycin gene as a selection marker both under the control of Ascomycetes specific promoters was used. Although the pNIIST plasmid was constructed as an E. coli–Aspergillus shuttle vector, the pNIIST plasmid exhibited a stable GFP expression in the basidiomycetous fungus, S. scitamineum. GFP-tagging of basidiomycetes has been challenging because of the incompatibility with the Ascomycetes specific promoters that were commonly used [29]. However, few studies on the expression of GFP in basidiomycetes upon integration of a vector with genes under Ascomycetes promoter have also been reported [16]. Here, the expression level of GFP signals varied among the transformants, which could be attributed to the plasmid integration onto different chromosomal locations and partial transformations in multinucleated fungi [30].
Both MAT-1gfp and MAT-2gfp transformants expressing a stable and strong green fluorescence were fused to form recombinant GFP-expressing dikaryotic mycelia in vitro. The results clearly demonstrated that the phenotype, fungal growth and life cycle development were not affected by the integration of the transgene. The critical steps of mating process in vitro were monitored continuously under epifluorescence microscopy. Formation of conjugation hyphae was observed upon interaction with the opposite mating type, which was followed by hyphal fusion during 4–6 h incubation and morphological transition leading to the formation of dikaryotic mycelia during 8–12 h. These timings are in agreement with the mating process of the GFP-expressing haploid sporidia, erstwhile demonstrated by Yan et al. [23].
In the present study, we investigated in detail the infection process in a susceptible variety Co 97009 during initial and whip emergence stages in detail. The GFP-tagged strain of a highly virulent Ss97009 was able to successfully penetrate and colonize the sugarcane buds, and the colonization stages during 2 dpi to 10 dpi were examined. Generally, microscopic detection of S. scitamineum during plant colonization is relatively difficult at the initial stages of infection, because the fungus cannot be differentiated morphologically from other endophytic fungi [5]. In our study, GFP-tagging of S. scitamineum Ss97009 facilitated the specific visualization, so as to track the natural route of the fungus during in planta colonization of sugarcane tissues. Upon challenge inoculation in a susceptible variety, the mixture of GFP-expressing haploid sporidia (Ss97009 MAT-1gfp and MAT-2gfp) could attach successfully to the external bud surface. The compatible mating types fused to form dikaryotic mycelia and the infective dikaryotic mycelia were able to colonize the internal tissues at 10 dpi. This is in accordance with a previous report on infection process during in planta colonization using a GFP-tagged S. scitamineum obtained through ATMT method [17]. The colonization of vascular tissues was reported to provide a treading path to meristematic tissues [31]. On the other hand, Yan et al. [23] reported that the visualization of GFP-tagged S. scitamineum in the host was a failure due to interference by strong autofluorescence of sugarcane tissues. Intriguingly, the selection of fungal transformants with stronger fluorescence enabled us to differentiate fungal structures from plant tissues with autofluorescence.
Here, the GFP-tagged S. scitamineum Ss97009 was able to develop whips in a similar pattern compared to the wild type, showing no impact on colonization and pathogenicity, thus demonstrating that the pathogenicity was not affected by eGFP gene integration. Similarly, fungal transformants tagged with improved variants of fluorescent proteins were reported to substantially enhance the visualization process without any loss in pathogenicity [8]. However, variation in growth, colonization, reduced virulence and fitness of GFP transformants due to the insertional disruption of functional genes or due to the additional metabolic burden for overexpressing GFP genes were also reported in some other cases [32, 33]. In our study, different portions of the whip were analyzed microscopically to investigate the finer details of the infection process. Intercellular and intracellular hyphal colonization of parenchyma cells and vascular tissues were distributed systemically throughout the whip shaped sorus. The basal region of the whip with active sporogenesis exhibited green fluorescent sporulation pockets with abundant fragmented hyphae, sporogenous cells and non-melanized teliospores in a gelatinous matrix. Similar pattern of fungal colonization resulting in the development of whip-shaped sorus was reported by Marques et al. [31] using electron microscopic studies. As evident from our work, it is clear that eventually, the sporulation pockets enlarged to cover the entire peripheral portion with sporulating loci. During teliospore maturation, they were unable to fluoresce possibly due to melanogenesis or thickened cell wall [34]. Thus, the study demonstrated a more precise and direct visualization of in planta colonization stages during whip emergence using a GFP-tagged strain.
