Abstract
Dehydroascorbate reductase (DHAR, EC 1.8.5.1) maintains redox pools of ascorbate (AsA) by recycling oxidized AsA to reduced AsA. To investigate whether DHAR affects rice yield under normal environmental conditions, cDNA-encoding DHAR (OsDHAR1) was isolated from rice and used to develop OsDHAR1-overexpressing transgenic rice plants, under the regulation of a maize ubiquitin promoter. Incorporation and expression of the transgene in transgenic rice plants was confirmed by genomic polymerase chain reaction (PCR), semi-quantitative reverse transcription PCR (RT-PCR), western blot, and enzyme activity. The expression levels were at least twofold higher in transgenic (TG) rice plants than in control wild-type (WT) rice plants. In addition, OsDHAR1-overexpression in seven-independent homologous transgenic plants, as compared to WT plants, increased photosynthetic capacity and antioxidant enzyme activities under paddy field conditions, which led to an improved AsA pool and redox homeostasis. Furthermore, OsDHAR1 overexpression significantly improved grain yield and biomass due to the increase of culm and root weights and to enhance panicle and spikelet numbers in the same seven independent TG rice plants during the farming season (2010 and 2011) in South Korea. The OsDHAR protein contained the redox-active site (Cys20), as well as the conserved GSH-binding region, GSH-binding motif, glutathione-S-transferase (GST) N-terminal domain, C-terminal domain interface, and GST C-terminal domain. Therefore, our results indicate that OsDHAR1 overexpression, capable of functioning in AsA recycling, and protein folding increases environmental adaptation to paddy field conditions by the improving AsA pool and redox homeostasis, which enhances rice grain yield and biomass.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is a staple crop grown worldwide that satisfies the necessary daily caloric requirements of millions of people (Khush 1997). In agricultural systems, rice yield can be greatly decreased by land erosion and degradation, as well as in response to various environmental factors, including high salinity, nutrient depletion and excess, drought, flooding, extreme temperatures, UV radiation, and photochemical cycling (Gill and Tuteja 2010). These adverse environmental conditions limit crop production through reactive oxygen species (ROS)-induced oxidative stress (Boyer 1982; Owens 2001).
Plants have evolved a variety of cell rescue systems to adapt to natural environmental conditions by neutralizing toxic ROS. Ascorbate (AsA; vitamin C) is an abundant metabolite in plants, where it plays important roles in various aspects of their lifecycle. Specifically, AsA participates in photo protection as well as the stress response, regulates growth development such as cell division and cell expansion (Garcia et al. 2009; Kerk and Feldman 1995), and serves as a signal transduction molecule (Noctor et al. 2000). AsA pools in plants are controlled precisely by the synthesis pathway and recycling system within cells or between organs (Qin et al. 2011). In particular, the AsA regeneration system plays a particularly important role in cellular responses and adaptation to adverse conditions. As an antioxidant (Yin et al. 2010), AsA is oxidized into an unstable short-lived monodehydroascorbate (MDHA) radical in response to the production of excess ROS in unfavorable conditions, after which it is reduced to AsA by MDHA reductase (MDHAR). The remainder disproportionates non-enzymatically to AsA and dehydroascorbate (DHA) (Noctor and Foyer 1998). DHA must be converted to AsA by DHA reductase (DHAR) in the presence of glutathione (GSH) as a reducing agent because it undergoes irreversible spontaneous hydrolysis to 2,3-diketogulonic acid (Deutsch 2000). Thus, DHAR is a key factor in maintaining a reduced pool of AsA and is of paramount importance in the adaptation to environmental conditions, especially to paddy field conditions (Eltayeb et al. 2007).
In the past few years, global genes with recycling function have attracted a great deal of attention from plant physiologists and agronomists. Among these, the DHAR gene is of scientific and industrial interest owing to its role in AsA homeostasis. Plant DHAR cDNAs have been obtained from wheat (Chen et al. 2003), tomato (Zoua et al. 2006), spinach (Sakihama et al. 2000), rice (Urano et al. 2000), and Arabidopsis (Yoshida et al. 2006). Furthermore, there is growing evidence that DHAR overexpression enhances tolerance to environmental conditions. For example, the overexpression of wheat DHAR conferred protection against ozone in tobacco (Chen and Gallie 2005), while the expression of the human DHAR gene in tobacco increased tolerance to low temperature and salt stress (Kwon et al. 2003). In addition, the overexpression of Arabidopsis cytosolic DHAR increased tolerance to drought and ozone stress in tobacco (Eltayeb et al. 2006). As mentioned above, these physiological investigations provided knowledge and a better understanding of the effects of abiotic stress on plants. However, these results provide only limited information on how environmental conditions affect plant growth, and eventually agricultural yield in the paddy field where the conditions are complicated by such factors as the non-availability of time inputs, inappropriate growing seasons, pest outbreaks, and abiotic stresses (Lee et al. 2007; Tang et al. 2008). The extent of crop yield loss due to abiotic stresses can be reduced by manipulating plant metabolism, perhaps through the use of genetically engineered plants.
