Abstract
Lucerne (Medicago sativa) is an autotetraploid forage legume, whose breeding mainly relies on phenotypic recurrent selection to create synthetic populations. Allele mining could be a strategy to take advantage of the large genetic diversity of the species. This paper reports the polymorphism of five genes impacting the phenotype for selected traits: CAD1 and CCoaOMT (digestibility), CONSTANS-like (forage yield), NHX1 (salt tolerance), and WXP1 (drought tolerance). Complete genes were sequenced for 387 genotypes of 43 cultivated accessions and 20 genotypes of wild accessions. Wild versus cultivated polymorphism were compared, adaptive evolution was evaluated by comparing M. sativa and Medicago truncatula sequences and variants of the cultivated pool were characterized. We showed that the wild pool was more variable than the cultivated pool, with 36.6% and 8.4% of the variants that were specific to the wild pool and the cultivated pool, respectively. This result confirmed a bottleneck effect during domestication and selection. We also found that CAD1, CCoaOMT, and NHX1 were under a strong purifying selection and contained few non-synonymous variants (8, 5, and 8, respectively), while CONSTANS-like and WXP1 were under a less pronounced purifying selection and were more polymorphic (45 and 91 non-synonymous variants, respectively). This result suggests that adaptive evolution could be an indicator of expected polymorphism. The application of allele mining strategy for plant breeding in autotetraploid species is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lucerne (Medicago sativa) is one of the most widely cultivated legume crop worldwide, for ruminant feeding, given its high yield, nutritional properties, and adaptability (Li and Brummer 2012). Considering pure stands and legume-grass mixtures, this species is estimated to be the most cultivated forage legume crop in 15 countries in the eastern and south-western Europe (Julier et al. 2017). Lucerne is an allogamous perennial autotetraploid species, with 2n = 4x = 32. It belongs to a species complex that comprises two subspecies (Sinskaya 1950). Subspecies sativa contains the cultivated forms and rare wild populations, except in the region of origin (Middle East) and Spain. Subspecies falcata mostly contains wild forms and is expanded from the north of the Mediterranean regions to the north of Russia. Two ploidies are observed in both subspecies (2x and 4x). In addition to the wide diversity of this species, the within-population variation is very high in cultivars and feral lucerne populations, compared to the between-population variation (Julier et al. 2000; Flajoulot et al. 2005; Bagavathiannan et al. 2010; Li and Brummer 2012). As most outbreeding species, linkage disequilibrium (LD) is short in lucerne, it tends to decay after 1 kb (Herrmann et al. 2010; Sakiroglu et al. 2012). Lucerne breeding mainly relies on recurrent selection based on phenotype. The main breeding goals are forage yield, nutritive value (including digestibility and protein content), and tolerance to biotic and abiotic stresses. However, except for some disease and pest resistance, most traits are mostly under quantitative inheritance with many quantitative trait loci (QTL) that underlie a very small effect on phenotype (Li and Brummer 2012; Annicchiarico et al. 2015). Combined with the short linkage disequilibrium, this characteristic makes it difficult to apply marker-assisted selection. Direct manipulation of genes using biotechnology such as transgenesis, mutagenesis, and genome editing can be a solution to have a major impact on plant phenotype. For instance, lucerne digestibility have been significantly increased by means of downregulation of CCoaOMT (12% reduction of lignin content) (Meyermans et al. 2000) or CAD (5–14% of in vitro dry matter digestibility) (Jackson et al. 2008). Besides, overexpression of Medicago truncatula WXP1 in transgenic M. sativa increased leaf wax accumulation by 30–38% and consequently drought tolerance (Zhang et al. 2005). The major drawback of gene manipulation is that genetically modified organisms (GMO) cannot be cultivated in many countries (www.isaaa.org/gmapprovaldatabase/countrylist/default.asp) and that consumers tend to be suspicious about it or even to strongly reject it (Bredahl 2001; Bigot 2002; Wunderlich and Gatto 2015). Allele mining is a further option that may be appropriate for impacting gene expression and then plant phenotype. Allele mining consists in the detection of natural allele variation that may have a significant impact on phenotype (Kumar et al. 2010). Ecotilling was first proposed to identify allele variation in natural populations (Comai et al. 2004), but this method appeared to be inappropriate for lucerne because of its tetraploidy and high polymorphism. Allele mining is based on gene sequencing and dissects naturally occurring allele variation in candidate genes that regulate important agronomic traits. It has potential usage in crop improvement programs (Kumar et al. 2010). Allele mining was successfully used in Populus nigra to screen naturally defective alleles for a set of genes involved in lignin biosynthesis. Homozygous offspring with alleles carrying a premature codon stop in HCT gene exhibited a significant change in lignin composition (Vanholme et al. 2013). It has been suggested that this breeding strategy be called BRDA (breeding with rare defective alleles). Such a method has never been investigated on lucerne yet.
Evolutionary history of a gene can be inferred by analyzing DNA sequences, both among and within species (Parsch et al. 2013). Thus, the departure from neutral expectation may vary according to the different selective constraints applying on a gene. Numerous species belong to Medicago genus, besides M. sativa. Among them, the annual and diploid M. truncatula has been chosen in the 1990s as a model species for legume crops (Barker et al. 1990). A reference genome sequence has been published (Young et al. 2011) and a re-sequencing program has delivered sequence polymorphism in this species (Branca et al. 2011). M. sativa and M. truncatula are highly syntenic species (Choi et al. 2004).
The present paper investigates nucleotide sequence variation in various lucerne genes involved in agronomic traits. First, gene variation at evolutionary scale is evaluated by comparing M. sativa and M. truncatula gene sequences. The variability between cultivated and wild pools is then assessed. Finally, the frequency of variants, the proportion of non-synonymous variants, and the number of insertions/deletions (InDel) of each studied gene are examined. Depending on the gene, cultivated diversity appeared to be sufficient to find alleles with a probable impact on gene expression and phenotype. The potential use of allele mining strategy in breeding program is finally discussed.
Material and methods
Plant material and DNA extraction
Lucerne cultivated diversity was represented by 387 (a total of 1548 alleles) genotypes from 43 contrasted accessions (varieties and breeding material), each accession being represented by 9 genotypes. The wild diversity was represented by 12 genotypes of M. sativa subsp. sativa and 8 genotypes of M. sativa subsp. falcata from as many populations of the centers of origin. These genotypes were previously studied in a PhD thesis (Muller 2002). Seedling was grown in a greenhouse and DNA was extracted from young leaves using a protocol adapted from the CIMMYT large-scale DNA extraction protocol (Pauly et al. 2012). This extraction protocol can be summarized as a cellular lysis with CTAB and then a purification with chloroform/octanol (24:1) in 96-well plates. The concentrations of the DNA solutions were estimated using a NanoVue™ and adjusted between 20 and 30 ng/μL.