Comprehensively, for the first time, an improved and efficient protoplast-based PEG-mediated transformation has been established for GFP-tagging to elucidate the critical events during mating process of S. scitamineum in vitro, and to monitor the time course development of in planta colonization of the fungus using a high virulent Indian isolate Ss97009. The GFP-tagged S. scitamineum was able to infect the sugarcane buds, facilitating visualization of the critical stages of life cycle transition events during early phases of disease development and sporogenesis occurring at whip emergence stage. The GFP-tagged transformants can also be utilized as a helpful tool for characterization and identification of environmental stress affecting the fungal growth in vitro and in planta [35]. Moreover, the identification of transformants with altered virulence would help in studying the effects of ectopic transgene integration and in identification of virulence-related genes from a functional genomics perspective [36]. The protoplast-based PEG-mediated transformation method developed in this study will also serve as a tool for functional characterization of genes through homologous recombination to decipher molecular mechanisms of plant-pathogen interactions [37]. In particular, the method would be useful for functional characterization of genes required for mating/filamentation and pathogenicity of S. scitamineum [38], thereby paving way for developing sugarcane varieties with smut resistance.
Conclusion
GFP-tagged transformants of the basidiomycetous fungus, S. scitamineum were developed by employing an efficient protoplast-based PEG-mediated transformation. Both MAT-1 and MAT-2 haploid sporidia of a highly virulent isolate, Ss97009 was transformed using pNIIST plasmid harboring eGFP gene. Using these GFP-tagged mating compatible strains, we visualized and monitored morphological transitions in vitro that occur during mating of haploid sporidia to form dikaryotic mycelia. Precise and direct visualization of in planta colonization stages of S. scitamineum in sugarcane tissues also enabled better understanding of infection process of the smut pathogen. The development of transformants with altered virulence by an efficient and improved protoplast-based PEG-mediated transformation would be a powerful tool for functional characterization of genes to elucidate the molecular mechanisms of S. scitamineum—sugarcane interaction. Overall, the development of an efficient transformation system and such critical analysis on infection process would unwind finer details of pathogenesis, and eventually lead to opening up of a broader range of disease management strategies.
Data availability
Not applicable.
References
Zekarias Y, Dejene M, Tegegn G, Yirefu F (2010) Importance and status of sugarcane smut (Ustilago scitaminea) in the Ethiopian sugar estates. Ethiop J Agric Sci 20:35–46
Magarey RC, Sheahan TJ, Sefton M, Kerkwyk RE, Bull JI (2011) Yield losses from sugarcane smut: indications from Herbert mill data and from individual crop assessments. In: Bruce RC (ed) Proceedings of the 2011 Conference of the Australian Society of Sugar Cane Technologists. Australian Society of Sugar Cane Technologists, Australia, pp. 1–9
Sundar AR, Barnabas EL, Malathi P, Viswanathan R (2012) A mini-review on smut disease of sugarcane caused by Sporisorium scitamineum. In: Mworia JK (ed) Botany. InTech Rijeka, Croatia, pp 107–128
Alexander KC, Srinivasan KV (1966) Sexuality in Ustilago scitaminea Syd. Curr Sci 35(23):603–604
Izadi MB, Moosawi-Jorf SA (2007) Isolation and identification of yeast-like and mycelial colonies of Ustilago scitaminea using specific primers. Asian J Plant Sci 6:1137–1142
Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ (1992) Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111(2):229–233
Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263(5148):802–805
