Abstract
In Tunisia, Capparis spinosa L. is widely distributed in different geographic areas. Although it has been extensively studied morphologically, the phylogenetic relationships by using molecular markers among Capparis taxa are still unclear. The Inter-Simple Sequence Repeats (ISSR) molecular markers were used to assess the genetic relationship of this species cultivated in the North and the South of Tunisia. Fifteen ISSR primers were analyzed to characterize the genetic profiles of 67 different genotypes. The ISSR markers produced 108 bands ranging from 4 (ISSR8) to 11 (IAM12 and ISSR16) with an average of 7.2. The observed heterozygosity ranged from 0.43 to 0.95 for ISSR7 and IAM12, respectively. The Polymorphic Information Content (PIC) ranged from 0.48 at the UBC808 to 0.85 at IAM12 and eight loci could be classified as useful for genetic mapping (PIC > 0.7). The genetic diversity within a population was high and varied according to the subspecies and bioclimatic zones. Both UPGMA (Unweighted Pair Group Method with Arithmetic mean) and PCoA (Principal Coordinate Analysis) analyses showed that populations from each subspecies grouped together. However, the structure analysis generated more groups than the PCoA plot and UPGMA, which revealed the mixed allelic of the species Capparis spinosa in Tunisia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Capparis L. genus includes more than 250 species growing in wild subtropical and tropical areas of the world [1,2,3,4]. Capparis spinosa L. is a characteristic shrub within the Mediterranean region and it is used as medicinal herbs. It consists of vivacious and ornamental plants [5]. A growing interest in the utilization of caper has been recently signalized for industrial, pharmaceutical and chemical domains [6, 7]. However, natural populations, through their over-use, become endangered. Studies of the Capparis preservation strategies and the population genetic structure according with the geographical origin, the large diversity of existing varieties and the lack of an accurate taxonomy, remain rather little compared to those evaluating their chemical composition and biological properties [8, 9].
In Tunisia, a high variability has been reported for wild populations [10, 11] and the systemic revisions between species, subspecies and varieties remain unclear. According to Pottier-Alapetite [12], Tunisian flora and anterior studies that had interest in this species revealed only one single specie with four varieties: var. aegyptica (Lamk) Boiss., var. genuine Boiss., var. coriacea Coss. and var. rupestris (S & Sm.). However, Higton and Akeroyd [13] divided C. spinosa into subsp. spinosa (thorny caper) and subsp. rupestris (inerm caper) (Sm.) Nyman. Later, Le Floc’h et al. [14] raised these four varieties to species level. The two subspecies could be allopathic in a wide part of the country [15]. The subspecies rupestris extended throughout the country, whereas the subspecies spinosa characterized the Northern regions. Thus, the provenances of these two subspecies are described by various ecological and environmental conditions [15].
Capparis spinosa L. known in Tunisia under the vernacular name of “Kabbar”, is used as medicinal herbs and spice. The species has a large natural distribution, mainly in dry and sunny areas. Populations are extended from the sub-humid to the saharan on sandy and often rocky soils, under a rainfall varying from 150 to 1000 mm/year and at altitudes ranging from 150 to 500 m [16]. The species occurs as mosaic local populations in small land along the landscape. The caper populations have been sharply reduced and fragmented due to overuse and habitat destruction caused by pasturing, clearing, soil quality and irregularity of rainfall. The habitat fragmentation and the spatial isolation of populations increase genetic drift and diversity between populations, and reduce their future adaptation to environmental changes. Thus, understanding the patterns of the genetic within and among populations crucial to assess the present status of these populations and to develop conservation programs.