Choice of the genes
Genes were selected for having a proven impact on different lucerne agronomic traits (quality, forage yield, stress tolerance). They were also not part of large multigenic family. They were chosen as possible targets to speed up genetic progress for the major breeding criteria in lucerne breeding. Five genes were thus retained; their biological roles are either enzymatic activity (CAD1 and CCoaOMT), transcription factor (CONSTANS-like, WXP1), or vacuolar exchangers (NXH1). CAD1 is the gene encoding cinnamyl alcohol dehydrogenase (E.C. 1.1.1.195), a monolignol biosynthetic enzyme of lignin pathway. This enzyme catalyzes the last step of monolignol synthesis that is the enzymatic reduction of coniferyl aldehyde, p-coumaryl aldehyde, and sinapyl aldehyde to coniferyl alcohol, p-coumaryl alcohol, and sinapyl alcohol, respectively (McKie et al. 1993). A significant impact on lucerne digestibility has been observed after a downregulation of CAD1 gene, without any growth and development difference compared to untransformed control (Baucher et al. 1999; Chen et al. 2002). CCoaOMT encodes the caffeoyl-CoA O-methyltransferase (E.C. 2.1.1.104), involved in methylation steps of caffeoyl and 5-hydroxyferuloyl CoA thioesters of the pathway for monolignol biosynthesis (Parvathi et al. 2001). Downregulation of CCoaOMT in lucerne induced a decrease in the number of guaiacyl units in lignin and an increase in digestibility (Guo et al. 2001; Jung et al. 2012). Sequence variation in a CONSTANS-like gene has been linked to flowering date in M. truncatula and M. sativa (Pierre et al. 2010; Herrmann et al. 2010; Julier 2012), as CONSTANS gene of Arabidopsis thaliana which plays a central role in a pathway that controls flowering in response to photoperiod (Putterill et al. 1995; Suárez-López et al. 2001). CONSTANS-like also impacts stem height, and hence yield, in lucerne (Herrmann et al., 2010; Julier 2012). NHX1, firstly described in yeast, is an homologous gene to the mammalian NHE family that codes for Na+/H+ antiporters (Nass et al. 1997). The plant homolog of NHX1 antiporter presents similar mechanism by which cations are detoxified by Na+ sequestration in vacuoles, removing potentially toxic ions from cytoplasm and contributing to increase cellular osmolarity to counteract osmotic stresses (Gaxiola et al. 1999; Shi and Zhu 2002). Overexpression of M. sativa NHX1 stimulated salt tolerance by increasing Na+ accumulation in vacuoles (Yang et al. 2005). WXP1 is an ethylene-responsive element-binding transcription factor, which was firstly described in M. truncatula (Zhang et al. 2005). Its overexpression in transgenic M. sativa and M. truncatula induced not only expression of a number of genes related to wax production, conferring a reduction of water loss and chlorophyll leaching, a better recovery after water stress, but also an increased freezing tolerance (Zhang et al. 2005, 2007; Jiang et al. 2010).
Primer design and PCR amplification
Primers were designed for each gene from a M. sativa provisional assembly (J. Gouzy et al., INRA, France, non-published data) on which reads of two M. sativa genotypes from a previous study (I. Litrico et al., INRA, France, non-published data) were aligned to determine conserved regions. Genes of interest were located on M. sativa scaffolds by aligning published M. truncatula sequences (version 4.0; www.medicagogenome.org) of each gene on the provisional assembly. Primers were defined approximately 1000 to 2000 bp before and after the gene, using the Primer3web program (version 4.0.0; http://bioinfo.ut.ee/primer3). Primers lengths were between 20 and 23 bp, with a melting temperature (Tm) between 51 to 58 °C with less than 2 °C of difference between two primers of the same pair (Online Resource 1A). Each primer pair was tested for amplification with DNA of six M. sativa genotypes from different varieties (Tequila, Paréva, Volta, Comète, Marais de Challans, and Regen), selected from a previous diversity study (S. Flajoulot, unpublished) (Online Resource 1B and 1C). Amplified fragment size was estimated by electrophoresis migration on agarose gel (Online Resource 1A). A second pair of primers was designed in each gene, to ensure the right gene was amplified. Those primers were used on the previous PCR products (Online Resource 1D and 1E). Once amplification tests were achieved, genes were amplified by PCR, for each genotype with a high-fidelity Platinium™ Taq DNA Polymerase (Invitrogen™). Amplified fragment concentrations were evaluated by electrophoresis migration on agarose gel. Minimal DNA concentration for sequencing was 20 ng.
Sequencing and trimming
Amplification products were sequenced at INRA of Montpellier, France (UMR/AGAP), in series of 48 genotypes. PCR product concentrations of each gene were determined by PicoGreen® dosage. Concentrations of each genotype were adjusted to obtain similar values for a given gene. Amplified DNA of the 5 genes of each genotype were pooled in such a way that mixes contains a similar number of molecules for each gene. These mixes were then purified using Agencourt® AmPure® XP system (1 X concentration) in order to retain DNA products only. DNA was randomly fragmented in a Bioruptor® over 6 cycles of 10 s of exposure every 90 s. This treatment resulted in a fragmentation of approximately 500 bp DNA fragments. A purification Agencourt® AmPure® XP (0.6 X) was performed to eliminate DNA fragments under 500 pb. The end of DNA strands were repaired and 5′ phosphorylated by using the Fast DNA End Repair (Thermo Scientific™ Part # K0771). A clean-up step was performed with 1.25× volume of AmPure® XP magnetic beads.
A PCR amplification was undertaken to enrich library fragments, extend the adaptor sequence, and incorporate an index to the P7 adaptor specific to each pool of 48 samples. The PCR reaction used the KAPA® HiFi HotStart ReadyMix PCR Kit (KAPABiosystems, Part # KR0370) with 500 nM PE-PCR_F (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTC) primer and 500 nM MPE-INDX_R (CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGT) indexed primers. This MPE-INDX_R primer carried 6 bases of the official Illumina Index. Thermocycling parameters were 2 min at 98 °C, followed by 18 cycles of 20 s at 98 °C, 30 s at 62 °C, and 30 s at 72 °C, with a final elongation of 5 min at 72 °C. The reaction volume was 50 μL. A final clean-up step was performed with 1.8× volume AmPure® XP magnetic beads before sequencing by Illumina sequencing technology (MiSeq, 2 × 250 bp). Sequencing was performed in three runs of 192 genotypes (i.e., 4 series of 48 genotypes prepared as explained above), the last one including 57 genotypes whose data quality was low after their first run (low number of reads).