Rice has two DHAR isoform genes, DHAR1 and DHAR2, which are responsible for AsA homeostasis by regenerating DHA to AsA in the cytosol and chloroplasts; however, it is likely that DHAR1 is a cytoplasmic isozyme because it has no transit peptide for its import into plastids. Recently, we demonstrated that the overexpression of Oryza sativa DHAR1 (OsDHAR1) increases acquired tolerance to ROS-induced oxidative stress in transgenic (TG) Escherichia coli (Shin et al. 2008), suggesting that DHAR has an important role in cellular protection against abiotic stresses. To support this evidence, it should be possible to elevate AsA levels artificially through the manipulation of DHAR and thus assess directly the in vivo function of AsA. To date, DHAR studies in higher plants have concentrated mainly on Arabidopsis, tobacco, and agricultural crops such as spinach and rice under environmental stresses (Urano et al. 2000). In contrast, there is virtually no information on the molecular characteristics of DHAR on agronomic traits under paddy field conditions. On the basis of these facts, we cloned OsDHAR1 cDNA encoding cytosolic DHAR from rice leaf and developed transgenic rice plants expressing OsDHAR1 under the control of the constitutive maize ubiquitin promoter. Our results show that OsDHAR1-expressing transgenic rice plants exhibit better growth development, phenotypes, and rice yield, including grain yield and biomass. These are achieved via improved redox homeostasis and AsA pool through the enhancement of photosynthetic capacity and antioxidant enzyme activity when compared to segregative control wild-type (WT) rice plants under natural paddy field conditions.
Materials and methods
Vector construction and rice transformation
Full-length cDNA (accession no. AY074786) encoding cytosolic dehydroascorbate reductase (DHAR) was synthesized from rice seedlings (Oryza sativa L. japonica cv. Ilmi) by reverse transcription polymerase chain reaction (RT-PCR) using the DHAR-CF and DHAR-CR primer set (Supplementary Table 1). The cDNA was then cloned between the maize ubiquitin promoter and nos terminator of pGA1611 (Kim et al. 2003) using HindIII and KpnI restriction endonucleases. Next, the approximately 3.6 kb blunted SacII fragment, including the ubiquitin promoter::OsDHAR1::Tnos terminator, was cloned into the SmaI site of pCAMBIA3300 and designated as pOsDHAR1. The nucleotide sequence of the promoter and OsDHAR1 were subsequently determined to ensure that the open reading frame was combined without any frame shifts or nucleotide conversions. The pOsDHAR1 binary vector was introduced into Agrobacterium strain LBA 4404, which was used to transform rice calli induced from the scutella of mature seeds of the Ilmi variety according to a method described previously (Hiei et al. 1994; Kang et al. 1998).
Plant materials and growth conditions
Oyrza sativa L. japonica cv. Ilmi was used as a host for OsDHAR1 overexpression. To examine genotypes and phenotypes, rice seeds of seven-independent homologous TG plants and segregative control WT plants were germinated at 28–32 °C in a growth chamber, transplanted into soil pots that were 4.5 or 12.5 cm in diameter, and maintained for 30 days at 28–32 °C under a 16 h light/8 h dark cycle. Next, they were transplanted into a normal paddy field located on the campus (Gunwi) of Kyungpook National University in 2010 (T2) and 2011 (T3), and then cultivated during the cultivation period (from June to October) in Korea. TG and WT plants consisted of 16 lines per plant. After planting, all leaves of rice seedlings grown for 60 days under paddy field conditions were sampled and used for the subsequent experiments.
Genomic DNA isolation and PCR
Genomic DNA was isolated from leaves using the alkali treatment method (Klimyuk et al. 1993). Integration of the Ubi::OsDHAR1 transgene into the TG rice genome was verified by PCR using the Ubi-FC and OsDHAR-RC primer set (Supplementary Table 1). PCR was carried out using the PCR PreMix kit (Bioneer, Daejeon, Korea) according to the manufacturer’s instructions. PCR conditions were as follows: initial denaturation for 3 min at 94 °C, followed by 30–32 cycles at 94 °C for 30 s, 54 °C for 30 s, 72 °C for 1.5 min, and a final extension at 72 °C for 5 min.
