Abstract
Monodehydroascorbate reductase (MDHAR), which is responsible for growth, development and stress response in plants, is a key enzyme in the maintenance of the ascorbate (AsA) pool through the AsA–glutathione (AsA–GSH) cycle and is induced by abiotic stresses. It has highly conserved regions containing FAD- and NAD(P)H-binding domains. In particular, NAD(P)H is a significant electron donor in the AsA–GSH pathway. In this context, we introduced RNA interference (RNAi) to determine the functional role of Oryza sativa L. japonica MDHAR isoform 3 (OsMDHAR3) and developed transgenic (mdhar3) rice plants in which the NAD(P)H domain was silenced. The mdhar3 rice plants were more sensitive to salt stress than the wild-type (WT) plants. In addition, the mdhar3 rice plants showed decreased ability for environmental adaptation because of an imbalance in the redox homeostasis and reduced AsA pool. These plants showed increased hydroperoxide levels and ion leakage, and decreased chlorophyll content and ascorbate/dehydroascorbate ratio under the paddy field conditions; they also exhibited a reduction in the total biomass and grain yield. Furthermore, the activity of a purified E196A mutant of the OsMDHAR protein decreased to approximately 70% of the activity of the WT protein. These results suggest that OsMDHAR3 plays a critical role in the intrinsic resistance, as well as in the sensitivity of seed maturation and productivity, of rice plants to environmental stresses, thereby indicating the functional importance of NADH in MDHAR activity, in vivo and in vitro.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice, a staple crop plant worldwide, whose complete genome has been sequenced, has been used as an important monocot model system for conducting molecular and genetic studies in crops (Xing and Zhang 2010). Currently, crop plants including rice are exposed to climate changes as a consequence of global warming. Change in global variables, rated as the most serious threat to the environment, has been at the center of debate among environmentalists and policy makers, as it has become not only an environmental, political, or economic issue but also a global problem affecting agriculture. At the plant or field scale, climate change is likely to interact with the rising CO2 concentrations and other environmental changes, including abiotic stresses, such as temperature, precipitation (associated with flooding and drought), salinity, and ultraviolet radiation, as well as biotic stresses, such as pathogen infection, to affect the physiological processes in crops, leading to reduced agricultural productivity (Zhu 2001; Wang et al. 2003; Munns 2005; Bae et al. 2013; Kim et al. 2013).
Most crop plants are affected by various environmental stresses under natural paddy fields; they induce the production of reactive oxygen species (ROS) that cause cellular damage, such as membrane instability, enzyme inactivation, and DNA oxidation (Mittler 2002). High ROS accumulation inhibits respiratory, metabolic, and photosynthetic processes, which limit plant growth and development, and cell rescue systems in the absence and presence of exogenous stimuli (Price and Hendry 1991). To overcome the problems posed by environmental stresses, plants have developed non-enzymatic and enzymatic antioxidant defense mechanisms (Asada 1996). Ascorbic acid (AsA) is a major antioxidant molecule that directly neutralizes ROS. In addition, AsA also functions as a substrate in the AsA–glutathione (AsA–GSH) cycle in plants (Halliwell 2000). The detoxification of ROS by the AsA–GSH cycle is controlled by pathways associated with their synthesis and recycling within the cells or across the organs in plants (Potters et al. 2002; Qin et al. 2011). In the antioxidant mechanism involving the AsA–GSH cycle, AsA is oxidized to monodehydroascorbate (MDHA) and MDHA is oxidized to dehydroascorbate (DHA) by spontaneous auto-oxidation. To maintain the AsA pool, the MDHA and DHA produced must be regenerated to AsA by monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR), respectively (Asada 1996). MDHAR is a well-known flavin adenine dinucleotide (FAD) enzyme, which uses different organic radicals as substrates, and its activity is widespread and upregulated by abiotic stresses, including temperature, ozone, and drought in plants. It is also known as AsA free radical reductase (Leterrier et al. 2005). Most of the MDHARs have three highly conserved domains, namely the ADP-, FAD-, and NAD(P)H-binding regions (Yoon et al. 2004). Among these domains, the relationship between NAD(P)H and MDHAR is very important for the antioxidant mechanism because NAD(P)H is the electron donor for MDHAR that catalyzes the conversion of the MDHA radical to AsA (Hossain et al. 1984). This system is critical in maintaining the AsA concentration in plant cells. However, to date, no in vivo or in vitro functional studies have been reported on MDHAR. Nonetheless, some previous studies have focused on conferring stress tolerance through the overexpression of MDHAR in transgenic tobacco (Eltayeb et al. 2007), pea (Leterrier et al. 2005), Arabidopsis (Li et al. 2010), and rice (Sultana et al. 2012) plants.
Functional genomics aims at discovering the biological function of a particular gene and deciphering how sets of genes and their products work together. Transgenic plants are powerful tools for studying various aspects of plant sciences. Functional genomics involves the following techniques: functional annotation for genes based on comparison of genomes and proteomes, gene-targeted and site-directed mutagenesis (loss of function), overexpression of a normal gene in transgenic plants (gain of function), study of gene expression using DNA-RNA hybridization, gene silencing, analysis of spatial and temporal expression of the gene under study, microarrays, and next-generation sequencing (Smith et al. 2000; Earley et al. 2006; Jain et al. 2007; Batista et al. 2008; Morozova and Marra 2008; Kim et al. 2014). One of the most popular functional genomics methods to understand the function of a specific gene is RNA-induced gene silencing (also known as RNA interference, RNAi); transgene-induced RNAi has been effective in silencing one or more genes in a wide range of plants (Hannon 2002). RNAi is a progressive, conserved, post-transcriptional gene silencing mechanism in which double-strand RNAs act as a trigger for RNA breakdown, and short-interfering RNAs lead to the degradation of a homologous target mRNA (Fire et al. 1998; Hannon 2002). The impairment of gene expression, which has been investigated in various phenotypic analyses, is a powerful functional genomics tool to determine gene function across the plant kingdom (Rassouli and Matin 2009). Since the discovery of this phenomenon, most gene silencing studies have focused on non-crop plants such as Arabidopsis.
