Abstract
A series of transgenic lines of alfalfa (Medicago sativa) were generated in which either one of the two potentially terminal enzymes of the monolignol pathway, cinnamoyl CoA reductase (CCR) or cinnamyl alcohol dehydrogenase (CAD) was down-regulated by expression of antisense transgenes. Levels of CCR enzymatic activity were reduced to between 10% to 65% of the control level, and levels of CAD activity were similarly reduced to between 5% to 40% of the control. Biomass yields were reduced in the most strongly down-regulated lines for both transgenes, but many of the lines exhibited reduced lignin levels but normal biomass and flowering time. In vitro dry matter digestibility was increased for most transgenic lines compared to controls. Saccharification efficiency was determined by measuring the release of sugars from cell walls directly, or after sulfuric acid pre-treatment and subsequent digestion with a mixture of cellulase and cellobiase. Several CCR down-regulated lines had significantly enhanced saccharification efficiency with both pre-treated and untreated tissues, whereas CAD down-regulation had less impact on sugar release when compared to that from CCR lines with similar lignin contents. One CCR line with a 50–60% improvement in saccharification efficiency exhibited normal biomass production, indicating the potential for producing high yielding, improved feedstocks for bioethanol production through genetic modification of the monolignol pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The accessibility of plant cell wall polysaccharides to chemical, enzymatic and microbial degradation is limited by many factors, including the presence of lignin [6, 42, 51]. Lignin is a polymer of hydroxylated and methoxylated phenylpropane units (monolignols) linked via oxidative coupling. It accumulates in secondary cell walls during the development of vascular tissues and fibers [5], and imparts both mechanical strength and hydrophobicity. The composition and structure of lignified cell walls has a dramatic impact on the technological value of raw materials, and has been identified as a key factor limiting effective biomass to biofuel conversion in processes where the sugar components of polysaccharides are released (saccharification) prior to fermentation to ethanol. In addition to physically preventing access of microbial enzymes to cellulose, lignin may also exert a negative impact on bioethanol production through its partial degradation to inhibitors of the microbial fermentation system [35].
The lignin biosynthetic pathway has been under extensive investigation for more than thirty years. Genes encoding most of the enzymes leading to the hydroxyphenyl (H), guaiacyl (G) and syringyl (S) monolignol building blocks of lignin have now been identified [25, 26]. Relationships between lignin content/composition and efficiency of lignocellulose utilization in the pulping of trees to make paper [37, 40] and in the digestibility of forages for ruminant animals [1, 21, 44] have been revealed by down-regulating some of these genes in transgenic plants. Furthermore, studies utilizing natural variation in hybrid poplar progeny suggested that the lignin S/G ratio might impact the acid pre-treatment step, which releases primarily pentose sugars from hemicellulose [13], and impacts of lignin content on both acid pre-treatment and enzymatic saccharification have been reported as a function of developmental stage in alfalfa (Medicago sativa), reed canarygrass (Phalaris arundinacea), and switchgrass (Panicum virgatum) [14].
Alfalfa has recently been promoted as a potential bioenergy crop [17, 28]. Currently, alfalfa is the world’s major forage legume, with an average annual value of more than $8 billion in the USA alone, supported by extensive agronomic improvement programs in the private sector. Although its yields do not approach those of the currently favored bioenergy crops switchgrass, Miscanthus or poplar, alfalfa is a high yielding perennial with the added benefits of nitrogen fixation, well defined agronomic practices, seed industry support, and extensive translational genomics resources through studies on the closely related model species Medicago truncatula [38]. We have previously identified the majority of the genes involved in the biosynthesis of monolignols in Medicago and generated stably transformed alfalfa lines independently down-regulated in seven different enzymatic steps of the monolignol biosynthetic pathway (l-phenylalanine ammonia-lyase, cinnamate 4-hydroxylase, hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase, coumaroyl shikimate 3-hydroxylase, caffeoyl CoA 3-O-methyltransferase, ferulate (coniferaldehyde) 5-hydroxylase, or caffeic acid 3-O-methyltransferase), resulting in lines with wide differences in both lignin content and composition in the same genetic background [11, 20, 21, 44, 48]. Using these materials, we have demonstrated the relationships between lignin content/composition and the efficiency of saccharification (release of sugars from lignocellulosic material) by chemical pre-treatment and enzymatic hydrolysis [10]. Genetic modification of lignin biosynthesis in alfalfa can improve enzymatic hydrolysis efficiency by up to twofold and possibly eliminate the requirement for costly chemical pre-treatment in biofuel production [10]. However, depending upon the lignin pathway gene targeted, improvements in bioprocessing efficiency may be compromised by negative impacts on yield and performance. For example, some transgenic alfalfa lines showed delayed flowering and biomass reduction, while in other cases there was no visible phenotype [21, 44, 48]. The transgenic lines with the greatest improvements in saccharification efficiency (those down-regulated in hydroxycinnamoyl transferase (HCT)) showed the greatest reduction in yield (an approximately 20% reduction in dry matter at flowering), and also exhibited increased branching [48]. In order to capture the full value of lignin modification technology for bioethanol processing, it is important to de-couple the changes in cell wall chemistry from potential reductions in yield.
In general, down-regulation of the later enzymes in monolignol biosynthesis has less effect on plant growth and yield than down-regulating the earlier enzymes. This may result from avoidance of pleiotropic growth effects due to perturbations in metabolites that feed into other pathways. Cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) catalyze the last two steps in monolignol biosynthesis, although CCR may also act earlier in the pathway [39]. Both enzymes have been targeted in a number of species in studies aimed at assessing effects on lignin content and composition [3, 12, 19, 22, 30, 37, 41, 43], and it has been reported that CAD down-regulation in alfalfa results in small improvements in in situ digestibility of forage in sheep [1]. Strong down-regulation of CCR has been shown to cause collapse of vascular tissue in tobacco, but, surprisingly, this can be overcome by simultaneously down-regulating CAD [8].