Nowadays, the morphological characterization of the species of Capparis is insufficient to make a perfect discrimination among the Capparis subspecies [3, 17]. Recently, other approaches, including application of appropriate molecular markers, have been adopted to resolve the problems in Capparis taxonomy [18]. Moreover, analyses based on genetic diversity and relationship among the species of Capparis could provide useful information for the conservation of genetic resources and the establishment of a Capparis breeding program [18]. Additionally, the Amplified Fragment Length Polymorphism (AFLP) molecular markers, the Inter-Simple Sequence Repeats (ISSRs) and Random Amplified Polymorphic DNAs (RAPDs) molecular analyses have been the most commonly used tools in wild and cultivated dorms of Capparis species in Iran [19], Trans-Himalayan region [20], Syria [21], Turkey [22], Egypt [23], Italy [24] and Morroco [25]. In the other hand, among the different molecular tools, the Inter-Simple Sequence Repeats (ISSRs) method has been extensively used to study the genetic diversity and geneticrelationships in the plants. ISSR markers are based on the amplification of regions between DNA adjacent microsatellite sequences via PCR (polymerase chain reaction). This technique has the benefit of discriminating between closely related genotypes, since it require no prior DNA sequence information, have a high degree of polymorphism, high reproducibility and can achieve results in a timely and cost-effectively manner [26, 27]. Several studies on genetic diversity in medicinal plants have been carried out using ISSR molecular markers [28, 29]. In Tunisia the genetic variation among genotypes/ species and/ or collected sites were analyzed by RAPD markers and AFLP markers.
The aim of the present study is to investigate the genetic diversity of the C. spinosa species and then to explore to the best of our knowledge for the first time in Tunisia the genetic relationships between the two subspecies of C. spinosa by using ISSR markers. This work will clarify the polymorphism rate within and among populations, resolve the intra-specific taxa discrimination and estimate the overall level of genetic variation of caper species in Tunisia. In particular, the genetic polymorphism study based on the molecular markers is fundamental for the effective use of genetic resources in breeding program of the C. spinosa in Tunisia and the design of its molecular patterns in the database of the Tunisian National Gene Bank.
Materials and methods
Plant material
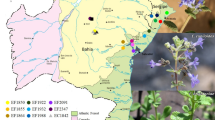
A total of 67 samples representing the two species of the Tunisian Capparis spinosa (subsp. spinosa and subsp. rupestris) were arbitrarily collected from eight different sites. The different caper populations are localized in different bioclimatic stage ranging from lower humid to lower arid bioclimatic areas (Fig. 1). The altitude of the sites varied from 8 m (Ichkeul) to 449 m (Djebel Naouali) and the average of annual rainfall ranged from 100 mm (Medenine) to 550 mm (Icheukl, Joumine and Djebel Naouali) (Table 1) localized in the North and the South of Tunisia (Table 1). Samples were placed on ice in plastic bags and transported to the laboratory for molecular analyses and to keep the voucher specimens (C. spinosa subsp. spinosa and C. spinosa subsp. rupestris) at the herbarium of the National Gene Bank of Tunisia (NGBT).
Geographic location of the 8 populations of C. spinosa. 1: Ichkeul, 2: Park Nahli, 3: Mrabaa, 4: El Arima, 5: Medenine, 6: Choaabat Jmal, 7: Dj Naouali. Filled circle: C. spinosa subsp. rupestris (inerm caper); filled square: C. spinosa subsp. spinosa (thorny caper). (https://www.google.tn/cartebioclimatiquetunisie) [52]
Genomic DNA extraction
Young leaves (0.50 g) from each plant were grind with liquid nitrogen, and total genomic DNA was extracted according to a modified CTAB (Cetyl Trimethyl Ammonium Bromide) method [30]. DNA concentration and quality were assessed on 1.5% (W/V) agarose gel using a NanoDrop spectrophotometer (NanoDrop Technologies Wilmington, DE, USA).
ISSR amplification
Fifteen ISSR primers (Table 2) were used for the total genomic DNA amplification [21, 25]. The PCR (Polymerase Chain Reaction) reaction was performed in 25 µl reaction volume containing 50 ng template DNA, 2.5 µl PCR buffer, 0.5 µl (20 mM) dNTPs, 0.5 µl MgCl2 50 mM, 2.5 mM primer, 0.4 U Taq polymerase (Bio-Basic-Canada INC) and bi-distilled water. Amplifications were performed using a BioRAD C1000 Thermal Cycler (Applied Biosystems, California, America) with the following PCR program: 3 min initial denaturation at 94 °C, 35 cycles of 94 °C for 1 min, 1 min for annealing at the primer-specific melting temperature, and 72 °C for 2 min, followed by a final extension of 3 min at 72 °C. The PCR products were analyzed by electrophoresis on 2.5% agarose gel with 0.5X TBE buffer.