Research on the physiological response of plants to changing levels of key factors involved in productivity and stress combat is complicated by the fact that these variables are likely to change simultaneously. We need to improve our understanding of the relationship between environmental factors and stress-responsive genes to increase crop productivity. In the present study, Oryza sativa L. japonica MDHAR3 (OsMDHAR3) cDNA was isolated from rice leaves and OsMDHAR3-silenced rice plants (mdhar3) were developed, in which the OsMDHAR3 gene under the control of the ubiquitin promoter was silenced by RNAi using the gateway cloning system. For determining the cell rescue function of OsMDHAR3 in rice plants, the antioxidant capacity and agronomic characteristics were evaluated under paddy field conditions. Stress sensitivity to abiotic stresses was tested under the same natural paddy field conditions. We observed a decrease in salt tolerance, components involved in antioxidant mechanisms (redox homeostasis and AsA pool), chlorophyll content, and grain productivity in the mdhar3 rice plants compared to the wild-type (WT) rice plants under the environmental stresses operating in the paddy fields. Our results suggest that OsMDHAR3 plays an important role in the redox homeostasis and affects the AsA pool through coactivation of the systems involved in antioxidant mechanisms and in determining the photosynthetic capacity of rice plants under natural paddy field conditions.
Materials and methods
Multiple alignment of amino acid sequences
The OsMDHAR sequence was aligned with the known MDHAR amino acid sequences using the Basic Local Alignment Search Tool (BLAST) available at the website of the National Center for Biotechnology Information (NCBI). The five MDHAR sequences that were aligned were from Oryza sativa (OsMDHAR; accession no. BAA77214), Zea mays (ZmMDHAR; accession no. AFW73040), Triticum aestivum (TaMDHAR; accession no. AFU52947), Arabidopsis thaliana (AtMDHAR; accession no. AEE34171), and Brassica rapa (BrMDHAR; accession no. AAK72107).
Generation of OsMDHAR3 knocked-down rice plants
The full-length cDNA encoding OsMDHAR3 (accession no. D85764) was synthesized from rice seedlings (O. sativa L. japonica ‘Ilmi’) by reverse transcription polymerase chain reaction (RT-PCR). For RNA interference, the sequences of the forward and reverse oligonucleotide primers were 5′-TTGCAACTGGCTCCTCAGTC-3′ and 5′-GCAGTTAACCTGAAGAATGGCA-3′, respectively. A 385-bp fragment from the coding region (458 to 842 bp) of OsMDHAR3 containing the NAD(P)-binding domain was PCR-amplified and cloned into a Gateway pENTR/SD/D-TOPO cloning vector (Invitrogen, USA), which carries two recombination sites (attL1 and attL2) for the LR Clonase reaction. Subsequently, the fragment derived from the target gene was transferred into a pANDA destination vector (Miki and Shimamoto 2004) by the recombinase reactions to form the recombinant vector carrying the MDHAR3-RNAi construct. In these reactions, the PCR-derived fragments are inserted into two regions flanked by two recombination sites (attB1 and attB2) in opposite directions, and a gus linker sequence is flanked by two inverted repeats (Miki and Shimamoto 2004). The pANDA vector was developed for Agrobacterium-mediated transformation and carried the kanamycin and hygromycin resistance markers for plant transformation. The recombinant binary pANDA vector was subsequently introduced into the Ilmi cultivar by Agrobacterium-mediated transformation to generate transgenic rice plants containing the OsMDHAR3-RNAi construct (Hiei et al. 1994). The hygromycin resistance assay of T0 transformants, inheritance analysis of T1 transgenic plants, DNA isolation, and PCR analysis were performed as reported previously (Lin et al. 2009). In brief, the inheritance of hygromycin from T0 to T1 generation of transgenic plants was examined. The seeds derived from the individual hygromycin-resistant T0 plants were soaked in water for 1 day and placed on wet filter papers for germination. After germination, a solution consisting of half-strength Murashige and Skoog (MS) Basal Medium salts and 150 mg L−1 hygromycin was poured into the plates for selection. The selection solution was added frequently to ensure that the roots of the seedlings were always immersed. Two weeks into the selection, the hygromycin-resistant transgenic plants remained green and continued to grow, whereas the untransformed plants showed inhibited growth, became necrotic, and finally died (Lin et al. 2009).
DNA extraction and genotyping
Genomic DNA was isolated from the leaves of rice plants by the alkali treatment method (Klimyuk et al. 1993). PCR was conducted using the genomic DNA and the PCR PreMix kit (Bioneer, South Korea). The transgenic rice plants were verified by PCR using the primers for ubiquitin (forward: 5′-CATCTTCATAGTTACGAGTTTAAGATGG-3′, reverse: 5′-CCAAGCAAATAAATAGGGTATGAAGGCA-3′) and the gus linker (forward: 5′-CGTTCTACTTTACTGGCTTTGGTCG-3′, reverse: 5′-CGCGATCAAAGACGCGGTGATACATATCC-3′). The PCR conditions were as follows: initial denaturation for 3 min at 94 °C, followed by 25–30 cycles at 94 °C for 30 s, 54 °C for 30 s, 72 °C for 40 s, and a final extension at 72 °C for 5 min.