Previous studies have shown that the very high gene sequence identity between orthologous genes in M. truncatula and alfalfa allows the use of antisense M. truncatula sequences for efficient down-regulation of the corresponding ortholog in M. sativa [44]. Using this approach, we here report the impacts of independent down-regulation of CCR and CAD on plant biomass yield, fiber properties, lignin composition and cell wall deconstruction by acid and enzymatic saccharification in alfalfa. Our results indicate that CCR is a better target than CAD for improving the saccharification efficiency of alfalfa stem biomass.
Results
Generation of Transgenic Alfalfa Plants with Reduced Expression of CCR and CAD
Searching the current Medicago gene index available at http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb = medicago revealed eight genes annotated as encoding CCR, and three genes annotated as encoding CAD. Phylogenetic analysis indicated that two Medicago CCR genes, represented by TC (tentative consensus sequence) 106830 (MtCCR1) and TC 100678 (MtCCR2), and one CAD gene, represented by TC 100737 (MtCAD1), were most closely related to functionally characterized CCR and CAD genes from other species (Fig. 1), and therefore targets for the modification of monolignol biosynthesis in alfalfa. The biochemical properties of the enzymes encoded by MtCCR1 and MtCCR2 will be described elsewhere (R. Zhou and R.A. Dixon, unpublished results). MtCCR1 was chosen for down-regulation in the present work.
Phylogenetic relationships between Medicago genes annotated as encoding CCR (a) and CAD (b) and functionally related genes encoding these enzymes from other species. The trees were drawn from ClustalW using the neighbor-joining method. The GeneBank numbers of the various CCR genes are: Arabidopsis thaliana, AAG46037; Codonopsis lanceolata, BAE48787; Eucalyptus cordata, AAT74875; Eucalyptus globules, AAT74879; Hordeum vulgare, AAN71760; Leucaena leucocephala, CAK22319; Oryza sativa, CAD21520; Picea abies, CAK18610; Pinus taeda, AAL47684; Populus tomentosa, AAR83344; Populus tremuloides, AAF43141; Populus trichocarpa, CAA12276; Solanum tuberosum, AAN71761; Zea mays, CAA74071. The GeneBank numbers of the various CAD genes are: Arabidopsis thaliana, AAM65761; Citrus sinensis, ABM67695; Eucalyptus botryoides, P50746; Eucalyptus globules, O64969; Lolium perenne, O22380; Medicago sativa, P31656; Nicotiana tabacum, P30360; Pinus radiata, Q40976; Pinus taeda, P41637; Populus deltoides, P31657; Populus tomentosa, AAR83343; Populus trichocarpa, CAC07423; Saccharum officinarum, O82056; Zea mays, O24562; Zinnia elegans, BAA19487
The complete open reading frames of MtCCR1 and MtCAD1 were amplified by PCR, verified by sequencing, and introduced into the pCAMBIA2200 binary vector in the antisense orientation. Transgenes were driven by the bean phenylalanine ammonia-lyase promoter, which exhibits vascular tissue-specific expression in alfalfa [20]. Constructs were transferred to Agrobacterium tumefaciens for alfalfa transformation and regeneration by somatic embryogenesis as described previously [20, 44].
Transcript levels for the target genes were determined by RNA gel blot analysis (Fig. 2A) or RT-PCR (data not shown) using pooled stem internodes 1–5. In the case of the CCR antisense lines, at least 10 lines showed strong down-regulation of the target transcripts when analyzed by RT-PCR. In the case of the CAD antisense lines, 15 out of the 17 lines analyzed had significantly reduced CAD transcript levels when compared to those in un-transformed or empty vector-transformed control lines (Fig. 2A).
Down-regulation of CAD and CCR in transgenic alfalfa. a RNA gel blot analysis of total RNA from the first to fifth internodes of mature alfalfa stems probed with the MtCAD cDNA or 18s rRNA (loading control). Lines designated Ck are untransformed controls; Vec denotes empty vector transformed lines. All other lines harbor the MtCAD1 antisense construct. b Extractable CCR activities from pooled first to sixth internodes of mature alfalfa stems. Ck represents the average value of two untransformed controls and one empty vector line set at 100%. c Extractable CAD activities from pooled first to sixth internodes of mature alfalfa stems. Determinations of CAD activity were performed in duplicate (technical replicates)
Twelve CCR and 11 CAD antisense transgenic lines with verified reductions in target transcript levels were selected for determination of enzymatic activity. Among the 12 CCR lines, levels of extractable CCR activity in pooled internodes 1–6 varied from 65% to 10% of the control value (Fig. 2B). In the case of the CAD antisense transgenics, one line (32a) showed less than a 20% reduction in activity, with the remaining 10 lines showing from 40–5% of wild-type activity (Fig. 2C).
Phenotypic Consequences of Down-regulating CCR or CAD in Alfalfa
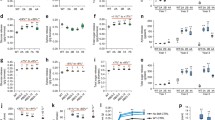
Some of the CCR and CAD down-regulated alfalfa lines exhibited reduced growth rate compared to controls, although others appeared normal (e.g., Supplementary Figure 1). Overall biomass was determined by weighing the total aerial material of plants at the early flowering stage, and the time taken from cutback to flowering was also determined for each of the lines. Three of the CCR down-regulated lines (CCR4, CCR13, and CCR37) had similar fresh and dry weights to controls, whereas two lines (CCR8 and CCR29) were more than 50% impaired in biomass production (Fig. 3A). Only one of the CCR down-regulated lines (CCR4) flowered significantly later than controls (Supplementary Table 1).