Data analysis
ISSR bands were scored for their presence (1) or absence (0) and then transformed into a binary matrix. Each marker band was assumed to represent a single locus. These data were used by GenAlEx 6.5 software [31] to determine the number of bands per locus, the observed heterozygosity (Ho), the expected heterozygosity (He) and the number of discriminated genotypes. The probability of null alleles was estimated according to the formula of Brookfield (1996) [32]:
The polymorphic information content (PIC) was calculated for each primer pair according to the formula of Tessier et al. [33]:
where: pi is the frequency of the genotype for the locus.
The genetic variation within and among populations also was estimated by analysis of molecular variance (AMOVA) test [34, 35]. The principal coordinate analysis (PCoA) based on the matrix of pairwise ФST (Nei’sun biased pairwise) was evaluated by the GenAlEx 6.5 software and the genetic identity between populations was estimated using the Unweighted Pair-Group Method (UPGMA).
Moreover, STRUCTURE 2.3.1 software [36] was used to provide the most reliable grouping of the 67 caper genotypes, which was analyzed using a Bayesian method (100.000 burn-ins, 100.000 Markov chain Monte Carlo). The estimate of the most likely number of genetic groups (K = 3) was performed with Structure Harverster according to Evanno et al. [37].
Results
ISSR genetic diversity
For all the populations, a total of 108 fragments were amplified, in a size range of 100–1000 bp, with 87 polymorphic bands (Table 2). The number of bands produced by each primer varied from 4 for ISSR8 to 11 for IAM12 and ISSR16. The average number of polymorphic alleles per locus was 7.2 alleles per ISSR primer. The observed heterozygosity (Ho) ranged from 0.43 for ISSR7 to 0.95 for IAM12 primer (Fig. 2), with an average of 0.69 (Table 2). The polymorphic information content (PIC) ranged from a minimum of 0.48 for UBC808 to a maximum of 0.85 for IAM12 primer, with an average of 0.68. All the studied ISSR markers are highly polymorphic (PIC > 0.5) except for UBC808 and ISSR7, and eight loci (IAM12, ISSR1, ISSR8, ISSR13, ISSR15, ISSR16, ISSR22 and ISSR47) could be considered as useful for genetic mapping (PIC > 0.7). The ФST (Nei’sun biased pairwise) value was 0.212 and the AMOVA test revealed that 79% of the total genetic variation occurred within groups and only 21% occurred among groups (Table 3). The high level of genetic differentiation within populations could be justified by the high degree of genetic recombination as a result of the open pollination between caper genotypes.
Genetic diversity and genetic structure investigated by ISSR markers
Genetic diversity levels
The data generated by ISSR study were analyzed using the Nei’s identity index (Table 4). All populations (Pop) were readily separated from each other. The identity value ranged from 0.7 to 0.96. The smallest genetic similarity value (GS = 0.7) was observed between the population Pop4 and Pop6, two populations which present two different taxa and differ greatly in their origin and their morphological parameters. The maximum of the genetic similarity value (GS = 0.96) was found between Pop3 and Pop7. These populations, growing in the North of Tunisia, showed very similar morphological characteristics. It is likely that they have a common genetic origin.
Cluster analysis and principal component analysis
The dendrogram based on Jaccard’s similarity matrix divided the eight populations into two main clusters at the genetic distance GS = 0.73 (Fig. 3). The first group (CL1) includes caper genotypes characterized by multi-ramified branches up to 2 m, thorny stipules with different sizes, recurved, in some cases horizontal or straight and leaves with different form passed from elliptic to oval. This group represents the C. subsp. spinosa which includes the populations growing in the North of Tunisia: Pop1 (Ichkeul), Pop2 (Park Nahli), Pop3 (El Mrabaa), Pop4 (El Arima), Pop7 (Djebel Naouali) and Pop8 (Joumine). The second cluster (CL2) represents the C. subsp. rupestris. These genotypes are localized in the South of Tunisia (Pop5 of Medenine and Pop6 of Choaabat Jmal) and they are essentially characterized by the orbicular form of their leaves and the absence or the small size of their stipules (< 2.5 mm) which are mostly setaceous or caduceus.