Analysis of mRNA expression
Total RNA was extracted from the leaf tissue using the TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. First-strand cDNA was synthesized from DNase-treated total RNA using SuperScriptIII (Invitrogen, USA). Semi-quantitative RT-PCR was performed using RT-PCR PreMix (Bioneer, South Korea). Quantitative real-time PCR was performed on the StepOne-Plus™ Real-Time System (Applied Biosystems, USA) using SYBR Green PCR master mix (Applied Biosystems, USA) and specific primers for the NAD(P)H-binding domain of OsMDHAR3. The normalization and relative quantification were conducted using the 2-ΔΔCt method (Ferreira et al. 2006). The sequences of the oligonucleotide primers used were as follows: 5′-TTGTTGGTGTTGGTGTTGGG-3′ (forward primer) and 5′-GCAGGCTGTAAAGGCAATCA-3′ (reverse primer).
Plant growth conditions and salt tolerance assay
O. sativa L. japonica ‘Ilmi’ was used as the host for the knockdown of OsMDHAR3. Genotypes and phenotypes were screened by germinating the seeds from non-transgenic WT and mdhar3 rice at 28 °C for 3 days. Subsequently, the seedlings were transplanted to soil and grown for 4 weeks in a greenhouse at 28–32 °C under a 16 h-light/8 h-dark photoperiod. Thereafter, the plantlets were cultivated in a natural paddy field at the Kyungpook National University, located in Gunwi, South Korea, during the cultivation season (June to October). For the salt tolerance assay, 4-week-old seedlings were exposed to 200 mM NaCl and were then allowed to recover on NaCl-free irrigated soil. The plant weight was measured using about ten mdhar3 and non-transgenic WT rice plants each.
Measurement of chlorophyll content and ion leakage
Chlorophyll content was measured using the previously reported ethanol method (Lichtenthaler 1987). About 0.1 g of leaf sample was soaked in 100% ethanol and incubated at 75 °C for 20 min. For determination of the chlorophyll content, the supernatant was transferred to a new tube. Subsequently, the absorbance was measured using a spectrophotometer at 470, 648, and 664 nm. For the measurement of ion leakage, ten leaf discs were punched out from the leaves of mdhar3 and WT plants and immediately floated on a solution containing 10 μM methyl viologen (MV) in deionized H2O. The rice leaf discs were incubated in dark for 12 h at 25 °C to allow MV to diffuse into the leaves. After pre-incubation, the leaf discs were placed under continuous white light. The effect of MV on leaf discs was analyzed by monitoring the phenotypic changes. The extent of cellular membrane damage was quantified by ion leakage from 0 to 72 h by using a conductivity meter (Isteck). At the end of the specified period, the samples were autoclaved for 15 min at 121 °C. Thereafter, the conductivity of the solution was measured again, and this value was considered to be the 100% ion leakage in subsequent calculations for the relative ion leakage at different time points. The experiment to assess the visible damage caused by MV application was repeated thrice (Lim et al. 2007).
Measurement of redox states
The redox states were measured using the method described previously (Ma et al. 2012), with minor modifications. The leaf samples were homogenized in a protein extraction buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM sucrose, 100 mM KCl, 1 mM PMSF, and 10 mM EDTA-free protease inhibitor cocktail (Roche, Swiss). The homogenate was centrifuged at 13,000 rpm for 15 min at 4 °C, after which the protein concentration was determined using the protein assay reagent (Bio-Rad, USA) with bovine serum albumin as a standard. The cleared supernatant was recovered for the determination of H2O2. The intracellular H2O2 contents were determined by ferrous ion oxidation in the presence of a ferric ion indicator, xylenol orange. The crude cell extract (50 μL) was added to 950 μL of FOX reagent [100 μM xylenol orange (water-soluble form), 250 μM ammonium ferrous sulfate, 100 mM sorbitol, and 25 mM sulfuric acid]. The mixture was incubated at room temperature for 30 min and centrifuged to remove any flocculated material before measuring the absorbance at 560 nm.
Measurement of AsA/DHA ratio
The total AsA (tAsA), AsA, and DHA contents were determined by the spectrophotometric method described previously (Gillespie and Ainsworth 2007), with minor modifications. The frozen leaf samples were ground with inert sand and 10% trichloroacetic acid (TCA) solution using a mortar and pestle. The homogenate was centrifuged at 12,000 rpm for 20 min. The tAsA contents were determined in a reaction mixture consisting of crude extract and 150 mM KH2PO4 buffer (pH 7.4) containing 5 mM EDTA and 10 mM dithiothreitol (DTT) for the reduction of DHA to AsA. The reaction mixtures were incubated at room temperature for 10 min and 0.5% N-ethylmaleimide (NEM) was added. AsA was assayed in a similar manner, except that deionized H2O was substituted for DTT and NEM. The color was developed in both the reaction mixtures by the addition of 10% TCA, 44% o-phosphoric acid, α,ά-dipyridyl in 70% ethanol, and 30% FeCl3. The reaction mixtures were incubated at 37 °C for 1 h and quantified spectrophotometrically at 525 nm.
Analysis of agronomic traits and grain length
The effects of OsMDHAR3 on the agronomic traits were evaluated by conducting field trials for the mdhar3 and WT rice plants during the cultivation periods in a natural paddy field. For determining the effect on the yield parameters, the rice plants were grown from vegetative to late reproductive stages and the phenotypic and agronomic traits were evaluated for about 6 months under natural paddy field conditions. The rice plants were then harvested and threshed by hand to separate the seeds; subsequently, the unfilled and filled seeds were separated, counted, and weighed (Bae et al. 2013). The following agronomic traits were evaluated: total plant biomass fresh weight (TPW), culm weight (CW), number of tillers (NT), and total grain weight (TGW). The results obtained for the mdhar3 rice plants were compared with those of the control WT rice plants. The grain length was measured for 10 grains on a line, and the average of each 10-grain length was obtained. One hundred-grain weight was evaluated and the average was calculated.