After sequencing, 17.62 Go of raw data were obtained. CLC Genomics Workbench 8.5.1 (https://www.qiagenbioinformatics.com/) was used for data processing. Reads were firstly trimmed on quality score (minimal value of 0.01), reads below 100 bp length were then discarded, and finally, the remaining reads were demultiplexed.
Definition of consensus sequences
No M. sativa reference sequences are available for any of the studied genes. Consequently, the gene sequences available on M. sativa provisional assembly (Debellé et al. 2012) were used as a template to build an improved sequence of each gene from newly sequenced data. More precisely, a subset of 16,750 reads from all cultivated genotypes and genes was randomly selected to theoretically cover sequenced genes at a 100× coverage. This value corresponded to the total length of target sequences (rounded to 36,000 bp), multiplied by the targeted coverage (100×) and then divided by mean read length (215 bp), rounded up to the tens of units. For each gene, the subset of reads was aligned on the provisional M. sativa genome assembly using CLC Genomics Workbench and the alignment was manually improved. Consensus sequence was extracted for each gene, corresponding to the most frequent nucleotide at each position and was called consensus sequence in this study. The gene length is given in Fig. 1.
Those M. sativa consensus sequences were annotated to define exon positions by aligning exons from M. truncatula reference sequence on these M. sativa consensus sequences (Fig. 1).
Variant detection
Consensus sequences were used as references for variant detection for each gene and each genotype, using the CLC Genomics Workbench. In this paper, the term “variant” is inclusive of single nucleotide polymorphism (SNP), multiple nucleotide polymorphisms (MNP), and insertion or deletion (InDel). Reads were firstly aligned against the consensus sequences (“Map Reads to Reference” tool) and then locally realigned to optimize alignment for InDels (“Local Realignment” tool). Variants were then detected from the “basic variant detection” tool of Genomics Workbench with the following criteria: minimal coverage of 30× at the considered position, at least 4 variants must be observed and their frequency must reach at least 10%. For each gene, a table was thus obtained. It contained various information on each detected variant, including variant position, reference nucleotide, variant nucleotide, variant frequency, and amino acid change if appropriate. The possibility to score allele frequency was assessed by plotting the distribution of variant frequency across all genotypes and variants. A discontinuous distribution, with breaks centered around frequencies of 0.25, 0.5, and 0.75, was expected (Julier et al. 2018). However, as the observed distribution was continuous (not shown), allele dosage could not be evaluated.
Gene coverage was generally heterogeneous along the sequence. Thus, some genotypes contained less covered parts where variants were not detected because coverage was lower than the 30× criterion descripted above. The following step was then to eliminate data from not enough covered genotypes. The minimal read number necessary to ensure a coverage to at least 30×, and thus to ensure all variants were detected, was estimated for each gene. For this purpose, sets of different numbers of reads were sampled in five contrasted genotypes, the number of sampled reads being a function of mean coverage as follows:
With gene length a constant for each gene (Table 1) and mean read length a constant of 215 bp. The different sets of reads corresponded to a mean coverage ranging from 20× to 140×.
Variant detection was executed for each set of reads, in ascending order. When the number of detected variants reached a plateau, the minimal mean coverage was considered as reached. The minimal coverage for variant detection was: 80× for CAD1, CONSTANS-like and WXP1 (2207, 1552, and 735 reads, respectively, for each genotype), 100× for NHX1 (3286 reads), and 120× for CCoaOMT (1858 reads for each genotype). Data from genotypes with lower mean read coverage were discarded from the study. Finally, data were collected from a variable amount of genotypes depending on the gene: 355 for CAD1, 345 for CCoaOMT, 362 for CONSTANS-like, 344 for NHX1, and 366 for WXP1, including 19 wild genotypes for CAD1 and WXP1 and 20 wild genotypes for the other genes.
Adaptive evolution of the studied genes
Gene variation was studied in an evolutionary perspective for each of the 5 genes, by calculating neutrality index (NI) and Ka/Ks ratio, by using M. sativa and M. truncatula coding sequences.
The NI evaluates the signature of natural selection for a given gene by comparing polymorphism within a species (P) to the divergence between species (D), at synonymous and non-synonymous sites. The NI was calculated as:
where Pn and Ps are the numbers of non-synonymous and synonymous polymorphic sites, respectively and Dn and Ds are the numbers of non-synonymous and synonymous divergent sites respectively. NI gives an estimation of the direction and degree of departure from neutral model according to which Dn/Ds = Pn/Ps, or NI = 1 (Kimura 1983; McDonald and Kreitman 1991; Bierne and Eyre-Walker 2004). A NI < 1 presumed an excess of fixation of non-synonymous substitutions relative to polymorphism, which indicates that positive selection, also known as Darwinian selection, is at work. However, a NI > 1 reflected that negative selection operated, preventing the fixation of maladaptive recessive mutations. Negative selection is also called purifying selection or background selection. To proceed with NI calculation, sequences of 21 M. truncatula genotypes from the HapMap project (http://www.medicagohapmap.org) were compared to two M. sativa sequences: the consensus sequence and a theoretical sequence carrying all the variants of the cultivated pool. The effect of slightly deleterious mutations was partially removed by excluding variants segregating at a frequency below 1% (Fay et al. 2001). NI was calculated on the MKT website (mkt.uab.es/mkt/; Egea et al. 2008). The statistical significance of the contingency tables obtained for NI calculation were tested using Fisher’s exact test rather than χ2 given the small sample size.
The other tool used to evaluate the evolution of the studied genes was the Ka/Ks ratio. This value is a way to evaluate the relative importance of selection and genetic drift in causing amino acid substitution by comparing two coding sequences of different species. Formally, Ka and Ks correspond to the mean number of nucleotide differences between the sequences per non-synonymous site and per synonymous site respectively. Ka/Ks ratio was calculated as:
where Sd is the number of synonymous substitutions, Sn the number of non-synonymous substitutions, S the number of non-synonymous substitutions, and N the number of potential non-synonymous substitutions. A ratio close to 1 indicates that amino acid substitutions are assumed to be largely neutral. Otherwise, a ratio below 1 implies that purifying selection mostly operated and a ratio over 1 suggests that positive selection mainly occurred. For each studied genes, M. sativa consensus sequence and a consensus sequence constructed from the 21 M. truncatula sequences from the HapMap project, were compared for the computation of Ka/Ks ratios. Those ratios were computed using the SNAP tool (v2.1.1) of HIV sequence database website (www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html; Korber 2001). In this program, Sd, Sn, S, and N were calculated based on the Nei and Gojobori method (Nei and Gojobori 1986).