The PCoA based on Nei’sunbiased pairwise ФST matrix for the 67 genotypes revealed that the first three principal axes accounted for 50.97% of the total variation. The plot according to the first two PCoA axes (47.33% of the total variation) revealed two major genotypes groups (Fig. 4). The first group, projected at the negative side of axis 1 and 2, clusters only the C. subsp. spinosa genotypes and includes all the samples of Pop8 and some genotypes of Pop1, Pop3 and Pop7. The second group, situated at the positive side of axis 1 and axis 2, includes the majority of the C. spinosa subsp. rupestris genotypes and some subsp. spinosa populations; this can be explained by a putative hybridization between the two taxa. Nevertheless, within each group, populations were not strictly clustered together according to bioclimatic and/or geographic proximity, but according to their taxonomic group.
Principal Coordinates Analysis (PCoA) plot of the eight populations of Tunisian Capparis spinosa L. (C. spinosa subsp. spinosa and C. spinosa subsp. rupestris) based on the first two principal coordinates (coord. 1 = 50.97% and coord. 2 = 14.86%). POP1 population 1, POP2 population 2, POP3 population 3, POP4 population 4, POP5 population 5, POP6 population 6, POP7 population 7, POP8 population 8, Gp1 groupe 1, Gp2 groupe 2
Structure analysis
ISSR data were studied by using the STRUCTURE 2.3.1 software [36] to provide the most reliable discrimination of the 67 caper genotypes. The data were analyzed based on Bayesian method (100.000 burn-ins, 100.000 Markov chain Monte Carlo). The estimation of the most likely number of genetic groups (K = 3) was performed with Structure Harverster according to Evanno et al. [37]. The highest H’ value was observed for K = 3 (H’ = 0.97) indicating the stability of the results for this model (Fig. 5). According to the model at K = 3, the different groups were defined by three colors: red, green and blue. The red color predominates the genetic profile of the tested genotypes, followed by the blue color and the green color. The first group includes the genotypes 6, 7, 15, 29, 39 40, 43, 45, 50, 56, 57 and 67 which present a genetic profile dominated by the red color (membership value > 0.8). These genotypes belong to the populations Pop3 and Pop7 and present morphological parameters specific to the taxa C. spinosa subsp. spinosa. The second group includes the genotypes 9, 12, 13, 14 19, 23, 25, 26, 27, 28, 41 and 49 which are predominated by the blue color with a common membership value (> 0.8). These genotypes, collected from the North and the South of Tunisia, presented the characteristics of C. spinosa subsp. spinosa such as the presence of thorny stipules, shoots ramification and architecture whereas they were related to C. spinosa subsp. rupestris for its habitat preference and the oval form of their leaves (Fig. 6). The third group includes only the caper genotypes 31, 32, 33, 34, 35 and 42 characterized by the predominance of the green color (membership value > 0.8), these genotypes grow essentially in the South of Tunisia and presented characteristics of the species C. spinosa subsp. rupestris. However, the majority of the tested genotypes could not be distinguished by a unique color, which revealed the mixed allelic of these genotypes.
Structure barplots for 67 different individuals presenting eight populations of the species of Capparis spinosa L. in Tunisia based on allele frequency and the most likely number of genetic groups K (K = 3).Vertical bars represent each individual analyzed in this study and bars are divided into several colours when there is evidence of admixture
Photos showing details of C. spinosa subsp. spinosa (A, B, C, D) such as the shoots ramification and the architecture, the colour and the small size of the leaf and the stem (A1, C1, D1), the small numbers of stamens (A1) and the presence of thorny stipules (A1, C1, D1) and Capparis spinosa subsp. rupestris (E) related to its habitat preference, the oval form and the large size of their leaves (E1) and the high numbers of stamens (E2)
Discussion
Molecular markers have been used extensively to determine genetic diversity and genetic relationships in plant science [38,39,40]. In our study, ISSR markers method was utilized to assess the genetic diversity of C. spinosa in Tunisia. The studied species maintained a high genetic diversity inside populations (H = 0.79) and otherwise a low diversity among population (H = 0.21). The variation level could be explained by finding of Hamrick and Godt [41] who explicated this variation by the persistence of numerous genotypes through generations derived from large populations before fragmentation. Our results are similar to that obtained by Saifi et al. [25] and Gristina et al. [24]. Furthermore, the band size 300 bp of the primer IAM12 and the two band sizes 400 bp and 500 bp of the ISSR22 primer exhibited the highest variation (100%) within population. In fact, several ISSR loci were revealed as specific to populations. However, the presence of these loci might not reflect the adaptability to ecological factors since they were not detected in all populations from the same bioclimatic zone. Their variation might not necessarily reflect the pattern of variation in adaptive genes.