Lorang JM, Tuori RP, Martinez JP, Sawyer TL, Redman RS, Rollins JA, Wolpert TJ, Johnson KB, Rodriguez RJ, Dickman MB, Ciuffetti LM (2001) Green fluorescent protein is lighting up fungal biology. Appl Environ Microbiol 67:1987–1994
Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY (1995) Understanding, improving and using green fluorescent proteins. Trends Biochem Sci 20:448–455
Spellig T, Bottin A, Kahmann R (1996) Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet 252:503–509
Sexton AC, Howlett BJ (2001) Green fluorescent protein as a reporter in the Brassica-Leptosphaeria maculans interaction. Physiol Mol Plant Pathol 58(1):13–21
Jansen C, Von Wettstein D, Schafer W, Kogel KH, Felk A, Maier FJ (2005) Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc Natl Acad Sci 102(46):16892–16897
Crespo-Sempere A, Lopez-Perez M, Martinez-Culebras PV, Gonzalez-Candelas L (2011) Development of a green fluorescent tagged strain of Aspergillus carbonarius to monitor fungal colonization in grapes. Int J Food Microbiol 148(2):135–140
Nandakumar M, Malathi P, Sundar AR, Viswanathan R (2020) Use of green fluorescent protein expressing Colletotrichum falcatum, the red rot pathogen for precise host-pathogen interaction studies in sugarcane. Sugar Tech 22(1):112–121
Ruiz-Diez B (2002) Strategies for the transformation of filamentous fungi. J Appl Microbiol 92(2):189–195
Sun L, Yan M, Ding Z, Liu Y, Du M, Xi P, Liao J, Ji L, Jiang Z (2014) Improved dominant selection markers and co-culturing conditions for efficient Agrobacterium tumefaciens-mediated transformation of Ustilago scitaminea. Biotechnol Lett 36:1309–1314
Carvalho G, Quecine MC, Longatto DP, Peters LP, Almeida JR, Shyton TG, Silva MML, Crestana GS, Creste S, Monteiro-Vitorello CB (2016) Sporisorium scitamineum colonisation of sugarcane genotypes susceptible and resistant to smut revealed by GFP-tagged strains. Ann Appl Biol 169(3):329–341
Li D, Tang Y, Lin J, Cai W (2017) Methods for genetic transformation of filamentous fungi. Microb Cell Fact 16(1):168
Madhavan A, Pandey A, Sukumaran RK (2017) Expression system for heterologous protein expression in the filamentous fungus Aspergillus unguis. Bioresour Technol 245:1334–1342
Barnabas L, Ashwin NMR, Sundar AR, Malathi P, Viswanathan R (2017) Putative orthologs of Ustilago maydis effectors screened from the genome of sugarcane smut fungus-Sporisorium scitamineum. Australas Plant Pathol 46(2):147–156
Abu Almakarem AS, Heilman KL, Conger HL, Shtarkman YM, Rogers SO (2012) Extraction of DNA from plant and fungus tissues in situ. BMC Res Notes 5:266. https://doi.org/10.1186/1756-0500-5-266
Yu J, Zhang Y, Cui H, Hu P, Yu X, Ye Z (2015) An efficient genetic manipulation protocol for Ustilago esculenta. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnv087
Yan M, Cai E, Zhou J, Chang C, Xi P, Shen W, Li L, Jiang Z, Deng YZ, Zhang LH (2016) A dual-color imaging system for sugarcane smut fungus Sporisorium scitamineum. Plant Dis 100(12):2357–2362
Morocko-Bicevska I, Fatehi J (2011) Infection and colonization of strawberry by Gnomonia fragariae strain expressing green fluorescent protein. Eur J Plant Pathol 129(4):567–577
Liu XM, Qi YX, Zhang X, Xie YX, Zhang H, Wei YX, Cao SW, Pu JJ (2014) Infection process of Corynespora cassiicola tagged with GFP on Hevea brasiliensis. Australas Plant Pathol 43(5):523–525
Deng YZ, Zhang B, Chang C, Wang Y, Lu S, Sun S, Zhang X, Chen B, Jiang Z (2018) The MAP kinase SsKpp2 is required for mating/filamentation in Sporisorium scitamineum. Front Microbiol 9:2555. https://doi.org/10.3389/fmicb.2018.02555
Sun S, Deng YZ, Cai E, Yan M, Li L, Chen B, Chang C, Jiang Z (2019) The farnesyltransferase β-Subunit Ram1 regulates Sporisorium scitamineum mating, pathogenicity and cell wall integrity. Front Microbiol 10:976. https://doi.org/10.3389/fmicb.2019.00976
Van De Rhee MD, Graca PM, Huizing HJ, Mooibroek H (1996) Transformation of the cultivated mushroom, Agaricus bisporus, to hygromycin B resistance. Mol Gen Genet MGG 250(3):252–258