Comparison of sequence diversity in cultivated and wild pools
In order to compare diversity level and to evaluate the impact of bottleneck effect during domestication and selection, the numbers of variants in the cultivated pool and in the wild pool were compared. However, the number of cultivated genotypes (324 to 346) was much higher than the number of wild genotypes (19 to 20). Hence, for each gene, 10,000 successive samplings of 19 cultivated genotypes for CAD1 and CCoaOMT, and 20 for CONSTANS-like NHX1 and WXP1 were executed. The mean number of variants in cultivated M. sativa and the standard deviation were calculated on basis of 10,000 samples and compared to the number of variants detected in the wild genotypes, for introns and exons of each gene.
The number of variants specific to cultivated pool and wild pool, in introns and exons, was calculated in order to evaluate the potential loss of diversity during domestication and selection. However, cultivated pool was represented by about 350 genotypes and wild pool was represented by 19 to 20 genotypes. In order to compare the number of variants specific to each pool without suffering the consequences of sampling effect due to the difference in population size, the probability P of an allele A of frequency Pa of being represented in a sample of n draws was calculated as follows (Crossa 1989):
By choosing P = 95% and n = 80 (the number of alleles in the wild pool), the frequency Pa was calculated to be 3.7%. As a consequence, only variants of the cultivated pool with a frequency above 3.7% were considered.
Variant characterization in the cultivated pool
Finally, all detected variants were further detailed for each gene in the cultivated pool. The number of variants in introns and exons was compared, as well as the mean base number between variants in introns and exons (sequence length divided per variant number). The numbers of non-synonymous variants, InDels and reading frame changes that may have an impact of the phenotype, were also calculated.
Results
Sequences were obtained for 324 to 346 cultivated genotypes and 19 to 20 wild genotypes depending on the gene.
Gene sequence variation at evolutionary scale
Both synonymous and non-synonymous sites have been identified in the five genes (Online Resource 2). The number of polymorphic variants evaluated within species (Ps + Pn) tended to be larger than the number of divergent variants evaluated between species (Ds + Dn). In most cases, more synonymous than non-synonymous variants was observed (Ps > Pn and Ds > Dn). For all the studied genes, Fisher’s exact test resulted in non-significant NI values. Thus, NI values could not be used to evaluate the signature of natural selection. However, the Ka/Ks ratio was below one for each studied genes (Table 2), indicating that purifying selection mostly operated and the genes tend to be conserved at an evolutionary scale. With Ka/Ks ratios of 0.021 and 0.033, NHX1 and CCoAOMT appeared to be the most conserved genes. CAD had also a relatively low ratio value (0.076). CONSTANS-like had a Ka/Ks ratio of 0.181 and WXP had the highest result, with a value of 0.329.
Comparison of sequence diversity in cultivated and wild populations
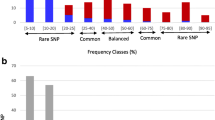
In order to overcome the large size difference between the wild and the cultivated pools, subsamples of the cultivated pool with the same number of genotypes as the wild pool were randomly selected and compared to the wild pool. For the five genes, the number of variants observed in the wild pool was significantly higher (on average 37.4%) than in the cultivated pool (Fig. 2a). The difference was even more pronounced in introns than in exons.
The number of variants in introns and exons of five genes in the wild and the cultivated lucerne genotypes. a For the wild pool: total number of variants; for the cultivated pool: mean and standard error calculated from 10,000 samplings of 20 genotypes. b The number of variants specific to the wild or the cultivated pool
As a total, 39 (8.4%) variants were specific to the cultivated pool and 171 (36.6%) were specific to the wild one over a total of 467 variants (Fig. 2b). The number of variants that were specifically detected in the wild pool was higher than those detected in the cultivated pool for each gene, in exons as well as in introns. The difference in the number of variants detected only in the wild and in the cultivated pool was less pronounced for CONSTANS-like than for the other genes. No specific variants were detected in CAD1 and in CCoaOMT exons.
Variant characterization in the cultivated pool
The variability, estimated by the mean number of bases between two consecutive variants, observed in the cultivated pool was more important in introns than in exons (on average, 8.8 and 32.1 bases, respectively), except for WXP1 which had no intron (Table 1, Fig. 1). Many more variants were observed in the exons of CONSTANS-like and WXP1 genes, especially considering non-synonymous variants, than in the three other genes. NHX1 was the least variable gene in both introns and exons. In exons, CAD1 was almost as little variable as NHX1. WXP1 was the most variable gene in exons. Numerous InDels were detected in WXP1 exon. Among them, 11 were constantly found together on the same allele by groups of 2 or 3, 1 to 3 bp apart. Combined together, they reconstituted the reading frame (not shown).
Discussion
In this study on 5 genes with a demonstrated impact on lucerne agronomic traits, we sequenced wild and cultivated M. sativa individuals. Our protocol was designed for a broad-scale survey of sequence polymorphism in candidate genes. Genotyping was successful and we obtained reliable gene sequences for most of the genotypes.
Comparison of sequence diversity in the cultivated and wild populations
DNA polymorphism in wild and cultivated genotypes has been documented in lucerne for neutral loci of nuclear genome (Muller et al. 2006) and mitochondrial genome (Muller et al. 2001, 2003). A demographic bottleneck that took place during the domestication was evidenced (Muller et al. 2003, 2006). However, there is a lack of information on the effect of domestication for genes which are likely to impact phenotype. In our study on 5 genes involved in agronomic traits, one third of the variants (36.6%) were specific to the wild pool, whereas only 8.4% were specific to the cultivated pool. Although, the number of wild genotypes was much smaller than that of cultivated genotypes, the number of variants was superior in the wild pool. Taking into account the difference of genotype number between the two pools, the number of variants observed per genotype in the cultivated pool was on average 37.4% lower than that the one observed in the wild pool.
Contrastingly, in a transcriptome study, 95% of SNP identified in wild lucerne relatives were also present in cultivated lucerne (Li et al. 2012). In this survey, only five wild genotypes were studied, four of them being diploid. This low number of allele copies may contribute to explain the difference with the results obtained in our study. Indeed, our findings are consistent with the results obtained in assumed neutral loci (glutamate synthase and pectate lyase genes) where 31% less diversity was found in the domesticated pool than in the wild pool (Muller et al. 2006). A diversity loss of the same order (around 30%) of magnitude was discovered in mitochondrial DNA, in the wild compared to the cultivated pools (Muller et al. 2003). Such a reduction of diversity is an indicator of a bottleneck effect that occurred during domestication (Frankel et al. 1995). Thus, the results obtained here seem to confirm the occurrence of such an event during lucerne domestication. Despite a selection pressure linked to artificial selection applied to agronomic traits, this study indicates that the loss of diversity in the candidate genes appears to be similar to the loss observed in neutral genes and mitochondrial DNA.