Therefore, the variation within populations could be more possibly related to the environmental factors than to geographic distances [3]. These findings are in line with the previous results of Saifi et al. [25] and Gristina et al. [24]. Furthermore, the genetic variation of the Tunisian C. spinosa species studied by ISSR markers showed discriminative power, enabled classification of the studied accessions into two main taxa spinosa and rupestris. The highest polymorphic information content was observed between the accessions of the thorny caper, this group is mainly characterized by erect shoots, thorny stipules, relatively small leaves and a small numbers of stamens. Whereas, the group of rupestris capers presents the lowest polymorphism. The accessions of this type were distinct by their shoots, relatively large leaves and high numbers of stamens. Our results proved the findings of Pottier-Alapetite [12], Barbera [2], Skouri H [42], National Institute of Research in Rural Engineering, Waters, and Forests [42] and Rivera et al. [43], witch approve that the genus Capparis represented in Tunisia by a one species (C. spinosa) subdivided into two mains subspecies (C. spinosa ssp. spinosa and C. spinosa ssp. rupestris). Over the past 20 years, molecular techniques, such as PCR and sequencing, have been developed that have resulted in a large number of DNA sequences. The use of these sequences induced profound changes in the systematic classification. The taxonomy of Capparis L. has undergone real changes with the introduction of molecular methods and their evolution. The inter-simple sequence repeats (ISSRs), Amplified Fragment Length Polymorphism (AFLP) and random amplified polymorphic DNAs (RAPDs) analysis have been the most commonly used techniques in wild and cultivated forms of Capparis species revealing genetic variation among capper genotypes in Syria [21], Iran [19], Trans-Himalayan region [20], Egypt [23], Morocco [25], Turkey [22] and Italy [24]. In Spain, Morocco, and Syria, the genetic analysis in Capparis was performed and showed that C. aegyptia and C. ovata are isolated from the other taxa. AFLP method has been extensively used to investigate the population genetic structure in C. spinosa and revealing the genetic variations among six different subspecies in Tunisia [18]. The Amplified Fragment Length Polymorphism (AFLP) method has been used for a wide range of species. The confirmation of the real existence of six caper subspecies in Tunisia need the use of specific molecular tools as ISSR especially that SSRs markers are not yet developed for the species Capparis spinosa L.. The ISSR markers are very useful in studies that aim at determining the genetic distance between parents, as well as the performance of the hybrids for the construction of genetic linkage maps, and for the characterization of accessions and cultivars of several species [45, 46]. The inter-simple sequence repeats (ISSRs) tested for the first time in Tunisia on the local wild caper patrimony revealed a high polymorphism level and separated this species into two main subspecies (C. spinosa subsp. spinosa and C. spinosa subsp. rupestris) which confirm the results of Khouildi et al. [47] and Ghorbel et al. [10]. This genetic result was in line with the morphological classification of Pottier-Alapetite [12], Saadaoui et al. [11], Saadaoui et al. [48] and Saadaoui et al. [15].
The H’ values are indicating the stability of the results for this model. This result can be explained by the hybridization between the two taxa, due to the out crossing pollination [19] and the geographic distribution that enhances gene flow through pollination and seed dispersal. Therefore, we can explain the structure analysis results that generated more groups than those generated by PCoA plot and UPGMA (Unweighted Pair-Group Method), by showing the identification of three main clusters and the admixed genetic profiles of the C. spinosa. in Tunisia.