Biomass production by CCR- and CAD-down-regulated alfalfa lines. a Seven CCR antisense lines, and average value of two untransformed controls and one empty vector control line (Ck). b Eleven CAD antisense lines. Control as above. Open bars, fresh weight; shaded bars, dry weight. All determinations were performed in duplicate (technical replicates) using pooled samples of six plants per line
The population of CAD antisense transgenics appeared similar to the CCR lines in that two lines (CAD32 and CAD54) had similar biomass yields to controls, whereas three lines (CAD6, CAD44, and CAD63) had around 50% reductions in both fresh and dry weights (Fig. 3B). One line (CAD 3) had increased biomass compared to controls. Most of the lines attained flowering within the same time frame as the control lines (Supplementary Table 1).
Unlike the observations of coloration observed in the xylem of tobacco, poplar and tomato down-regulated in CCR, [34, 41, 50], no coloration was observed in the CCR down-regulated alfalfa plants Microscopical analysis of cross sections of CAD transgenics revealed the presence of red pigmentation in the vascular cells of lines CAD3 (weak), CAD17 (strong), CAD26, and CAD56 (strong) (Supplementary Figure 2). This is consistent with previous reports of CAD down-regulation in tobacco [24] and Arabidopsis [49]; the pigmentation has been shown to be primarily the result of accumulation of monolignol aldehyde units [2, 22, 24].
Lignin Content and Composition of Alfalfa Plants with Reduced CCR or CAD Activity
For preliminary, semi-quantitative screening of lignin content/composition, cross sections of CCR and CAD transgenics were stained with phloroglucinol (to detect total lignin) (Supplementary Figure 3A,B) and with Mäule reagent (to detect S lignin) (Fig. 4A,B). Generally, phloroglucinol staining did not reveal major differences between the lines, with the possible exception of reduced staining in CCR4 and stronger staining of xylem fibers of CAD downregulated lines. However, Mäule staining revealed decreases in S lignin content, particularly in CAD lines 3, 17, and 26 (replacement of the red staining with an orange-brown coloration) and in CCR lines 4, 8 and 13.
Total lignin content in mature internodes (internodes 8 and above) of control and transgenic lines was first determined by the acetyl bromide method (Fig. 5A,B). Apart from CCR lines 8 and 29, all CCR transgenics appeared to have lower AcBr lignin levels than controls (Fig. 5A). Seven CAD transgenics also appeared to have reduced AcBr lignin levels (Fig. 5B), and there appeared to be a correlation between the reduction in lignin levels and the appearance of red pigmentation in the vascular tissue (Supplementary Figure 2).
Lignin content and composition of mature stem internodes of CCR- and CAD-down-regulated alfalfa plants. a Acetyl bromide lignin content of control and CCR-down-regulated lines (SD of ck = 5.62). b Acetyl bromide lignin content of control and CAD-down-regulated lines (SD of ck = 8.66). c Total lignin monomer thioacidolysis yields of control and CCR-down-regulated lines (SD of ck = 10.90). d Total lignin monomer thioacidolysis yields of control and CAD-down-regulated lines (SD of ck = 34.38). e Lignin S/G ratios of control and CCR-down-regulated lines. (SD of ck = 0.058). f Lignin S/G ratios of control and CAD-down-regulated lines (SD of ck = 0.037). Control (Ck) represents the average value from two wild-type lines and one empty vector control line grown in parallel
Lignin composition was determined by the thioacidolysis method, which relies on gas chromatography/mass spectrometry to quantify monomeric units primarily released by cleavage of β-ether bonds; the method is therefore affected by lignin monomer linkage patterns [29, 46]. Total thioacidolysis monomer yields were reduced in several of the CCR transgenics, particularly lines CCR4, CCR41 and CCR48 (Fig. 5C); these changes reflected the changes recorded for total lignin using the acetyl bromide method. Similar relationships were seen in the case of the CAD transgenics (Fig. 5B,D). Lines CAD3 and CAD17 had the lowest thioacidolysis yields among the CAD down-regulated plants (Fig. 5D). Plotting the thioacidolysis total monomer yield per gram of lignin (Supplementary Figure 6) showed that the CCR transgenics had similar yields compared to the control while CAD3 and CAD17 had the lowest yields. This clearly shows that the CCR and CAD transgenics have different lignin structures.
Down-regulation of CCR resulted in an increase in S/G ratio in most lines, (Fig. 5E), and there was an inverse correlation between thioacidolysis yield and S/G ratio. In contrast, S/G ratios were decreased in several of the CAD antisense lines, particularly CAD3 and CAD17 (Fig. 5F), consistent with the loss of staining by Mäule reagent in stem cross sections of these lines. In CAD transgenic lines, sinapaldehyde and coniferaldehyde markers [30] were also detected using GC/MS during the thioacidolysis analysis, however, we were not able to quantify them due to the low amount and partial overlap with other compounds.
Similar differences in lignin content and composition between the various lines were also recorded when younger tissues (internodes 1–6) from other cuttings were analyzed (Supplementary Figure 4).