In other crops, genome-wide loss of diversity appears to be commonly around or over 30% (28% in soybean, 30% in rice, 33% in sunflower, 38% in maize, 69% in bread wheat, and 84% in durum wheat) but lower reduction has also been observed in some crops (14% in sorghum and no reduction of diversity in einkorn) (Buckler et al. 2001; Tenaillon et al. 2004; Casa et al. 2005; Hyten et al. 2006; Haudry et al. 2007; Kilian et al. 2007; Li et al. 2009; Mandel et al. 2011). Thus, the loss of diversity observed in lucerne appears to be average compared to other species.
Adaptive evolution and gene polymorphism
Polymorphism of cultivated M. sativa germplasm was assessed for 5 genes in relation with the signature of natural selection. Interspecific sequence comparison is a way to measure the intensity of purifying selection by evaluating the part of mutations that have been fixed or eliminated by selection after the speciation event (Halligan and Keightley 2006). Thus, the direction and degree of departure from neutral evolution was evaluated by comparing synonymous and non-synonymous mutation rate (Ka/Ks) in M. sativa and M. truncatula coding DNA sequences, the NI values being non-significant for the five genes. We have found that the three genes with the lowest Ka/Ks ratio (CCoAOMT, NHX1, and CAD1) were also the genes with the lowest variability in exons. Conversely, the genes with the highest ratio (CONSTANS-like and WXP1) had the largest variability in exons. These results suggest that polymorphism might be linked to the intensity of selection pressure.
For future studies on other genes, the NI and the Ka/Ks ratio could be firstly calculated using sequence data from a few dozens of genotypes of a target and a relative species. This information could be a way to make an idea of the expected polymorphism, and thus to estimate the number of genotypes necessary for allele mining. In that respect, the size of the studied population could be smaller for a gene under a low selection pressure than for a gene under negative selection.
The results obtained in this study about gene polymorphism were mostly consistent with previous studies conducted in other species or in genes with similar functions. For example, CONSTANS-like family genes have been demonstrated as evolving rapidly in Brassicaceae family, and the non-synonymous substitution rates were higher in CONSTANS-like family genes than in other genes (Lagercrantz and Axelsson 2000). The authors of this study assumed that the higher substitution rate of CONSTANS-like genes could be explained by the frequent occurrence of duplication events that allows divergence and new function acquisition. For genes involved in lignin biosynthesis, such as CAD1 and CCoaOMT, a low number of variants was detected in coding DNA of various species (Grima-Pettenati et al. 1993; Poke et al. 2003; Guillet-Claude et al. 2004; Fariza et al. 2008; Wang et al. 2015). For example, approximately one SNP every 37 bases were detected in Larix olgensis CAD (Wang et al. 2015) and one SNP every 39 bases in Acacia mangium CAD (Fariza et al. 2008), which is similar to one variant every 41 bases in our study. WXP1, a transcription factor of ERF gene family group, was very polymorphic in our study. This is in accordance with the high level of polymorphism observed in a region of SmERF gene of Slavia miltiorrhiza, with about one SNP every 13 base pairs (Cui et al. 2010). However, 8 and 9 among 10 Na+/H+ antiporters including NHX copies have been shown as carrying an excess of non-synonymous mutations in Populus euphratica and Populus. pruinosa respectively, suggesting a positive selection (Wu et al. 2017). The latter result contrasts with the result of this study where NHX1 appears to be under strong negative selection and the least variable of the five studied genes.
Prospect of allele mining for breeding of autotetraploid species
The results obtained on variant detection can be exploited in breeding. Since the proteins arising from genes carrying non-synonymous variants have a chance to have a different function, we will focus on non-synonymous variants.
A gene submitted to positive selection, or even to moderate negative selection, is likely to cumulate many mutations (e.g., CONSTANS-like and WXP1). In such cases, genotyping a few dozen of genotypes must be enough to detect a large amount of variants. Working with elite genotypes would be an interesting strategy in order to quickly obtain diverse and suitable material for breeding. Contrastingly, a gene under a strong negative selection (e.g., CAD1, CCoaOMT, and NHX1) should not contain many variants. In this situation, genotyping of a large number of genotypes, taken from diverse origins, will be necessary to detect enough variants. Promising alleles could be absent from breeding material, meaning that non-adapted, wild material should be prospected to find those alleles. However, the introgression of diversity from wild germplasm to elite material implies several backcross generations in order to minimize the genetic load and to obtain a suitable genetic background for breeding. Knowing the selection pressure a gene is submitted to, the budget allocated to genotyping and the effort agreed to subsequent selection steps will be different. In all cases, several generations, preferably marker-assisted, will be necessary to fix a mutation in elite breeding material.
For an allele mining approach, the impact of alleles of interest on the phenotype must be evaluated. This phase may be more or less costly and easy to execute, depending on the trait. Once variants have been identified, alleles of interest should be determined according to their probability to impact plant phenotype. When the gene tends to be under a strong negative selection pressure, any non-synonymous variants are likely to impact phenotype. However, if the gene tends to cumulate many variants, it could be interesting to focus on variants inducing a reading frame shift or premature stop codon. Indeed, those variants are most likely to have an impact on phenotype given that the encoded protein would be truncated. The closest the shift of frame or the premature stop codon will be to the beginning of the gene, the greater likelihood the protein change has to impact the protein function.
A way to evaluate the impact of a mutated allele (B) on the phenotype of an autotetraploid species such as lucerne is to compare not only homozygous genotypes with (BBBB) and without (AAAA) the mutation but also genotypes carrying various dosages of the mutated allele (AAAB, AABB, ABBB). However, attention must be paid to avoid the confusion between the effect of the mutation itself and that of the genetic background. Firstly, if the targeted mutation is fairly common, a large number of genotypes with contrasted genetic background could be assessed (Fig. 3a). Such situation may occur if the mutation has a strong effect, in favor of the selected trait, and tends to be selected by phenotypic selection since plant domestication. Otherwise, a number of genotypes carrying the mutation can be polycrossed in order to obtain a population with a homogeneous genetic background and carrying different dosages of the mutated alleles. If duplex genotypes (AABB) are identified, one generation will be enough to obtain all the possible dosage classes (Fig. 3a). Alternatively, if only simplex genotypes (AAAB) are detected, a first generation will be necessary to obtain duplex genotypes, the latter could be polycrossed in order to get every dosage class. A trial could then be established to compare 20 to 50 genotypes of each dosage class. The comparison of the different dosage classes offers the possibility to evaluate the contribution of the mutation to the phenotype (Fig. 3b). If no significant phenotypic difference is observed regardless of the dosage, the mutation has no effect on phenotype for the considered trait. If only two main phenotypic profiles are identified, it means that one of the alleles (A or B) is dominant. If heterozygotes have the same phenotype than homozygous genotypes without the mutation (AAAA), the allele B is recessive to the allele A. Otherwise, the allele B is dominant to the allele A. Finally, if intermediate phenotypes are observed for the heterozygotes according to their variant dosage, this means that genetic effects are additive. Once the impact of a mutation on the phenotype is established, the mutation could be either selected or eliminated by selection in the breeding program, with the help of molecular markers. If the mutant is recessive but has a desired effect on phenotype, the aim should be to obtain a population that is homozygous for this allele. However, if the favorable allele has a dominant or an additive genetic effect, maintaining a high proportion of this allele could be sufficient.