On the other hand, our results are in line with the work of Gristinaa et al. [24] which revealed a high hybridization level in Capparis spinosa L. originated from the Mediterranean countries. Similarly, the current study proved the highest level of the intra-population variation at the Mediterranean area, and suggested that ecological adaptations to different environments could have been driven genetic differentiation between the two taxa by isolation [24, 25]. Inocencio et al. [48] explained that the distribution of the subgenus Capparis in the Mediterranean countries have been influenced by human involvement, seed dispersion and pollination which explain the high level (79%) of polymorphism within population. In addition, AMOVA analysis showed that the most of the total variation was find among genotypes within populations in their corresponding subspecies. Subsequently, the population’s structure could be explained by geographic isolation combined to ecological factors such as altitudes and rainfall influencing flowering time and longevity [49].
The present work based on ISSRs analyses, corroborates with previous morphological studies, which discriminates C. spinosa into two distinct subspecies: C. spinosa spinosa and C. spinosa rupestris. Indeed, morphological studies of vegetative and reproductive apparatus showed that in Tunisia there was only one species with two different subspecies: C. spinosa subsp. spinosa (thorny caper) and C. spinosa subsp. rupestris (inerm caper) [11]. According to the two research groups, Pottier-Alapetite [12], Tunisian flora description and previous publications interested in C. spinosa species, revealed only one single species with four varieties. Floch et al. [14] raised these varieties to species level. Moreover, ISSR marker analyses demonstrated in this present study the existence of one caper species Capparis spinosa L. in Tunisia with two taxa subspecies (spinosa and rupestris), and on the other hand revealed the presence of several hybrid forms between the two subspecies. The majority of these hybrids are looked very similarly to the subsp. spinosa subspecies especially by the presence of thorny stipules, shoots ramification and architecture, whereas were related to subsp. rupestris by habitat preference, leaf morphology and leathery consistency which confirmed the results obtained by Gristina et al. [24].
Conclusion
To the best of our knowledge, this is the first study that reveals the ISSR molecular characterization of Tunisian caper populations. Our study showed that ISSR analysis is an efficient method to investigate the genetic diversity and the relationship between Capparis species. The obtained data showed that caper germplasm resources present a wide range of diversity based on molecular traits allowing the distinction of the C. spinosa subsp. spinosa (thorny caper) and C. spinosa subsp. rupestris (inerm caper) and their grouping into two main genetic clusters emphasizing the existence of recognizable genetic similarity within species and genetic heterogeneity between them. This investigation showed that the grouping pattern of caper populations was not related to geographical origin. Therefore, this study is very important to identify the Tunisian caper species. The SSR markers would be our next step for characterizing the great diversity of varieties and to give an accurate taxonomy for caper genetic resources in Tunisia.
References
Dogan H, Ercisli S, Temim E, Hadziabulic A, Tosun M, Yilmaz SO, Zia-Ul-Haq M (2014) Diversity of chemical content and biological activity in flower buds of a wide number of wild grown caper (Capparis ovate Desf.) genotypes from Turkey. Comptes Rendus De L Acad Bulgare Des Sci 67:1593–1600
Barbera G (1991) Programme de recherche agrimed: le câprier (Capparis spp) CEE Rapport EUR13617 FR
Fici S (2001) Intraspecific variation and evolutionary trends in Capparis spinosa L. (Capparaceae). Plant Syst Evol 228:123–141
Raja P, Moorthy ND, Kala A, Soosai RS (2013) Extended distribution of Capparis shevaroyensis sund-ragh (Capparaceae) an endemic and vulnerable shrub in peninsular India to southern eastern ghats of tamilnaidu. Indian J Fundam Appl Life Sci 3:137–140