Total RNA isolation and semi-quantitative RT-PCR
Total RNA was isolated from leaf tissues using RNeasy Plant Mini kit (Qiagen, Frankfurt, Germany). RT-PCR was performed with a One-Step RT-PCR PreMix kit (Intron, Seoul, Korea) using each primer set (OsDHAR-F and OsDHAR-R; Supplementary Table 1) according to the manufacturer’s instructions. Each PCR reaction contained 50 ng of total RNA and 1 μg of primers in a total reaction volume of 20 μL. PCR was conducted by subjecting the samples to 22–25 cycles at 94 °C for 30 s, 54 °C for 30 s, 72 °C for 40 s, and a final extension at 72 °C for 5 min. Each PCR amplicon was resolved on a 1.2 % agarose gel. The PCR product of the tubulin (Tub) gene, obtained using the Tub-F and Tub-R primer set, was used as a housekeeping control.
Crude protein extraction, SDS-PAGE, and western blot analysis
Rice leaves (0.2 g) were ground in liquid nitrogen and homogenized for 20 min at 4 °C in an extraction buffer containing 0.5 M Tris–HCl (pH 7.5), 10 mM EDTA, 0.7 M sucrose, 0.1 M KCl, 1 mM PMSF, and protease inhibitor cocktail. The homogenate was then clarified by centrifugation at 15,000 rpm for 20 min at 4 °C, after which the protein concentration was determined using protein assay reagent (Bio-Rad, Hercules, CA) with bovine serum albumin as a standard. Crude protein (20 μg) was separated by 12 % SDS-PAGE and transferred electrophoretically to a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membranes were then blocked for 1.5 h at room temperature with a blocking solution [5 % non-fat skim milk in Tris/HCl-buffered saline containing 0.05 % Tween 20 (TBST) and 0.02 % sodium azide] and incubated overnight at 4 °C with each primary antibody diluted in the blocking solution. A polyclonal antibody against OsDHAR1 was raised in rabbits after DHAR protein purification in E. coli BL21. The blots were washed four times for 40 min with TBST, and then incubated for 1.5 h at room temperature with a secondary antibody. After washing four times every 10 min with TBST, the immunoreactive proteins were visualized using ECL western blotting detection reagent (GE Healthcare, Piscataway, NJ).
Measurement of ascorbate content
The contents of total ascorbate (tAsA), reduced AsA (AsA), and dehydroascorbate (DHA) were determined by spectrophotometry according to a method reported previously (Gillespie and Ainsworth 2007). Briefly, approximately 0.2 g of frozen leaf sample was ground with inert sand and 2 mL of 5 % (v/v) m-phosphoric acid by using a mortar and pestle. The homogenate was then centrifuged at 12,000 rpm for 20 min, after which the total AsA content was determined in a reaction mixture consisting of 100 μL of crude extract, 500 μL of 150 mM KH2PO4 buffer (pH 7.4) containing 5 mM EDTA, and 100 μL of 10 mM dithiothreitol (DTT) for reduction of DHA to AsA. The reaction mixtures were incubated at room temperature for 10 min and 100 μL of 0.5 % (w/v) N-ethylmaleimide (NEM) was then added to remove excess DTT. AsA was assayed in a similar manner, except that 200 μL of deionized H2O was substituted for DTT and NEM. Color developed in both series of reaction mixtures upon the addition of 400 mL of 10 % (w/v) trichloroacetic acid, 400 μL of 44 % (v/v) o-phosphoric acid, 400 μL of α,α′-dipyridyl in 70 % (v/v) ethanol, and 200 μL of 30 % FeCl3. The reaction mixtures were incubated at 40 °C for 1 h and quantified spectrophotometrically at 525 nm. Finally, the concentration of DHA was estimated on the basis of the difference in the concentrations of total AsA and reduced AsA.
Enzyme activity of DHAR, monodehydroascorbate reductase, ascorbate peroxidase, and glutathione reductase
Rice-seedling leaves were used to assess the enzyme activity of ascorbate peroxidase (APX), MDHAR, DHAR, and glutathione reductase (GR). For DHAR and MDHAR activity, crude protein extracts were prepared in a lysis buffer containing 50 mM sodium phosphate (pH 7.5), 3 mM MgCl2, 1 mM EDTA, 1 mM PMSF, and protease inhibitor cocktails. For GR and APX activity, crude protein extracts were prepared in a lysis buffer containing 50 mM sodium phosphate (pH 7.0), 7 mM 2-mercaptoethanol, 1 mM PMSF, and protease inhibitor cocktail. In particular, crude extract protein for APX activity was prepared in the presence of 2 mM AsA under the same buffer (Kausar et al. 2012). DHAR activity was measured at room temperature in a 1 mL reaction mixture containing 50 mM potassium phosphate (pH 7.0), 0.1 mM EDTA, 0.5 mM DHA, 2.5 mM GSH, and crude extract. The absorbance was measured at 265 nm and the activity was calculated using an absorbance coefficient of 14.6 mM−1 cm−1 (Yoshida et al. 2006). One unit of DHAR activity was defined as the amount of enzyme that produces 1 nmol AsA/min at 25 °C. APX activity was assayed using ascorbic acid as a substrate (Kausar et al. 2012). The oxidation of AsA was initiated by using H2O2; the decrease in absorbance was monitored at 290 nm and activity was calculated using an extinction coefficient of 2.8 mM−1 cm−1. One unit of APX was defined as the amount of enzyme oxidizing 1 nmol AsA/min. GR activity was performed at room temperature in a 1 mL reaction mixture containing 50 mM phosphate buffer (pH 7.6), 2.5 mM EDTA, 2.5 mM GSSG, 1.0 mM NADH, and crude protein. The absorbance was monitored at 340 nm. The activity was calculated using an absorbance coefficient of 6.2 mM−1 cm−1 (Foyer and Halliwell 1976). The MDHAR activity assay was performed at room temperature with a 1 mL reaction mixture containing 50 mM sodium phosphate (pH 7.2), 0.2 mM NADH, 2 mM AsA, 1 U AsA oxidase, and crude extract. The absorbance was measured at 340 nm. The activity was calculated using an absorbance coefficient of 6.2 mM−1 cm−1. One unit was defined as the amount of enzyme that oxidizes 1 nmol NADH/min at 25 °C (Eltayeb et al. 2007; Kausar et al. 2012). The specific enzyme activity was expressed as U/mg protein.