For a wide-scale application of allele mining in plant breeding, it would be valuable to consider not only a few genes with demonstrated effect on phenotype, but rather a large amount of genes likely to impact selected traits. However, this approach implies to mobilize substantial resources not only for genotyping but also for gene impact evaluation on phenotype. Moreover, it would be necessary to find a way to automate data processing, particularly to detect and determinate which variants are the most likely to have an impact on protein function.
References
Annicchiarico P, Barrett B, Brummer EC, Julier B, Marshall AH (2015) Achievements and challenges in improving temperate perennial forage legumes. Crit Rev Plant Sci 34:327–380. https://doi.org/10.1080/07352689.2014.898462
Bagavathiannan MV, Julier B, Barre P, Gulden RH, van Acker RC (2010) Genetic diversity of feral alfalfa (Medicago sativa L.) populations occurring in Manitoba, Canada and comparison with alfalfa cultivars: an analysis using SSR markers and phenotypic traits. Euphytica 173:419–432. https://doi.org/10.1007/s10681-010-0156-5
Barker DG, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S, Flament P, Gallusci P, Génier G, Guy P, Muel X, Tourneur J, Dénarié J, Huguet T (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Report 8:40–49. https://doi.org/10.1007/BF02668879
Baucher M, Bernard-vailhé MA, Chabbert B et al (1999) Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol Biol 39:437–447. https://doi.org/10.1023/A:1006182925584
Bierne N, Eyre-Walker A (2004) The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol 21:1350–1360. https://doi.org/10.1093/molbev/msh134
Bigot R (2002) L’opinion et les comportements des français en matière d’environnement. CREDOC-IFEN https://www.credoc.fr/publications/lopinion-et-les-comportements-des-francais-en-matiere-denvironnement. Accessed 2 Nov 2018
Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti AK, Woodward JE, May GD, Gentzbittel L, Ben C, Denny R, Sadowsky MJ, Ronfort J, Bataillon T, Young ND, Tiffin P (2011) Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc Natl Acad Sci 201104032:E864–E870. https://doi.org/10.1073/pnas.1104032108
Bredahl L (2001) Determinants of consumer attitudes and purchase intentions with regard to genetically modified food—results of a Cross-National Survey. J Consum Policy 24:23–61. https://doi.org/10.1023/A:1010950406128
Buckler ES, Thornsberry JM, Kresovich S (2001) Molecular diversity, structure and domestication of grasses. Genet Res 77:213–218. https://doi.org/10.1017/S0016672301005158
Casa AM, Mitchell SE, Hamblin MT, Sun H, Bowers JE, Paterson AH, Aquadro CF, Kresovich S (2005) Diversity and selection in sorghum: simultaneous analyses using simple sequence repeats. Theor Appl Genet 111:23–30. https://doi.org/10.1007/s00122-005-1952-5
Chen L, Auh C, Chen F, Cheng X, Aljoe H, Dixon RA, Wang Z (2002) Lignin deposition and associated changes in anatomy, enzyme activity, gene expression, and ruminal degradability in stems of tall fescue at different developmental stages. J Agric Food Chem 50:5558–5565. https://doi.org/10.1021/jf020516x
Choi H-K, Kim D, Uhm T, Limpens E, Lim H, Mun JH, Kalo P, Penmetsa RV, Seres A, Kulikova O, Roe BA, Bisseling T, Kiss GB, Cook DR (2004) A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics 166:1463–1502. https://doi.org/10.1534/genetics.166.3.1463
Comai L, Young K, Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Henikoff S (2004) Efficient discovery of DNA polymorphisms in natural populations by ecotilling. Plant J 37:778–786. https://doi.org/10.1111/j.0960-7412.2003.01999.x
Crossa J (1989) Methodologies for estimating the sample size required for genetic conservation of outbreeding crops. Theor Appl Genet 77:153–161
Cui GH, Feng H, Li WY, Wang WY, Huang LQ (2010) Cloning and polymorphism analysis of SmERF in Salvia miltiorrhiza. Yao Xue Xue Bao 45:1188–1193
Debellé F, Belser C, Aury J, Labadie K, Roux B, Sallet E, Carrere S, Wincker P, Schiex T, Julier B, Gamas P, Gouzy J (2012) Genomic resources for functional analysis in Medicago truncatula and related crop species. VI International Conference on Legume Genetics and Genomics. Hyderabad, October 2–7, 2012, P-NGG09
Egea R, Casillas S, Barbadilla A (2008) Standard and generalized McDonald–Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Res 36:W157–W162. https://doi.org/10.1093/nar/gkn337
Fariza MSN, Pang SL, Choong CY, Wickneswari R (2008) Extensive DNA sequence variations in two lignin genes, cinnamate 4-hydroxylase and cinnamyl alcohol dehydrogenase from Acacia mangium and Acacia auriculiformis. J Biol Sci 8:687–690. https://doi.org/10.3923/jbs.2008.687.690
Fay JC, Wyckoff GJ, Wu C-I (2001) Positive and negative selection on the human genome. Genetics 158:1227–1234
Flajoulot S, Ronfort J, Baudouin P, Barre P, Huguet T, Huyghe C, Julier B (2005) Genetic diversity among alfalfa (Medicago sativa) cultivars coming from a breeding program, using SSR markers. Theor Appl Genet 111:1420–1429. https://doi.org/10.1007/s00122-005-0074-4
Frankel OH, Brown AHD, Burdon JJ (1995) The conservation of plant biodiversity. Cambridge University Press
Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci 96:1480–1485. https://doi.org/10.1073/pnas.96.4.1480
Grima-Pettenati J, Feuillet C, Goffner D, Borderies G, Boudet AM (1993) Molecular cloning and expression of a Eucalyptus gunnii cDNA clone encoding cinnamyl alcohol dehydrogenase. Plant Mol Biol 21:1085–1095. https://doi.org/10.1007/BF00023605
Guillet-Claude C, Birolleau-Touchard C, Manicacci D, Fourmann M, Barraud S, Carret V, Martinant JP, Barrire Y (2004) Genetic diversity associated with variation in silage corn digestibility for three O-methyltransferase genes involved in lignin biosynthesis. Theor Appl Genet 110:126–135. https://doi.org/10.1007/s00122-004-1808-4