Le Floc ‘h E (1983) Contribution à une étude ethnobotanique de la flore tunisienne. - PubI. Sci. Tunisiennes. Programme «Flore et végétation tunisiennes». Imprimerie Officielle de la République Tunisienne, Tunisia, p 402
Bailey LH (1950) The Standard Cyclopedia of Horticulture. 658 p. Macmillan Company, New York
Tlili N, Nasri N, Saadaoui E, Khaldi A, Triki S (2009) Carotenoid and tocopherol Composition of Leaves, Buds and Flowres of Capparis spinosa Grownl. Wild in Tunisia. J Agric Food Chem 57:5381–5385
Ali-Shtayeh MS, Abu Ghdeib SI (1999) Antifungal activity of plant extracts against dermatophytes. Mycoses 42:665–672
Eddouks M, Lemhadri A, Michel JB (2005) Hypolipidemic activity of aqueous extract of Capparis spinosa L. in normal and diabetic rats. J Ethnopharmacol 98:345–350
Ghorbel A, Ben-Salem FA, Khouildi S, Skouri H, Chibani F (2001) Le câprier; caractérisation et multiplication. Des modèles biologiques à l’amélioration des plantes 157–172
Saadaoui E, Khaldi A, Khouja ML, El-Gazzah M (2007) Etude- de la variabilité morphologique du câprier (Capparis spp.) en Tunisie. Revue des Régions Arides 2:73–527
Pottier-Alapetite G (1979) Flore de la Tunisie: Angiiospermes - Dicotyledones Apétales Dialypétales, Première partie. Imprimerie officielle de la république, Tunisienne, p 651
Higton RN, Akeroyd JR (1991) Variation in Capparis spinosa L. in Europe. Flora Eur 106(2):104–112
Le Floc’h E, Boulos L, Vela E (eds) (2010) Catalogue synonymique commenté de la flore de Tunisie. p 500
Saadaoui E, Guetat A, Tlili N, El Gazzah M, Khaldi A (2011) Subspecific variability of Tunisian wild populations of Capparis spinosa L. J Med Plants Res 5:4339–4348
Rhizoploulou S, Psaras GK (2003) Development and structure of drought-tolerant leaves of the Mediterranean shrub Capparis spinosa L. Ann Bot 92:377–383
Rhimi A, Hannachi H, Hjaoujia S, Boussaid M (2012) The use of morphological descriptors to study variability in wild populations of Capparis spinosa L. (Capparaceae) in Tunisia. Afr J Ecol 51:47–54
Yousfi AH, Bahri AB, Medini M, Rouz S, Rejeb MN, Grabi GZ (2016) Genetic diversity and population structure of six species of Capparis in Tunisia using AFLP markers. CR Biol 339:442–453
Nosrati H, Hosseinpour Feizi MA, Mazinani M, Haghighi AR (2012) Effect of population size on genetic variation levels in Capparis spinosa (Capparaceae) detected by RAPDs. Eur Asian J BioSci 6:70–75
Bhoyar MS, Mishra GP, Naik PK, Murkute AA, Srivastava RB (2012) Genetic variability studies among natural populations of Capparis spinosa from cold arid desert of Trans-Himalayas using DNA markers. Natl Acad Sci Lett 35:505–515
Al-Safadi B, Faouri H, Elias R (2014) Genetic diversity of some Capparis L. species growing in Syria. Br Arch Biol Technol 57:916–926
Ozbek O, Kara A (2013) Genetic variation in natural populations of Capparis from Turkey, as revealed by RAPD analysis. Plant Syst Evol 299:1911–1933
Moubasher H, Abd El-Ghani MM, Kamel W, Mansi M, El-Bous M (2011) Taxonomic considerations among and within some Egyptian taxa of Capparis and related genera (Capparaceae) as revealed by RAPD fingerprinting. Collectanea Botanica 3:29–35. https://doi.org/10.3989/collectbot.2011.v30.003
Gristina AS, Fici S, Siragusa M, Fontana I, Garfi G, Carimi F (2014) Hybridization in Capparis spinosa L.: molecular and morphological evidence from a Mediterranean island complex. Flora 209(12):733–741. https://doi.org/10.1016/j.flora.2014.09.002
Saifi N, Ibijbijen J, Echchgadda D (2011) Genetic diversity of caper plant (Capparis ssp.) from North Morocco. J Food Agric Environ 9:299–304
González A, Coulson M, Brettell R (2002) Development of DNA markers (ISSRs) in mango. Acta Hortic 575:139–143
Borba RS, Garcia MAS, Kovalleski A, Oliveira AC (2005) Dissimilaridade genética de linhagens de Trichogramma Westwood (Hymenoptera: Trichogrammatidae) através de marcadores moleculares ISSR. Neotrop Entomol 34:565–569