Frandsen RJN (2011) A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciens-mediated transformation. J Microbiol Methods 87(3):247–262
Visser M, Gordon TR, Wingfield BD, Wingfield MJ, Viljoen A (2004) Transformation of Fusarium oxysporum f. sp. cubense, causal agent of Fusarium wilt of banana, with the green fluorescent protein (GFP) gene. Australas Plant Pathol 33(1):69–75
Marques JPR, Appezzato-da-Gloria B, Piepenbring M, Massola NS Jr, Monteiro-Vitorello CB, Vieira MLC (2017) Sugarcane smut: shedding light on the development of the whip-shaped sorus. Ann Bot 119(5):815–827
Dunn AR, Fry BA, Lee TY, Conley KD, Balaji V, Fry WE, McLeod A, Smart CD (2013) Transformation of Phytophthora capsici with genes for green and red fluorescent protein for use in visualizing plant-pathogen interactions. Australas Plant Pathol 42(5):583–593
Wu L, Conner RL, Wang X, Xu R, Li H (2016) Variation in growth, colonization of maize, and metabolic parameters of GFP-and DsRed-labeled Fusarium verticillioides strains. Phytopathology 106(8):890–899
Trione EJ (1980) Teliospore formation by Ustilago scitaminea in sugarcane. Phytopathology 70:513–516
Kato A, MiyakeT NK, Tateishi H, Teraoka T, Arie T (2012) Use of fluorescent proteins to visualize interactions between the Bakanae disease pathogen Gibberella fujikuroi and the biocontrol agent Talaromyces sp. KNB-422. J Gen Plant Pathol 78(1):54–61
Talhinhas P, Muthumeenakshi S, Neves-Martins J, Oliveira H, Sreenivasaprasad S (2008) Agrobacterium-mediated transformation and insertional mutagenesis in Colletotrichum acutatum for investigating varied pathogenicity lifestyles. Mol Biotechnol 39(1):57–67
Weld RJ, Plummer KM, Carpenter MA, Ridgway HJ (2006) Approaches to functional genomics in filamentous fungi. Cell Res 16(1):31–44
Zhang B, Cui GB, Chang CQ, Wang YX, Zhang HY, Chen BS, Deng YZ, Jiang ZD (2019) The autophagy gene ATG8 affects morphogenesis and oxidative stress tolerance in Sporisorium scitamineum. J Integr Agric 18(5):1024–1034
Acknowledgements
The authors are grateful to The Director, ICAR-Sugarcane Breeding Institute for providing facilities and continuous encouragement. The financial support received from Department of Biotechnology (DBT), Department of Science and Technology - Science and Engineering Research Board (DST-SERB) and Indian Council of Agricultural Research (ICAR), New Delhi are greatly acknowledged. We greatly acknowledge Dr. Rajeev Sukumaran, CSIR- National Institute for Interdisciplinary Science and Technology, Trivandrum, Kerala for sharing the pNIIST plasmid.
Funding
This work was financially supported by the Department of Biotechnology (DBT) (Sanction No. BT/PR12883/BPA/118/142/2015) and Department of Science and Technology - Science and Engineering Research Board (DST-SERB) (Sanction Order No. EMR/2016/006055) in the form of research grants to the corresponding author.
Author information
Authors and Affiliations
Contributions
V. N. A., N. M. R. A. and A. R. S. conceived and designed the experiments. V. N. A. performed the experiments and wrote the first draft of the manuscript. N. M. R. A., R. T. V. and K. N. are involved in analyzing the results and in revising the manuscript. A. R. S., N. M. R. A., P. M. and R. V. shared their expertise in execution of experiments, and were involved in revising and improving the intellectual content of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical approval
The present research work did not involve human participants and/or animals.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Agisha, V.N., Ashwin, N.M.R., Vinodhini, R.T. et al. Protoplast-mediated transformation in Sporisorium scitamineum facilitates visualization of in planta developmental stages in sugarcane. Mol Biol Rep 48, 7921–7932 (2021). https://doi.org/10.1007/s11033-021-06823-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06823-x