Guo DJ, Chen F, Inoue K et al (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88. https://doi.org/10.1105/tpc.13.1.73
Halligan DL, Keightley PD (2006) Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res 16:875–884. https://doi.org/10.1101/gr.5022906
Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, Hochu I, Poirier S, Santoni S, Glémin S, David J (2007) Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol 24:1506–1517. https://doi.org/10.1093/molbev/msm077
Herrmann D, Barre P, Santoni S, Julier B (2010) Association of a CONSTANS-LIKE gene to flowering and height in autotetraploid alfalfa. Theor Appl Genet 121:865–876. https://doi.org/10.1007/s00122-010-1356-z
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci 103:16666–16671. https://doi.org/10.1073/pnas.0604379103
Jackson LA, Shadle GL, Zhou R, Nakashima J, Chen F, Dixon RA (2008) Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. BioEnergy Res 1:180–192. https://doi.org/10.1007/s12155-008-9020-z
Jiang Q, Zhang J, Guo X et al (2010) Improvement of drought tolerance in white clover (Trifolium repens) by transgenic expression of a transcription factor gene WXP1. Funct Plant Biol 37:157–165
Julier B (2012) Alfalfa breeding benefits from genetic analyses on Medicago truncatula. In: Proceedings of the 7th International Symposium on the Molecular Breeding of Forage and Turf. USDA-ARS, Washington (Etats-Unis) 978–1–4675-4762-8, Salt Lake City, pp 17–19
Julier B, Huyghe C, Ecalle C (2000) Within- and among-cultivar genetic variation in alfalfa. Forage Quality, Morphology, and Yield Crop Sci 40:365–369. https://doi.org/10.2135/cropsci2000.402365x
Julier B, Gastal F, Louarn G et al (2017) Lucerne (Alfalfa) in European cropping systems. In: Legumes in cropping systems. D Murphy-Bokern, FL Stoddard and CA Watson (Eds), CABI Publishing, Wallingford, p 168–191
Julier B, Lambroni P, Delaunay S et al (2018) Use of GBS markers to distinguish among lucerne varieties, with comparison to morphological traits. Mol Breed 38:133
Jung H-JG, Samac DA, Sarath G (2012) Modifying crops to increase cell wall digestibility. Plant Sci 185–186:65–77. https://doi.org/10.1016/j.plantsci.2011.10.014
Kilian B, Özkan H, Walther A et al (2007) Molecular diversity at 18 loci in 321 wild and 92 domesticate lines reveal no reduction of nucleotide diversity during Triticum monococcum (Einkorn) domestication: implications for the origin of agriculture. Mol Biol Evol 24:2657–2668. https://doi.org/10.1093/molbev/msm192
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, New York, p 367
Korber B (2001) HIV signature and sequence variation analysis. In: GH Learn Jr. (Ed) Computational and evolutionary analysis of HIV. Kluwer Academic Publishers, Boston, AG Rodrigo, p 55–72
Kumar GR, Sakthivel K, Sundaram RM, Neeraja CN, Balachandran SM, Rani NS, Viraktamath BC, Madhav MS (2010) Allele mining in crops: prospects and potentials. Biotechnol Adv 28:451–461. https://doi.org/10.1016/j.biotechadv.2010.02.007
Lagercrantz U, Axelsson T (2000) Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol Biol Evol 17:1499–1507. https://doi.org/10.1093/oxfordjournals.molbev.a026249
Li X, Brummer EC (2012) Applied genetics and genomics in alfalfa breeding. Agronomy 2:40–61. https://doi.org/10.3390/agronomy2010040
Li X, Tan L, Zhu Z, Huang H, Liu Y, Hu S, Sun C (2009) Patterns of nucleotide diversity in wild and cultivated rice. Plant Syst Evol 281:97–106. https://doi.org/10.1007/s00606-009-0191-7
Li X, Acharya A, Farmer AD, Crow JA, Bharti AK, Kramer RS, Wei Y, Han Y, Gou J, May GD, Monteros MJ, Brummer E (2012) Prevalence of single nucleotide polymorphism among 27 diverse alfalfa genotypes as assessed by transcriptome sequencing. BMC Genomics 13:568. https://doi.org/10.1186/1471-2164-13-568
Mandel JR, Dechaine JM, Marek LF, Burke JM (2011) Genetic diversity and population structure in cultivated sunflower and a comparison to its wild progenitor, Helianthus annuus L. Theor Appl Genet 123:693–704. https://doi.org/10.1007/s00122-011-1619-3
McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. https://doi.org/10.1038/351652a0
McKie JH, Jaouhari R, Douglas KT et al (1993) A molecular model for cinnamyl alcohol dehydrogenase, a plant aromatic alcohol dehydrogenase involved in lignification. Biochim Biophys Acta Protein Struct Mol Enzymol 1202:61–69. https://doi.org/10.1016/0167-4838(93)90063-W
Meyermans H, Morreel K, Lapierre C, Pollet B, de Bruyn A, Busson R, Herdewijn P, Devreese B, van Beeumen J, Marita JM, Ralph J, Chen C, Burggraeve B, van Montagu M, Messens E, Boerjan W (2000) Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275:36899–36909. https://doi.org/10.1074/jbc.M006915200
Muller M-H (2002) Flux de gènes et domestication: l’histoire évolutive d’un complexe sauvage-cultivé. Approches expérimentale et théorique chez la luzerne pérenne (Medicago sativa L. ). PhD Thesis, Université de Montpellier II, p 103
Muller M-H, Prosperi JM, Santoni S, Ronfort J (2001) How mitochondrial DNA diversity can help to understand the dynamics of wild-cultivated complexes. The case of Medicago sativa in Spain. Mol Ecol 10:2753–2763. https://doi.org/10.1046/j.0962-1083.2001.01400.x
Muller M-H, Prosperi JM, Santoni S, Ronfort J (2003) Inferences from mitochondrial DNA patterns on the domestication history of alfalfa (Medicago sativa). Mol Ecol 12:2187–2199. https://doi.org/10.1046/j.1365-294X.2003.01897.x
Muller M-H, Poncet C, Prosperi J et al (2006) Domestication history in the Medicago sativa species complex: inferences from nuclear sequence polymorphism. Mol Ecol 15:1589–1602. https://doi.org/10.1111/j.1365-294X.2006.02851.x