Rocha TO, Freitas JS, Santos ESL, Scaldaferri MM (2016) Estimate of genetic diversity in cassutinga (Croton heliotropiifolius) based on molecular markers. Afr J Biotechnol 15:518–523
Trieu LN, Mien NT, Tien TV, Ket NV (2016) Genetic diversity of Panax stipuleanatus Tsai in North Vietnam detected by inter simple sequence repeat (ISSR) markers. Biotechnol Biotechnol Equip 30:506–511
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNAsepacer-lengtb polymorphism in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci 81:8014–8019
Peakall R, Smouse PE (2012) GENALEX6.5: genetic analysis in. Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Brookfield JFU (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:233–257
Tessier C, David J, This P, Boursiquot JM, Charrier A (1999) Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet 98:171 – 177
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss [Buchloe dactyloides (Nutt.) Engelm.]. Theor Appl Genet 86:927–934
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Halasz J, Pedryc A, Ercisli S, Yilmaz KU, Hegedus A (2010) S-genotyping supports the genetic relationships between Turkish and Hungarian apricot germplasm. J Am Soc Hortic Sci 135(5):410–417
Ercisli S, Ipek A, Barut E (2011) SSR marker-based DNA fingerprinting and cultivar identification of olives (Olea europaea). Biochem Genet 49:555–561
Caliskan O, Bayazit S, Oktem M, Ergul A (2017) Evaluation of the genetic diversity of pomegranate accessions from Turkey using new microsatellite markers. Turkish J Agric For 41:142–153
Hamrick JL, Godt MJW (1996) Conservation genetics of endemic plant species. In: Avise JL (ed) Conservation genetics case histories from nature. Chapman and Hall, London, pp 281–304
National Institute of Research in Rural Engineering, Waters, and Forests (INRGREF) (2001) Development of the culture of the caper in the West North of Tunisia. Final Scientific Report, Tunisia, p 84
Rivera D, Inocencio C, Obón C, Alcaraz F (2003) Review of food and medicinal uses of Capparis L. subgenus Capparis (Capparidaceae). Econ Bot 57:515–534
Skouri H (2000) Enzymatic polymorphism study at the caper (Capparis spinosa L.). Faculty of the Sciences, University of Tunis II, Tunis, p 96
Santos RC, Queiroz CM, Batista VGL, Silva CRC (2013) Variabilidade de progênies F2 de amendoim geradas por meio de seleção de genitores ISSR-divergentes. Rev Cienc Agron 44:578–586
Soares ANR, Vitória MF, Nascimento ALS, Ledo AS (2016) Genetic diversity in natural populations of mangaba in Sergipe, the largest producer State in Brazil. Genet Mol Res 15:1–12
Khouildi S, Pagnotta MA, Tanzarella OA, Ghorbel A, Porceddu E (2000) Suitability of RAPD (random amplified polymorphic DNA) technique for estimating the genetic variation in natural genotypes of Tunisian and Italian caper (Capparis spinosa L.). Agricoltura-Mediterranea 130:72–77
Saadaoui E, Khaldi A, Khouja ML, El Gazzah M (2009) Intraspecific variation of Capparis spinosa L. in Tunisia. J Herbs Spices Med Plants 15:9–15
Inocencio C, Cowan RS, Alcaraz F, Rivera D, Fay MF (2005) AFLP fingerprinting in Capparis subgenus Capparis related to the commercial sources of capers. Genet Resour Crop Evolut 52:137–144
Mhamdi TM, Ben Haj Ali I, Messaoud C, Boussaid M (2012) Genetic variability of Tunisian wild strawberry tree (Arbutus unedo L.) populations interfered from isozyme markers. Sci Hortic 146:92–98
Benzarti Z (2003) La pluviométrie en Tunisie. Analyse des années très pluvieuses. In: ENS (ed). EAU ET ENVIRONNEMENT - Tunisie et milieux méditerranéens. Lyon, pp. 63–77
Acknowledgements
The authors thank the National Genes Bank of Tunisia Institute and Mss Siwar Nsiri for her comments on an early version of manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies conducted on human or animal subjects.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rhimi, A., Mnasri, S., Ben Ayed, R. et al. Genetic relationships among subspecies of Capparis spinosa L. from Tunisia by using ISSR markers. Mol Biol Rep 46, 2209–2219 (2019). https://doi.org/10.1007/s11033-019-04676-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04676-z