Lignocellulosic Fiber Properties of Alfalfa Plants with Reduced CCR or CAD Activity
Standard forage quality wet chemistry techniques were employed to determine the fiber properties of the biomass from the CCR and CAD down-regulated lines. These analyses revealed that CCR down-regulation led to reductions in acid detergent lignin in all lines (Fig. 6A), and to reductions in neutral detergent fiber (NDF) and acid detergent fiber (ADF) in some lines (particularly CCR8 and CCR29) (Supplementary Figure 5A,B). Increases in in vitro dry matter digestibility (IVDMD), determined by digestion of dried biomass in rumen fluid, from 5–10% were recorded in lines CCR4, 8, 29, 41 and 48 (Fig. 6B), these being the lines with some of the lowest acid detergent lignin, acetyl bromide lignin and total thioacidolysis yield values. Similar results for fiber properties were observed with a number of the CAD antisense lines (Fig. 6C,D; Supplementary Figure 5C,D). Thus, CCR and CAD down-regulation in alfalfa results in increased accessibility of the biomass to microbial degradative enzymes, presumably associated with the changes in lignin content. This conclusion had been made earlier for CAD down-regulated alfalfa lines in the context of digestibility for sheep [1].
Acid detergent lignin (a, c) and in vitro dry matter digestibility (b, d) of CCR- (a, b) and CAD- (c, d) down-regulated alfalfa lines. Controls (Ck) are the average value from two wild-type lines and one empty vector control line grown in parallel. Standard deviations were determined for technical replicates
Saccharification of Cell Wall Material from CCR- and CAD-down-regulated Alfalfa Lines
To determine whether the altered lignin contents and compositions in CCR and CAD down-regulated alfalfa translate to improved enzymatic saccharification for bioethanol production, dry milled cell wall preparations were subjected to two treatments: (1) treatment with dilute (1.5%) sulfuric acid at 130°C for 40 min followed by exposure to a mixture of cellulase and cellobiase for 72 h (acid pre-treated tissues) or (2) direct exposure to a mixture of cellulase and cellobiase for 72 h (untreated tissues). Levels of carbohydrates in the cell walls, and released into solution, were determined before and after enzyme treatment, and the results expressed as saccharification efficiency (the % of available carbohydrate released from the cell walls) (Table 1). Levels of total available carbohydrates in cell walls from untreated biomass varied from 580 (CCR4) to 883 (CAD17) mg/g dry weight of wall material (Table 1). There was no correlation between these values and the extent of enzyme down-regulation. The amount of available sugar as a function of total cell wall dry mass in pre-treated walls was usually higher than from the corresponding untreated samples, presumably reflecting the removal of non-carbohydrate material during the pre-treatment. There was also no obvious relationship between the actual values for available carbohydrate and the subsequent saccharification efficiency (Table 1).
Clearly, down-regulation of CCR had a greater effect on saccharification efficiency than did down-regulation of CAD (Table 1). For example, the average value for enzymatic saccharification efficiency of acid pre-treated control cell walls was around 25% for the CCR experiment, but this value was up to 50% higher in two of the four lines analyzed in Table 1, and from 45% (CCR29) to 100% (CCR4) higher in seven transgenic lines analyzed in an independent replicate experiment performed on a subsequent cut-back of the material (data not shown). A similar pattern was seen for enzymatic hydrolysis of cell walls without acid pre-treatment (Table 1). In contrast, only two of the CAD transgenics (56, 17) appeared to have clearly increased enzymatic saccharification efficiency, and this increase was less than observed for the CCR down-regulated lines. In fact, several of the CAD transgenics exhibited reduced saccharification efficiency compared to the relevant control value (Table 1).
Discussion
Previous studies have validated the use of M. truncatula monolignol biosynthetic gene sequences for targeting down-regulation of their closely related orthologs in alfalfa [44]. Informatic analysis suggests that M. truncatula has two CCR genes and one CAD gene that are closely related to the CCR and CAD genes from other species that have been shown to function in lignin biosynthesis [1, 8, 18, 19, 31, 32]. The selection of MtCCR1 for antisense down-regulation of alfalfa CCR was justified by the strong reductions of up to 90% in CCR enzymatic activity observed.
The antisense strategy proved efficient for the generation of multiple, independent transgenic alfalfa lines exhibiting strong reductions in CCR or CAD activity. Overall, the level of enzyme down-regulation was somewhat stronger in the group of CAD transgenics, although we were able to select more than five independent lines for each transgene with reductions in enzymatic activity of 70% or more. In spite of the overall lower target enzyme activities in the CAD transgenics, total lignin levels, as measured by the acetyl bromide method, were overall lower in the CCR transgenics, although the extent of down-regulation of lignin levels determined as thioacidolysis yields was overall similar in the two sets of plants (with some of the CAD lines exhibiting no decrease or even an increase in thioacidolysis yield in spite of reduced AcBR lignin). This is most likely due to the differences in lignin composition (S/G ratios) in the CAD and CCR transgenics which can lead to changes in lignin structure (i.e., linkage pattern) that will affect the thioacidolysis yields of the different monomeric units [33]. Significant differences in cell wall ultrastructure have also been reported for vascular elements of CCR- and CAD-down-regulated plants, with the CCR transgenics showing distorted or swollen vessels [9, 36].
A previous study with transgenic alfalfa lines down-regulated independently in six enzymes upstream of CAD led us to conclude that, over the ranges studied, lignin content, rather than lignin composition, was the major factor affecting saccharification efficiency [10]. It was therefore surprising that a comparison of CCR and CAD down-regulated lines with similar lignin levels revealed greater saccharification efficiency in the case of CCR. The reason for this has yet to be determined, although CCR down-regulation does appear to have unique effects on vascular cell wall structure [9].