Purification and enzymatic activity of MDHAR3
The WT and E196A mutant proteins of OsMDHAR3 were purified from Escherichia coli NiCo21 (DE3), as reported previously (Do et al. 2014; Park et al. 2016). The OsMDHAR activity was assayed spectrophotometrically (UV-1650PC; Shimadzu Corp., Japan). The assay was performed at 25 °C in a reaction mixture (1 mL) containing 50 mM potassium phosphate, pH 7.2, 0.25 mM NADH or NADPH, 2 mM AsA, 1 unit AsA oxidase (Sigma-Aldrich, USA), and the purified protein (10 μg). The absorbance was measured at 340 nm by monitoring the NAD(P)H oxidation, and the activity was calculated using an absorbance coefficient of 6.2 mM−1 cm−1. One unit is the amount of enzyme that oxidizes 1 nmoL of NADH per min at 25 °C. The enzyme activity was represented relative to that of the WT proteins, which was defined as 100%.
Statistical analysis
The values for the relative agronomic traits were calculated with respect to those obtained for the WT rice plants (which were considered as 100%). The biochemical experiments were performed at least thrice. The enzymatic assays were performed at least three times independently and the results are expressed as means ± standard deviation (SD). The representative results of phenotypic and genotypic PCR are presented.
Results
Amino acid sequence alignment of OsMDHAR3 protein
The OsMDHAR3 protein contains 435 amino acids and has a predicted molecular weight of 47 kDa. It has three conserved putative amino acid domains involved in the binding of ADP, FAD, and NAD(P)H. One of the domains binds to the ADP moiety of NAD(P) and is known as the NAD(P)-binding domain (domain II; Yoon et al. 2004). NAD(P)H serves as an electron donor and has high affinity for MDHAR. The MDHAR converts MDHA, a product of the AsA–GSH cycle, to AsA in the presence of NADH. The MDHA is also produced by AsA oxidase-mediated AsA oxidation (Do et al. 2014). The multiple amino acid sequence alignment of OsMDHAR and other plant MDHARs was performed using the BLAST tool available at the NCBI website. OsMDHAR showed 56, 55, 47, and 78% identity and 98, 98, 95, and 99% similarity to ZmMDHAR (maize), TaMDHAR (wheat), AtMDHAR (Arabidopsis), and BrMDHAR (Chinese cabbage), respectively. The proteins from all the organisms had the conserved NAD(P)H-binding region (Fig. 1). This result indicates that the conserved NAD(P)H-binding domain is probably critical for the enzymatic activation in the MDHAR-mediated AsA recycling process.
Alignment of the MDHAR protein sequence. The alignment of MDHAR from Oryza sativa and four other plant species is shown. The asterisks indicate that the residues in that column are identical, colons indicate the presence of conserved substitutions, and dots indicate the presence of semi-conserved substitutions. The region of OsMDHAR that was used in the cloning is underlined. Three domains are boxed. Domain I (gray box) binds to the ADP moiety of FAD, domain II (yellow box) binds to the ADP moiety of NAD(P)H, and domain III (black box) binds to the flavin moiety of FAD
Development of mdhar3 transgenic rice plants and salt stress response
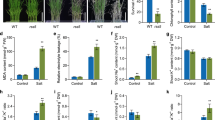
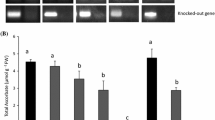
As mentioned above, we postulated the functional importance of the NAD(P)H-binding domain in the MDHAR-mediated reaction. To confirm this hypothesis, we generated OsMDHAR3-repressed (mdhar3) transgenic rice plants using the pANDA vector, which caused RNA interference in the rice plants (Fig. 2a). Approximately 20 transformants were screened in the presence of hygromycin. Four independent homozygous transgenic plants were identified in the T2 generation. Of these, only one mdhar3 transgenic plant was used for the subsequent experiments because the other three homozygous plants lost the RNAi activity because of the off-target effects or due to the inefficacy and instability during inheritance (Small 2007). The results of PCR used for the genotyping of mdhar3 transgenic rice plants are represented in Fig. 2b. On the basis of these results, the relationship between mdhar3 and salt stress was determined by exposing the 4-week-old seedlings to 200 mM NaCl for 7 days, followed by their recovery for 2 weeks under NaCl-free condition. Under the normal conditions, the mdhar3 rice plants showed weaker shoots compared to the WT plants. However, salt stress severely damaged the mdhar3 rice plants and they could not survive despite recovery under salt-free conditions (Fig. 2c). The total plant weight was almost comparable for the mdhar3 and WT rice plants, with a decrease of only 0.5 g observed under salt stress (Fig. 2d). The expression of OsMDHAR3 in the mdhar3 rice plants was lower than in the WT plants regardless of the salt stress even though a subtle difference in the expression was observed between the semi-quantitative RT-PCR (upper panel) and qRT-PCR (lower panel; Fig. 2e). Therefore, our results suggest that the RNAi-mediated gene silencing of OsMDHAR3 was effectively operating under the control of ubiquitin promoter in the mdhar3 rice plants, which lead to increased sensitivity of the transgenic plants to salt stress.