Nass R, Cunningham KW, Rao R (1997) Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase insights into mechanisms of sodium tolerance. J Biol Chem 272:26145–26152. https://doi.org/10.1074/jbc.272.42.26145
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426. https://doi.org/10.1093/oxfordjournals.molbev.a040410
Parsch J, Meiklejohn CD, Hartl DL (2013) Inferring evolutionary history through inter- and intraspecific DNA sequence comparison: the Drosophila janus and ocnus genes. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013. Available from: https://www.ncbi.nlm.nih.gov/books/NBK6313/. Accessed 07 Nov 2018
Parvathi K, Chen F, Guo D, Blount JW, Dixon RA (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25:193–202. https://doi.org/10.1111/j.1365-313X.2001.00956.x
Pauly L, Flajoulot S, Garon J, Julier B, Béguier V, Barre P (2012) Detection of favorable alleles for plant height and crown rust tolerance in three connected populations of perennial ryegrass (Lolium perenne L.). Theor Appl Genet 124:1139–1153. https://doi.org/10.1007/s00122-011-1775-5
Pierre J-B, Bogard M, Herrmann D, Huyghe C, Julier B (2010) A CONSTANS-like gene candidate that could explain most of the genetic variation for flowering date in Medicago truncatula. Mol Breed 28:25–35. https://doi.org/10.1007/s11032-010-9457-6
Poke FS, Vaillancourt RE, Elliott RC, Reid JB (2003) Sequence variation in two lignin biosynthesis genes, cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase 2 (CAD2). Mol Breed 12:107–118. https://doi.org/10.1023/A:1026006828721
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857. https://doi.org/10.1016/0092-8674(95)90288-0
Sakiroglu M, Sherman-Broyles S, Story A, Moore KJ, Doyle JJ, Charles Brummer E (2012) Patterns of linkage disequilibrium and association mapping in diploid alfalfa (M. sativa L.). Theor Appl Genet 125:577–590. https://doi.org/10.1007/s00122-012-1854-2
Shi H, Zhu J-K (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol 50:543–550
Sinskaya EN (1950) Flora of cultivated plants of the USSR XIII. Perennial leguminous plants. Part 1. Medic, sweetclover, fenugreek. Israel Program for Scientific Translations, Jerusalem, p 661
Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120. https://doi.org/10.1038/35074138
Tenaillon MI, U’Ren J, Tenaillon O, Gaut BS (2004) Selection versus demography: a multilocus investigation of the domestication process in maize. Mol Biol Evol 21:1214–1225. https://doi.org/10.1093/molbev/msh102
Vanholme B, Cesarino I, Goeminne G, Kim H, Marroni F, van Acker R, Vanholme R, Morreel K, Ivens B, Pinosio S, Morgante M, Ralph J, Bastien C, Boerjan W (2013) Breeding with rare defective alleles (BRDA): a natural Populus nigra HCT mutant with modified lignin as a case study. New Phytol 198:765–776. https://doi.org/10.1111/nph.12179
Wang Y, Jia Q, Zhang L, Zhang Z, Zhang H (2015) Allelic variation in cinnamyl alcohol dehydrogenase (LoCAD) associated with wood properties of Larix olgensis. Forests 6:1649–1665. https://doi.org/10.3390/f6051649
Wu Y, Meng K, Liang X (2017) Distinct patterns of natural selection in Na+/H+ antiporter genes in Populus euphratica and Populus pruinosa. Ecol Evol 7:82–91. https://doi.org/10.1002/ece3.2639
Wunderlich S, Gatto KA (2015) Consumer perception of genetically modified organisms and sources of Information123. Adv Nutr 6:842–851. https://doi.org/10.3945/an.115.008870
Yang Q, Wu M, Wang P, Kang J, Zhou X (2005) Cloning and expression analysis of a vacuolar Na+/H+ antiporter gene from alfalfa. DNA Seq 16:352–357. https://doi.org/10.1080/10425170500272742
Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, van de Peer Y, Proost S, Cook DR, Meyers BC, Spannagl M, Cheung F, de Mita S, Krishnakumar V, Gundlach H, Zhou S, Mudge J, Bharti AK, Murray JD, Naoumkina MA, Rosen B, Silverstein KAT, Tang H, Rombauts S, Zhao PX, Zhou P, Barbe V, Bardou P, Bechner M, Bellec A, Berger A, Bergès H, Bidwell S, Bisseling T, Choisne N, Couloux A, Denny R, Deshpande S, Dai X, Doyle JJ, Dudez AM, Farmer AD, Fouteau S, Franken C, Gibelin C, Gish J, Goldstein S, González AJ, Green PJ, Hallab A, Hartog M, Hua A, Humphray SJ, Jeong DH, Jing Y, Jöcker A, Kenton SM, Kim DJ, Klee K, Lai H, Lang C, Lin S, Macmil SL, Magdelenat G, Matthews L, McCorrison J, Monaghan EL, Mun JH, Najar FZ, Nicholson C, Noirot C, O’Bleness M, Paule CR, Poulain J, Prion F, Qin B, Qu C, Retzel EF, Riddle C, Sallet E, Samain S, Samson N, Sanders I, Saurat O, Scarpelli C, Schiex T, Segurens B, Severin AJ, Sherrier DJ, Shi R, Sims S, Singer SR, Sinharoy S, Sterck L, Viollet A, Wang BB, Wang K, Wang M, Wang X, Warfsmann J, Weissenbach J, White DD, White JD, Wiley GB, Wincker P, Xing Y, Yang L, Yao Z, Ying F, Zhai J, Zhou L, Zuber A, Dénarié J, Dixon RA, May GD, Schwartz DC, Rogers J, Quétier F, Town CD, Roe BA (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524. https://doi.org/10.1038/nature10625
Zhang J-Y, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42:689–707. https://doi.org/10.1111/j.1365-313X.2005.02405.x
Zhang J-Y, Broeckling CD, Sumner LW, Wang Z-Y (2007) Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol Biol 64:265–278. https://doi.org/10.1007/s11103-007-9150-2
Acknowledgements
We thank the Association Nationale de la Recherche et de la Technologie (ANRT) for support to the PhD grant of C. Gréard. Technical staffs of INRA Lusignan, INRA Montpellier, Jouffray-Drillaud and GIE GRASS are acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Gréard, C., Barre, P., Flajoulot, S. et al. Sequence diversity of five Medicago sativa genes involved in agronomic traits to set up allele mining in breeding. Mol Breeding 38, 141 (2018). https://doi.org/10.1007/s11032-018-0898-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0898-7