Large increases in saccharification efficiency of cell wall material were recorded for many of the CCR down-regulated lines, but fewer of the CAD down-regulated lines showed this improvement and, for those that did, the effects were much smaller. In a previous study that evaluated biomass from transgenic alfalfa independently down-regulated in seven enzymes preceding CCR and CAD in the monolignol pathway, the highest saccharification efficiencies, of 67–79% for pre-treated wall material, were observed in lines down-regulated in C3H and HCT expression, compared to 43% in controls [10]. In the present work, CCR line 41 exhibited the highest saccharification efficiency for pre-treated material, with a value of 71% (compared to a control value of 42%), approaching that of the previously analyzed HCT lines. Down-regulation of HCT or C3H resulted in cell wall biomass that yielded greater saccharification efficiencies for untreated material than seen for controls after acid pre-treatment [10]. This was not observed for CCR down-regulation, although line CCR4 came close to showing this phenomenon (39.7% efficiency for untreated wall material compared to 41.5% for the pre-treated control).
In vitro dry matter digestibility (IVDMD) was increased to similar extents (from 8–14%) in most of the CCR and CAD down-regulated transgenic lines. This represents the amount of biomass degraded or released by the action of rumen microorganisms in a fermentation that had been allowed to proceed to completion (see [21] for in vitro digestion kinetics of alfalfa biomass in this type of assay). However, the enzymatic saccharification efficiencies of the cell wall material isolated from the various transgenic lines did not parallel the IVDMD values. For example, CAD35 has a similar IVDMD value (81%) to that of CCR4 (82%), but the saccharification efficiency of pre-treated biomass from CAD35 was only 29.7% (lower than the control value), compared to 62.4% for CCR4. This could be due to effects of alterations in lignin monomer composition and/or linkage patterns in the transgenic plants impacting on rumen microbes and/or their hydrolytic enzymes.
Genetic modification of monolignol biosynthesis can result in negative pleitropic effects on biomass yield and plant architecture [4, 15, 27]. The molecular basis for these effects is still unclear although, in the case of Arabidopsis plants with reduced HCT expression, it may involve inhibited auxin transport as a result of spill-over of flux from monolignol to flavonoid biosynthesis [4]. Maintaining high biomass is critical for the economics of lignocellulose-derived bioethanol, and, although improvements in saccharification efficiency can outbalance reduced yields [10], it will be important to combine more efficient bioprocessing properties with maximum agronomic performance. In this respect, it is interesting that the alfalfa line with the greatest improvement in saccharification efficiency (CCR4) exhibited normal biomass yields; this line did not have the most strongly reduced target enzyme activity. CCR modification is clearly a good target as regards the development of alfalfa as a bioenergy crop. Nevertheless, it is likely that full optimization of biomass and bioprocessing potential will require independent improvement of both traits.
Methods
Plant Materials and Genetic Transformation
Alfalfa (Medicago sativa cv Regen SY) plants were grown in the greenhouse under standard conditions with a 16 h day from 06:00 to 22:00 hours facilitated by supplementary lighting. Genetic transformations were performed with clonally propagated material of one selected highly regenerable line of cultivar Regen SY. The entire coding region of M. truncatula CAD (TC100737) was amplified using ACATGGGTAGCATTGAAGCTGCAGAAAGAACAAC as forward primer and CACCCTTGTCCACACTCTTCATGATATTGTATGT as reverse primer. The corresponding forward and reverse primers for amplification of the full M. truncatula CCR (TC106830) open reading frame were TCCCTTTACAAGAAGAAGGAATATATGCCTGCC and CACCGATGGGTTTAGGATTTGACTGCTAGAG, respectively. pCAMBIA2200-MtCADas and pCAMBIA2200-MtCCRas constructs were made for bean PAL promoter-driven antisense-mediated down-regulation of CAD and CCR, respectively, following the procedure described previously [44]. The generation of transgenic alfalfa lines was as reported previously [20, 44]. Independent events were selected by taking embryos from separate individual leaf disc explants (one embryo per explant). Controls were either empty vector transformed lines, or untransformed lines that had been put through the regeneration regimen.
Phylogenetic Analysis
Phylogenetic analysis was carried out with the MEGALIGN program in the DNASTAR package (Madison, WI). The deduced amino acid sequences of M. truncatula proteins annotated as potentially encoding CCR and CAD were aligned with their corresponding counterparts from other plant species using the ClustalW method with default gap penalty of 10 and gap-length penalty of 0.2. The GeneBank numbers of these proteins are listed in the legend to Fig. 1. Phylogenetic trees were generated with ClustalW using the neighbor-joining method. The identity of CCR1 and CCR2 (77.9% at the nucleotide level) was determined by the DNASTAR protein alignment protocol using the Jotun-Hein algorithm [23]. Default parameters used for the Jotun–Hein method for multiple alignments included a gap penalty of 11 and a gap-length penalty of 3.
RT-PCR and RNA Gel Blot Analysis
RNA was isolated from 200 mg of ground stem tissue (internodes 1–5) using TRI Reagent solution (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s instructions. Any remaining contaminating DNA was removed with Turbo DNA-free (Ambion, Austin, TX). The RNA was further purified by lithium chloride precipitation (Ambion). Four μg of RNA were used to generate first strand products using the Superscript III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA) according to the product protocol. PCR was performed with the first strand products as template DNA using gene-specific primers or actin primers and TaKaRa ExTaq polymerase (Takara Bio, Inc.). PCR products were visualized on a 1% agarose gel.