Salt stress treatment and gene expression. a Schematic diagram of RNAi-mediated OsMDHAR3 gene silencing. RB right border; NPT II kanamycin resistance gene; Ubiquitin maize ubiquitin promoter; attR1 & attR2 LR clonase recombination sites; OsMDHAR Oryza sativa monodehydroascorbate reductase; NOS NOS terminator; HPT hygromycin resistance gene; LB left border b Homozygous transgenic rice plants were identified by genotyping PCR using the DNA extracted from the leaves of T2 generation plants. c Salt stress response of mdhar3 and WT rice plants. The phenotypes of 4-week-old seedlings showed changes in the morphological characteristics under normal, stressed (200 mM NaCl, 7 days), and recovered (salt-free water, 7 days) conditions. d Fresh weights of mdhar3 and WT rice plants recovered after salt treatment. e Analysis of the mRNA levels of OsMDHAR3 using semi-quantitative RT-PCR and real-time PCR. The amplicon from the tubulin gene was used as a housekeeping control
Comparative investigation of mdhar3 and WT rice plants under natural field conditions
We identified that silencing of OsMDHAR3 increased the sensitivity of the mdhar3 transgenic rice plants to high salinity. Based on this result, we postulated that OsMDHAR3 could affect the rice productivity. To support this idea, the agronomic traits were investigated under natural paddy field conditions. The expression level of OsMDHAR3 in the mdhar3 rice plants was considerably lower than in the WT plants (Fig. 3a). The mdhar3 plants showed a dwarf phenotype compared to the WT plants (Fig. 3b). In addition, biochemical assays were conducted to determine whether the silencing of OsMDHAR3 affected the MV-mediated ion leakage, hydroperoxide content, chlorophyll content, and AsA/DHA ratio in the plants. MV was introduced because it is known as a superoxide-generating agent, which disrupts the membrane stability and leads to cell death in plants (Bowler et al. 1991). There was no difference in ion leakage between the mdhar3 and WT rice plants under the non-MV conditions; upon administration of the MV condition, ion leakage in the mdhar3 rice plants was higher than in the WT plants, and its increase was time-dependent (Fig. 3c; upper panel). The leaf color of the mdhar3 rice plants became lighter than that of the WT rice plants after 72 h of the MV treatment (Fig. 3c; lower panel). The hydroperoxide content of the mdhar3 rice plants was approximately 26% higher than that of the WT rice plants under paddy field conditions (Fig. 3d). In contrast, the chlorophyll content of the mdhar3 rice plants was about 23% less than that of the WT rice plants under the same conditions (Fig. 3e). The AsA/DHA ratio was also measured in both the mdhar3 and WT rice plants under environmental conditions. The AsA/DHA ratio in the mdhar3 rice plants was 1.15, which was approximately 1.63-fold lower than the ratio (1.88) in the WT plants (Fig. 3f). These results indicate that OsMDHAR3 silencing in the mdhar3 rice plants increased the sensitivity to ROS-induced oxidative stress by causing an imbalance in the redox homeostasis and lowering of the AsA pool and photosynthetic capacity under the paddy field conditions, thereby, affecting the plant growth and development.
Analysis of the morphological phenotype and level of different components under paddy field conditions. a mRNA expression of OsMDHAR3 in both the mdhar3 and WT rice plants under the natural field conditions. Tubulin was used as a positive control. b The phenotypic changes in the mdhar3 and WT rice plants at 60 days after transplantation under paddy field conditions. c The relative ion leakage was measured in the leaf discs obtained from the rice plants that were floated on 10 μM MV solutions. The leaf discs were incubated at 25 °C for 72 h. The percentage of ion leakage was calculated considering the value obtained after autoclaving as 100%. Hydroperoxide content (d), chlorophyll content (e), and AsA/DHA ratio (f) were measured from the leaf tissue of rice seedlings grown for 60 days under the paddy field conditions after transplantation as described in materials and methods
Analysis of the agronomic traits and grain yield
We investigated whether the RNAi-mediated down-regulation of OsMDHAR3 in the mdhar3 rice plants affected the grain yield and biomass of the plants exposed to environmental stresses under natural paddy field conditions. The total length of the mdhar3 rice plants was shorter than that of the WT plants (Fig. 4a). The following rice yield parameters of the mdahr3 rice plants decreased with respect to those of the control WT plants under the paddy field conditions: TPW, CW, NT, and TGW. Overall, the values of TPW, CW, NPT, and TGW in the mdhar3 rice plants were reduced by 80, 20, 19, and 70%, respectively, compared to the values in the WT plants (Fig. 4b). Accordingly, the mdhar3 rice plants showed a distinguishable difference for TGW between the WT and mdhar3 rice plants owing to the larger number of empty grain heads despite of the high filling rate in the mdhar3 rice plants (Fig. 4c; upper panel). The grain length was shorter and the 100-grain weight was less in the mdhar3 rice plants compared to those in the WT plants (Fig. 4c; lower panel). Overall, our findings show that OsMDHAR3 has an important role in the environmental adaptation of rice plants and on their grain yield under paddy field conditions because the total biomass and grain yield of the mdhar3 rice plants were significantly decreased compared to the WT plants.
Comparison of the agronomic traits of mdhar3 and WT rice plants under natural paddy field conditions. a Phenotype analysis at the late reproductive stage in both mdhar3 and WT rice plants under the natural paddy field conditions in 2015. b Comparison of the agronomic traits between mdhar3 and WT rice plants. Each data point represents the percentage of the mean values for the WT rice plants that were assigned a reference value of 100%. TPW total plant biomass fresh weight; CW culm weight; NT number of tillers; TGW total grain weight. c Comparison of grain size (upper panel), average of each grain length (lower left panel), and 100-grain weight (lower right panel)
In vitro enzymatic activity of the purified MDHAR3 protein
We identified that the RNAi-mediated silencing of OsMDHAR3 affected the stress adaptation and productivity of rice plants. To investigate whether the NAD(P)H-binding domain plays a key role in the enzymatic activity of OsMDHAR3, the WT and E196A mutant forms of OsMDHAR3 were purified from E. coli by immobilized metal affinity chromatography. The quality of each purified protein was confirmed by SDS-PAGE (Fig. 5a). The OsMDHAR3 activity of the E196A mutant protein was approximately 80% lower than that of WT protein (Fig. 5b), which preferred NADPH to NADH (Fig. 5c). The enzymatic activity of the OsMDHAR3 protein was not detected in the presence of NADPH but was observed in the presence of NADH. These results suggest that E196, the residue of the OsMDHAR3 protein blocked by RNAi, plays an important role in the enzymatic activity of MDHAR through the binding of NADH, but not of NADPH, in vitro; this observation is consistent with the RNAi-mediated silencing of OsMDHAR3 in vivo.