For RNA gel blot analysis, 15 μg of total RNA was separated on a 1% formaldehyde-containing agarose gel and transferred onto a nylon membrane (Zeta-Probe Genomic Tested Blotting Membranes, Bio-Rad, Hercules, CA, USA) by standard procedures [47] and UV cross-linked using a Stratalinker (Stratagene, La Jolla, CA). The CAD coding sequence was labeled with α-32P-dCTP using Ready-To-Go labeling beads (GE Healthcare) and purified on Probe Quant G50 micro columns (Amersham, Piscataway, NJ). The purified probe was used for RNA gel blot hybridization as described previously [45].
Assay of CAD Enzyme Activity
Alfalfa stems (internodes 1–6) (six plants per line) were pooled and ground to powder in liquid nitrogen. One gram batches of fresh tissue were weighed into 12 ml tubes on ice, and 4 ml of extraction buffer containing 100 mM Tris–HCl (pH 7.5) and 5 mM DTT, along with 80 mg of polyethylene glycol 6000 and 80 mg of polyvinylpolypyrrolidone (to remove phenolic compounds) were added. The mixtures were homogenized with a polytron for 10 s on ice, vortexed and incubated on ice for 5 min. The supernatants were separated by centrifugation at 10,000×g at 4°C for 10 min, transferred to new 12 ml tubes chilled on ice, and centrifuged again at 10,000×g at 4°C for 10 min. CAD enzyme activity of the supernatants was determined by measuring the increase in absorbance at 400 nm as coniferyl alcohol was oxidized to coniferaldehyde [53]. The assay was performed for 30 min at 30°C in a total volume of 1.0 ml using 130 mM Tris–HCl (pH 8.8), 2.0 mM coniferyl alcohol, and 2.0 mM NADP. Protein concentrations were determined by the Bradford method [7].
Assay of CCR Enzyme Activity
Alfalfa stems (internodes 1–6, six plants per line) were pooled and ground to powder in liquid nitrogen. One gram batches of frozen powdered stem tissue were weighed into 12 ml tubes on ice, and 4 ml of extraction buffer containing 100 mM Tris–HCl (pH 7.6), 2% PEG 6000 and 5 mM DTT was added. The mixture was homogenized with a polytron for 10 s on ice, a small amount of polyvinylpolypyrrolidone was added, and the mixture was then vortexed and incubated on ice for 5 min. The supernatants were separated by centrifugation at 10,000×g at 4°C for 10 min, and 3 ml were transferred to new 12 ml tubes chilled on ice. Solid ammonium sulfate (0.7 g) was added and the supernatants stirred on a rotating shaker at 4°C for 1 h. After centrifugation at 15,000×g for 30 min, the supernatants were transferred to new tubes containing an additional 0.95 g of solid ammonium sulfate and stirred for 1 h at 4°C. The supernatants were removed and discarded after the mixtures had been centrifuged for 30 min at 15,000×g, and the remaining pellets were re-suspended in 2.5 ml of re-suspension buffer that contained 20 mM Tris–HCl (pH 7.5), 5 mM DTT, and 5% ethylene glycol, and the solutions recentrifuged for 15 min at 10,000×g at 4°C. After desalting by passage through Sephadex G-25 (PD10) columns, the collected protein was used for assay for CCR activity.
The reaction was carried out at 30°C in a final volume of 500 μl containing sodium phosphate buffer (100 mM pH 6.2), NADPH (2.5 mM), protein (10 μg) and feruloyl CoA (70 μM). The feruloyl CoA was added last, and CCR enzyme activity determined by measuring the decrease in absorbance at 366 nm between 2 and 15 min [37].
Determination of Lignin Content and Composition
Extractive-free cell wall samples were generated by extracting plant tissue with methanol (three times at 37°C for 1.5 h) and chloroform:methanol (2:1) (three times at 37°C for 1.5 h). The samples were then washed three times with water at 37°C for 1.5 h and lyophilized for 48 h. Lignin content and composition were measured in extractive-free cell wall material. The mature stem tissue (Fig. 5) from six plants per line was harvested when plants were grown to the late flowering stage and the first seven internodes from top were discarded leaving only internodes eight and higher. The young stem tissue (supplemental figure 4) was harvested when plants were grown to eight internodes, but only internodes 1–6 were used for analysis. Total lignin was determined by the acetyl bromide method (Fukushima and Hatfield, 2004) using ∼30 or 15 mg extractive-free material for young and mature tissues respectively. Molar extinction coefficients of 17.2 and 17.12 (determined for lignin from young and mature wild-type alfalfa stem tissue, respectively) were used.
Thioacidolysis methods [29, 33] were used for determination of lignin composition. Thioacidolysis was performed using ∼15 mg of extractive-free samples reacted with 5 ml of 0.2 M BF3 etherate in an 8.75:1 dioxane/ethanethiol mixture. Lignin-derived monomers were identified by gas chromatography mass spectrometry (GC/MS), and quantified by GC as their trimethylsilyl derivatives. GC/MS was performed on a Hewlett–Packard 5890 series II gas chromatograph with a 5971 series mass selective detector (column: HP-1, 60-m · 0.25-mm · 0.25-μm film thickness), and mass spectra were recorded in electron impact mode (70 eV) with 60–650 m/z scanning range.
Histochemical Staining of Lignin
Alfalfa stem cross-sections 100 μm thick (CAD transgenics) or 20 μm (CCR transgenics) were made using a vibratome (series 1000; Ted Pella Inc., Redding, CA) and subjected to Mäule or phloroglucinol staining as described previously [20]. Photographs were taken using an Olympus SZX stereomicroscope system with a SPOT RT color camera.
Determination of Fiber Quality
Vegetatively propagated alfalfa cuttings (six plants for each line were pooled for analysis) were grown in parallel in one gallon pots in the greenhouse. Aerial portions were harvested at the early bud stage to ensure material was matched developmentally, and dried in a 50°C oven for at least 72 h. The samples were then ground in a Thomas-Wiley model 4 Laboratory Mill (Lehman Scientific, Wrightsville, PA) with 1 mm sieves. Acid detergent fiber (ADF) and neutral detergent fiber (NDF) were estimated by standard protocols (Goering and Van Soest, 1970). For NDF analysis, 0.35 g of ground samples were transferred to a F57 ANKOM filter bag (ANKOM Technology Corporation, Fairport, NY) and heated at 100°C for 1 h in an ANKOM Fiber Analyzer, according to the manufacturer’s instructions. The samples were washed in near boiling water, dried at 105°C for 6 h, and weighed to determine fiber loss. ADF was estimated sequentially on the material remaining after NDF analysis. The residue was then used for determination of acid detergent lignin by incubation in 72% (v/v) sulfuric acid for 3 h, washing thoroughly and drying at 105°C for 6 h, prior to weighing.
For determination of in vitro digestibility, ground tissue samples were dried at 105°C for 6 h prior to determining pre-extraction dry weights. The same procedure was used to obtain post-extraction dry weights. Digestibility analysis (0.5 g samples) was performed using F57 filter bags and the DAISY II incubator (ANKOM Technology Corporation, Fairport, NY [52]), following the manufacturer’s instructions.
Determination of Saccharification Efficiency
Extractive-free stem cell wall material at a loading of 10% (w/v) was mixed with 1.5% (w/v) sulfuric acid and pre-treated in an autoclave at 130°C for 40 min. After pre-treatment, hydrolysates were separated and collected by filtration from residual biomass and the biomass residues were washed with water.
Enzymatic saccharification of biomass residues was according to the analytical procedure of the National Renewable Energy Laboratory (LAP-009) (http://www.nrel.gov/biomass/analytical_procedures.html). Biomass samples equal to 0.1 g equivalents of cellulose were hydrolyzed with a mixture of Celluclast 1.5 L (cellulase from Trichoderma reesei) and Novozyme 188 (cellobiase from Aspergillus niger) (Sigma, St Louis, MO, USA) in 10 ml sodium citrate buffer (0.1 M, pH 4.8) for 72 h. The enzyme cocktail was obtained by mixing equal volumes of Celluclast 1.5 L and Novozyme 188. The enzyme loadings were 21 FPU per g cell wall residue as determined by the method described by Ghose (Ghose, 1987). Enzyme blanks and Whatman #1 filter paper (0.1 g) were digested alongside the samples. Hydrolysis of filter paper was always more than 95%.
Total sugars (of cell wall preparations and hydrolysates) were analyzed spectrophotometrically using the phenol-sulfuric acid assay method [16]. Saccharification efficiency was determined as the ratio of sugars released by enzymatic hydrolysis to the amount of sugars present in the cell wall material prior to enzymatic hydrolysis for both acid pre-treated and non-pre-treated biomass.
Abbreviations
- CAD:
-
cinnamyl alcohol dehydrogenase
- CCR:
-
cinnamoyl coenzyme A reductase
- G:
-
guaiacyl
- HCT:
-
hydroxycinnamoyl transferase
- S:
-
syringyl
References
Baucher M, BernardVailhe MA, Chabbert B et al (1999) Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol Biol 39:437–447
Baucher M, Chabbert B, Pilate G et al (1996) Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol 112:1479–1490
Bernard-Vailhé MA, Besle JM, Maillot MP et al (1998) Effect of down-regulation of cinnamyl alcohol dehydrogenase on cell wall composition and on degradability of tobacco stems. J Sci Food Agric 76:505–514
Bessau S, Hoffmann L, Geoffroy P et al (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19:148–162
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Boudet AM, Kajita S, Grima-Pettenati J et al (2003) Lignins and lignocellulosics: a better control of synthesis for new and improved uses. Trends Plant Sci 8:576–581
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chabannes M, Barakate A, Lapierre C et al (2001a) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J 28:257–270
Chabannes M, Ruel K, Yoshinaga A et al (2001b) In situ analysis of lignins in transgenic tobacco reveals a differential impact of individual transformations on the spatial patterns of lignin deposition at the cellular and subcellular levels. Plant J 28:271–282
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotech 25:759–761
Chen F, Reddy MSS, Temple S et al (2006) Multi-site genetic modulation of monolignol biosynthesis suggests new routes for the formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J 48:113–124
Chen L, Auh C-K, Dowling P et al (2003) Improved forgage digestibility of tall fescue (Festuca arundinacea) by transgenic down-regulation of cinnamyl alcohol dehydrogenase. Plant Biotechnol J 1:437–449
Davison BH, Drescher SR, Tuskan GA et al (2006) Variation of S/G ratio and lignin content in a Populus family influences the release of fermentable sugars by dilute acid hydrolysis. Appl Biochem Biotechnol 129–132:427–435
Dien BS, Jung H-JG, Vogel KP et al (2006) Chemical composition and response to dilute-acid pretreatment and enzymatic saccharification of alfalfa, reed canarygrass, and switchgrass. Biomass Bioenergy 30:880–891
Do C-T, Pollet B, Thevenin J et al (2007) Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid 3-O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226:1117–1129
Dubois M, Gilles KA, Hamilton JK et al (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Ebert J (2007) Alfalfa’s bioenergy appeal. Ethanol Producer Magazine (BBI International), Sept 2007, pp 88–94
Eudes A, Pollet B, Sibout R et al (2006) Evidence for a role of AtCAD1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225:23–39