Discussion
Genetic approaches are strongly favored for identifying the functions of genes in plants because obtaining mutants of every gene using untargeted approaches, including the chemical methods or mutagenesis, requires massive efforts (Small 2007). Among the functional analysis methods, RNAi offers an easy and cost-effective approach to generate the phenocopies of genetic mutants despite the disadvantages and pitfalls in the technique. We investigated the functional role of the cytosolic OsMDHAR3 of rice under abiotic stress conditions using RNAi-mediated gene silencing. The OsMDHAR amino acid sequence showed high identity and similarity to the MDHAR sequences from several other plant species, such as maize, wheat, Arabidopsis, and Chinese cabbage. Furthermore, all the analyzed sequences had the conserved region that binds the ADP moiety of NAD(P)H (Fig. 1). The activity of OsMDHAR highly correlates with the NAD(P)H content in the plant antioxidant system; it contributes to the continuous regeneration of AsA for the detoxification of ROS by using NAD(P)H as an electron donor (Li et al. 2010; Sakihama et al. 2000). Thus, for understanding the role of OsMDHAR in oxidative stress, inactivation of the NAD(P)H-binding site is an effective strategy. Several studies have investigated the relationship between MDHAR overexpression and ROS-induced oxidative stress in transgenic plants (Eltayeb et al. 2007; Li et al. 2010; Sultana et al. 2012). However, functional analysis of OsMDHAR3 through its repression has not yet been attempted. In this study, an RNAi-based approach was used to elucidate the functional importance of OsMDHAR3 under salt stress and paddy field conditions.
The mdhar3 rice plants with the NAD(P)H-binding region of MDHAR3 blocked using RNAi-based gene silencing were generated (Fig. 2a, b) and their salt stress response, redox state, and agronomic characteristics were analyzed under environmental stresses in the natural paddy field conditions. The mdhar3 transgenic rice plants showed a lower survival rate (Fig. 2c) and plant weight (Fig. 2d) under salt stress compared to the WT rice plants. Furthermore, the OsMDHAR3 transcription in the mdhar3 rice plants was extremely low compared to that in the WT rice plants in the absence of stress (Fig. 2e). According to some previous reports, the regeneration of AsA by the antioxidant enzymes, including MDHAR and DHAR, is essential for the survival of plants under salt stress conditions (Kim et al. 2014; Zhu 2001). The overexpression of MDHAR plays a critical role in the plant growth by enhancing the AsA recycling under normal conditions (Ren et al. 2015). Interestingly, the mdhar3 rice plants showed poor growth, unlike the WT plants under normal conditions. Moreover, they were unable to survive under the salt stress conditions (Fig. 2c). Therefore, our results indicate that OsMDHAR3 has an important role in plant growth and development under normal conditions and in acquiring tolerance to salt stress because the mdhar3 rice plants showed the poor phenotype following reduced plant growth in the absence and presence of salt stress, which led to the increased sensitivity to high salinity.
Furthermore, under natural field conditions, the expression level of OsMDHAR3 in the mdhar3 rice plants was considerably lower than in the WT rice plants (Fig. 3a). In addition, the mdhar3 rice plants showed a lower growth rate (Fig. 3b) and higher ion leakage and ROS content, as well as stunted growth (Fig. 3c, d). These effects were attributed to the low chlorophyll content and AsA/DHA ratio (Fig. 3e, f). Although photosynthesis and detoxification processes require NAD(P)H as an electron donor, and the mechanism of MDHAR is similar to that of NADH-cytochrome b 5 reductase (Hossain and Asada 1985), we identified that the E196 residue of OsMDHAR3 protein plays a critical role in NADH binding, and the protein requires NADH rather than NADPH as the reducing power (Fig. 5). NAD(P)H is an essential factor for plant survival; therefore, the inactivation of the NAD(P)H-binding site results in the decrease of growth rate and ability to scavenge ROS under abiotic stress. In particular, the decline in the AsA/DHA ratio leads to the reduction of disposable AsA considerably more than that of DHA. The lower AsA contents in the leaf tissue influence the plant growth rate (plant height, root length, and leaf weight; Liu et al. 2011) and cell growth (Smirnoff and Wheeler 2000); this weakens the antioxidant system, and the plants cannot adapt well under the paddy field conditions. Therefore, our results suggest that OsMDHAR3 has an influence on the plant growth and salt stress tolerance, because the mdhar3 rice plants, with the NADH-binding region containing the E196 residue deleted, exhibited reduced salt tolerance and decreased growth, mediated by the imbalance of redox homeostasis, and reduced photosynthetic capacity and AsA pool compared to the WT plants.
The mdhar3 rice plants exhibited a dwarf phenotype, unlike the WT rice plants, under the natural field conditions (Fig. 4a). Furthermore, their agronomic traits, such as TPW, CW, NT, and TGW, were remarkably lower than those of the WT rice plants (Fig. 4b). The total length of the mdhar3 rice plants was less than that of the WT plants, whereas the number of panicles in the mdhar3 plants was similar to that of the WT plants at the vegetative growth stage (Fig. 4). In particular, there was a remarkable difference in the seed size and weight between the mdhar3 and WT plants; this is one of the significant factors that limit the rice grain productivity (Fig. 4c). The information on the importance of AsA in determining the rice yield is very limited. The overexpression of DHAR was reported to enhance the grain yield and biomass under environmental conditions (Kim et al. 2013). Also, an increase in the AsA pool was effective in expanding the tillers of the leaves of plants (Amin et al. 2008; Jafar et al. 2012), which are one of key factors to evaluate the rice grain yield (Wang and Li 2011). Moreover, the AsA-deficient rice plants exhibited a significantly reduced seed set and 1000-grain weight (Liu et al. 2011, 2013). RNAi is considered an evolutionarily-conserved mechanism for gene regulation that is critical for many aspects of growth and development. There are various pathways by which RNAi can inhibit the gene expression in plants, at both the transcriptional and post-transcriptional levels. These pathways are dependent on the specific mechanism of silencing. Recently, RNAi has emerged as one of most powerful tools for crop improvement by manipulation of the genes involved in biomass, grain yield, and enhanced shelf-life of fruits and vegetables. It has also been applied for developing resistance against biotic (pathogenic infection) and abiotic stresses (drought, salinity, and cold, etc.), and for nutritional improvements, including those of amino acids, fatty acids, antioxidants, and other nutrients beneficial for human health (Saurabh et al. 2014; Kamthan et al. 2015). In addition, the RNAi-based modifications include the reduced content of food allergens and toxic compounds, alteration in the morphology, crafting of male sterility, enhancement of secondary metabolite synthesis, and generation of seedless plant varieties (Saurabh et al. 2014). Overall, our findings suggest that OsMDHAR3 positively affects the various components (TPW and TGW) of grain yield and participates, at least functionally, in seed maturation and morphology.
In conclusion, the mdhar3 rice plants, in which the NADH-binding site of OsMDHAR3 was inactivated by RNAi, exhibited increased sensitivity to salt and environmental stresses. In addition, the yield of rice in the mdhar3 plants decreased due to the loss of the ability to eliminate ROS following the decrease in AsA content. These results indicate that OsMDHAR3, in particular, its NAD(P)H-binding region, plays an important role in the intrinsic resistance of rice plants to salt and environmental stresses. The MDHAR activity was observed to be associated with the grain productivity by detoxifying ROS through the maintenance of the AsA pool, and was also linked to seed maturation at the reproductive stage, a hitherto unknown phenomenon. Furthermore, our findings provide the basic information for OsMDHAR3-mediated transgenic studies for enhancing the crop productivity under environmental stresses.
Abbreviations
- AsA:
-
Ascorbic acid
- DHA:
-
Dehydroascorbic acid
- GSH:
-
Glutathione
- MDHAR:
-
Monodehydroascorbate reductase
- MV:
-
Methyl viologen
- ROS:
-
Reactive oxygen species
- RNAi:
-
RNA interference
References
Amin AA, Rashad EM, Gharib FAE (2008) Changes in morphological, physiological and reproductive characters of wheat plants as affected by foliar application with salicylic acid and ascorbic acid. Aust J Basic Appl Sci 2:252–261
Asada K (1996) Radical production and scavenging in the chloroplasts. In: Baker NR (ed) Photosynthesis and the environment. Kluwer, Dordrecht, pp 123–150
Bae MJ, Kim YS, Kim IS, Choe YH, Lee EJ, Kim YH, Park HM, Yoon HS (2013) Transgenic rice overexpressing the Brassica juncea gamma-glutamulcysteine synthetase gene enhances tolerance to abiotic stress and improves grain yield under paddy field conditions. Mol Breeding 31:931–945
Batista R, Saibo N, Lourenco T, Oliveira MM (2008) Microarray analyses reveal that plant mutagenesis may induce more transcriptomic changes than transgene insertion. Proc Natl Acad Sci U S A 105(9):3640–3645
Bowler C, Slooten L, Vandenbranden S, Rycke RD, Botterman J, Sybesma C, Montagu MV, Inze D (1991) Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J 10(7):1723–1732
Do H, Kim IS, Kim YS, Shin SY, Kim JJ, Mok JE, Park SI, Wi AR, Park H, Lee JH, Yoon HS, Kim HW (2014) Purification, characterization and preliminary X-ray crystallographic studies of monodehydroascorbate reductase from Oryza sativa L. japonica. Acta Crystallogr F Struct Biol Commun 70(Pt 9):1244–1248
Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45(4):616–629
Eltayeb AE, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Shibahara T, Inanaga S, Tanaka K (2007) Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225(5):1255–1264
Ferreira ID, Rosario VE, Cravo PV (2006) Real-time quantitative PCR with SYBR green I detection for estimating copy numbers of nine drug resistance candidate genes in plasmodium falciparum. Malar J 5:1
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811
Gillespie KM, Ainsworth EA (2007) Measurement of reduced, oxidized and total ascorbate content in plants. Nat Protoc 2(4):871–874
Halliwell B (2000) The antioxidant paradox. Lancet 355:1179–1180
Hannon GJ (2002) RNA interference. Nature 418(6894):244–251
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6(2):271–282
Hossain MA, Asada K (1985) Monodehydroascorbate reductase from cucumber is a flavin adenine dinucleotide enzyme. J Biol Chem 260(24):12920–12926