Goujon T, Ferret V, Mila I et al (2003) Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: effect on phenotype, lignins and cell wall degradability. Planta 217:218–228
Guo D, Chen F, Inoue K et al (2001a) Down-regulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa (Medicago sativa L.): impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88
Guo D, Chen F, Wheeler J et al (2001b) Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res 10:457–464
Halpin C, Knight ME, Foxon GA et al (1994) Manipulation of lignin quality by down-regulation of cinnamyl alcohol dehydrogenase. Plant J 6:339–350
Hein JJ (1990) Unified approach to alignment and phylogenies. Meth Enzymol 183:626–645
Hibino T, Yakabe K, Kawazu T et al (1995) Increase of cinnamaldehyde groups in lignin of transgenic tobacco plants carrying an antisense gene for cinnamyl alcohol dehydrogenase. Biosci Biotech Biochem 59:929–931
Hoffmann L, Maury S, Martz F et al (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278:95–103
Humphreys JM, Chapple C (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5:224–229
Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26:205–216
Lamb JFS, Sheaffer CC, Samac DA (2003) Population density and harvest maturity effects on leaf and stem yield in alfalfa. Agron J 95:635–641
Lapierre C, Monties B, Rolando C (1985) Thioacidolysis of lignin: comparison with acidolysis. J Wood Chem Technol 5:277–292
Lapierre C, Pilate G, Pollet B et al (2004) Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65:313–321
Lapierre C, Pollet B, MacKay JJ et al (2000) Lignin structure in a mutant pine deficient in cinnamyl alcohol dehydrogenase. J Agric Food Chem 48:2326–2331
Lapierre C, Pollet B, PetitConil M et al (1999) Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol 119:153–163
Lapierre C, Pollet B, Rolando C (1995) New insight into the molecular architecture of hardwood lignins by chemical degradative method. Res Chem Intermed 21:397–412
Leple JC, Dauwe R, Morreel K et al (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19:3669–3691
Luo C, Brink DL, Blanch HW (2002) Identification of potential fermentation inhibitors in conversion of hybrid poplar hydrolyzate to ethanol. Biomass Bioenergy 22:125–138
Nakashima J, Chen F, Jackson L et al (2008) Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa L.): effects on lignin composition in specific cell types. New Phytol 179:738–750
O’Connell A, Holt K, Piquemal J et al (2002) Improved paper pulp from plants with suppressed cinnamoyl-CoA reductase or cinnamyl alcohol dehydrogenase. Transgenic Res 11:495–503
Oldroyd GE, Geurts R (2001) Medicago truncatula, going where no plant has gone before. Trends Plant Sci 6:552–554
Parvathi K, Chen F, Guo D et al (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25:193–202
Pilate G, Guiney E, Holt K et al (2002) Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol 20:607–612
Piquemal J, Lapierre C, Myton K (1998) Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J 13:71–83
Ragauskas AJ, Williams CK, Davison BH et al (2006) The path forward for biofuels and biomaterials. Science 311:484–489
Ralph J, Hatfield RD, Piquemal J et al (1998) NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamyl-alcohol dehydrogenase and cinnamoyl-CoA reductase. Proc Natl Acad Sci USA 95:12803–12808
Reddy MSS, Chen F, Shadle GL et al (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci USA 102:16573–16578
Reddy MSS, Ghabrial SA, Redmond CT et al (2002) Resistance to bean pod mottle virus in transgenic soybean lines expressing the capsid polyprotein. Phytopathology 91:831–838
Rolando C, Monties B, Lapierre C (1992) Thioacidolysis. In: Lin SY, Dence CW (eds) Methods in lignin chemistry. Springer-Verlag, Berlin Heidelberg, pp 334–340
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, New York
Shadle G, Chen F, Reddy MSS et al (2006) Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxy cinnamoyl transferase in transgenic alfalfa impacts lignification, development and forage quality. Phytochemistry 68:1521–1529
Sibout R, Eudes A, Mouille G et al (2005) Cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17:2059–2076
van der Rest B, Danoun S, Boudet AM et al (2006) Down-regulation of cinnamoyl-CoA reductase in tomato (Solanum lycopersicum L.) induces dramatic changes in soluble phenolic pools. J Exp Bot 57:1399–1411
Vogel KP, Jung HJG (2001) Genetic modification of herbaceous plants for feed and fuel. Crit Rev Plant Sci 20:15–49
Vogel KP, Pederson JF, Masterson SD, Toy JJ (1999) Evaluation of a filterbag system for NDF, ADF, and IVDMD forage analysis. Crop Sci 39:276–279
Wyrambik D, Grisebach H (1979) Enzymic synthesis of lignin precursors. Further studies on cinnamyl-alcohol dehydrogenase from soybean-cell-suspension cultures. Eur J Biochem 97:503–509
Acknowledgements
We thank Drs Ajith Anand and Stephen Temple for critical reading of the manuscript, and Dennis Walker for assistance with fiber quality analysis. This work was supported by the US Department of Energy (award number DE-FG02-06ER64303), Forage Genetics International and the Samuel Roberts Noble Foundation. This report was prepared as an account of work partly sponsored by the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of the authors expressed herein do not necessarily reflect those of the United States Government or any agency thereof.
Author information
Authors and Affiliations
Corresponding author
Additional information
Lisa A. Jackson, Gail L. Shadle: These authors contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
ESM 1
Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. (DOC 410 KB)
Rights and permissions
About this article
Cite this article
Jackson, L.A., Shadle, G.L., Zhou, R. et al. Improving Saccharification Efficiency of Alfalfa Stems Through Modification of the Terminal Stages of Monolignol Biosynthesis. Bioenerg. Res. 1, 180–192 (2008). https://doi.org/10.1007/s12155-008-9020-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-008-9020-z