Hossain MA, Nakano Y, Asada K (1984) Monodehydroascorbate reductase in spinach chloroplasts and its participation in regeneration of ascorbate for scavenging hydrogen peroxide. Plant Cell Physiol 25:385–395
Jafar M, Farooq M, Cheema M, Afzal I, Bastra S, Wahid M, Aziz T, Shchid M (2012) Improving the performance of wheat by seed priming under saline conditions. J Agron Crop Sci 198:38–45
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143(4):1467–1483
Kamthan A, Chaudhuri A, Kamthan M, Datta A (2015) Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci 6:208
Kim YS, Kim IS, Bae MJ, Choe YH, Kim YH, Park HM, Kang HG, Yoon HS (2013) Homologous expression of cytosolic dehydroascorbate reductase increases grain yield and biomass under paddy field conditions in transgenic rice (Oryza sativa L. japonica). Planta 237:1613–1625
Kim YS, Kim IS, Shin S, Park TH, Park H, Kim YH, Lee GS, Kang HG, Yoon HS (2014) Overexpression of dehydroascorbate reductase confers enhanced tolerance to salt stress in rice plants (Oryza sativa L. japonica). J Agron Crop Sci 152:941–953
Klimyuk VI, Carroll BJ, Thomas CM, Jones JD (1993) Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J 3(3):493–494
Leterrier M, Corpas FJ, Barroso JB, Sandalio LM, del Rio LA (2005) Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol 138(4):2111–2123
Li F, Wu QY, Sun YL, Wang LY, Yang XH, Meng QW (2010) Overexpression of chloroplastic monodehydroascorbate reductase enhanced tolerance to temperature and methyl viologen-mediated oxidative stresses. Physiol Plant 139(4):421–434
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148:350–382
Lim S, Kim YH, Kim SH, Kwon S, Lee H, Kim JS, Cho KY, Peak KY, Kwak SS (2007) Enhanced tolerance of transgenic sweetpotato plants that express both CuZuSOD and APX in chloroplasts to methyl viologen-mediated oxidative stress and chilling. Mol Breeding 19:227–239
Lin J, Zhou B, Yang Y, Mei J, Zhao X, Guo X, Huang X, Tang D, Liu X (2009) Piercing and vacuum infiltration of the mature embryo: a simplified method for Agrobacterium-mediated transformation of indica rice. Plant Cell Rep 28:1065–1074
Liu Y, Yu L, Wang R (2011) Level of ascorbic acid in transgenic rice for L-galactono-1,4-lactone dehydrogenase overexpressing or suppressed is associated with plant growth and seed set. Acta Physiol Plant 33:1353–1363
Liu Y, Yu L, Tong J, Ding J, Wang R, Lu Y, Xiao L (2013) Tiller number is altered in the ascorbic acid-deficient rice suppressed for L-galactono-1,4-lactone dehydrogenase. J Plant Physiol 170(4):389–396
Ma B, Gao L, Zhang H, Cui J, Shen Z (2012) Aluminum-induced oxidative stress and changes in antioxidant defenses in the roots of rice varieties differing in al tolerance. Plant Cell Rep 31(4):687–696
Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45(4):490–495
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7(9):405–410
Morozova O, Marra MA (2008) Applications of next-generation sequencing technologies in functional genomics. Genomics 92(5):255–264
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167(3):645–663
Park AK, Kim IS, Do H, Jeon BW, Lee CW, Roh SJ, Shin SC, Park H, Kim YS, Kim YH, Yoon HS, Lee JH, Kim HW (2016) Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica. Sci Rep 6:33903
Potters G, Gara LD, Asard H, Horemans N (2002) Ascorbate and glutathione: guardians of the cell cycle, partners in crime? Plant Physiol Biochem 40:538–548
Price AH, Hendry GAF (1991) Iron-catalysed oxygen radical formation and its possible contribution to drought damage in nine native grasses and three cereals. Plant Cell Environ 14:477–484
Qin A, Shi Q, Yu X (2011) Ascorbic acid contents in transgenic potato plants overexpressing two dehydroascorbate reductase genes. Mol Biol Rep 38(3):1557–1566
Rassouli FB, Matin MM (2009) Gene silencing in human embryonic stem cells by RNA interference. Biochem Biophys Res Commun 390(4):1106–1110
Ren J, Duan W, Chen Z, Zhang S, Song X, Liu T, Hou X, Li Y (2015) Overexpression of the monodehydroascorbate reductase gene from non-heading chinese cabbage reduces ascorbate level and growth in transgenic tobacco. Plant Mol Biol Report 33(4):881–892
Sakihama Y, Mano J, Sano S, Asada K, Yamasaki H (2000) Reduction of phenoxyl radicals mediated by monodehydroascorbate reductase. Biochem Biophys Res Commun 279(3):949–954
Saurabh S, Vidyarthi AS, Prasad D (2014) RNA interference: concept to reality in crop improvement. Planta 239:543–564
Small I (2007) RNAi for revealing and engineering plant gene functions. Curr Opin Biotechnol 18:148–153
Smirnoff N, Wheeler GL (2000) Ascorbic acid in plants: biosynthesis and function. Crit Rev Biochem Mol Biol 35(4):291–314
Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407(6802):319–320
Sultana S, Khew CY, Morshed MM, Namasivayam P, Napis S, Ho CL (2012) Overexpression of monodehydroascorbate reductase from a mangrove plant (AeMDHAR) confers salt tolerance on rice. J Plant Physiol 169(3):311–318
Wang Y, Li J (2011) Branching in rice. Curr Opin Plant Biol 14(1):94–99
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218(1):1–14
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Yoon HS, Lee H, Lee IA, Kim KY, Jo J (2004) Molecular cloning of the monodehydroascorbate reductase gene from Brassica campestris and analysis of its mRNA level in response to oxidative stress. Biochim Biophys Acta 1658(3):181–186
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6(2):66–71
Acknowledgements
This work was supported by a grant from the Next-Generation BioGreen 21 Program (no. PJ011142012016), Rural Development Administration, Republic of Korea and this research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1A6A1A05011910), Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kim, JJ., Kim, YS., Park, SI. et al. Cytosolic monodehydroascorbate reductase gene affects stress adaptation and grain yield under paddy field conditions in Oryza sativa L. japonica . Mol Breeding 37, 118 (2017). https://doi.org/10.1007/s11032-017-0720-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0720-y