Redox state analysis
Leaf samples (0.2 g) were homogenized in a 50 mM sodium phosphate buffer (pH 7.2) containing 1 mM hydroxylamine as a catalase inhibitor, 5 % glycerol, 1 mM PMSF, and EDTA-free protease inhibitor cocktail. The homogenate was centrifuged at 13,000 rpm for 20 min at 4 °C. The cleared supernatant was recovered for the determination of H2O2. The intracellular hydroperoxide level was determined by ferrous ion oxidation in the presence of the ferric ion indicator, xylenol orange. Fifty microliters of crude extract were added to 950 μL FOX reagent [100 μM xylenol orange (water-soluble form), 250 μM ammonium ferrous sulfate, 100 mM sorbitol, and 25 mM sulfuric acid]. The mixture was incubated at room temperature for 30 min and then centrifuged to remove any flocculated material before measuring the absorbance at 560 nm (Gay et al. 1999).
Chlorophyll-fluorescence measurement
To estimate photosynthetic capacity in the plant leaves, chlorophyll-fluorescence was measured on the basis of the photochemical yield (Fv/Fm), which represents the maximum quantum yield of photosystem II, using a chlorophyll-fluorescence meter (Handy PEA, Hansatech, Kings Lynn, UK) after 30 min of dark adaptation. Measurements were conducted at room temperature after applying to the third leaves of the rice plants. Ten leaf flats (1 cm in diameter) from fully expended leaves of different rice plants grown under paddy field conditions were incubated for different times at 25 °C under continuous light intensity of 150 μM photons m−2 s−1. The Fv/Fm values were measured at the indicated time (Oh et al. 2005).
Amino acid sequence alignment and molecular building
Oryza sativa DHAR (OsDHAR) was aligned with known DHAR sequences using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) software (http://clustalw.ddbj.nig.ac.jp/). The amino acid sequences were as follows: O. DHAR (OsDHAR; accession no. NP_00154470.1), Homo sapiens DHAR (HsDHAR; accession no. AAD26137.1), A. thaliana DHAR (AtDHAR; accession no. NP_177662.1), Glycine max DHAR (GmDHAR; accession no. NP_285857.1), Zea mays DHAR (ZmDHAR; accession no. NP_001151414.1), and Cucumis sativus DHAR (CsDHAR; accession no. ABO64438). To predict the OsDHAR three-dimensional (3D) structure based on this homology, we used bioinformatics tools, including the Protein Data Bank (PDB; http://www.rcsb.org/pdb) and Chimera software (http://www.cgl.ucsf.edu/chimera/index.html) from the University of California, San Francisco (UCSF). Homology modeling was performed using the target sequence and the 3D structure of a human DHAR isoform (accession no. AAD26137.1). The FASTA sequence of the query (accession no. NP_001054470.1) and PDB template (1KOO_A) were uploaded to the ESyPred3D Web Server 1.0 (http://www.fundp.ac.be/sciences/biologie/urbm/bioinfo/esypred/) to construct an OsDHAR PDB file. The PDB files for the query and homologous target sequence were further utilized for 3D model building. Visualization of 3D protein structures was conducted using UCSF Chimera, which enabled us to assess the positions of different amino acids present in the active sites of the proteins.
Estimation of agronomic characteristics of rice plants grown under paddy field conditions
To examine the rice yield components of transgenic rice plants under natural paddy field conditions, seven-independent (T2) and (T3) homozygous TG plants and their respective segregative WT rice plants were transplanted into a normal paddy field located on the campus (Gunwi) of Kyungpook National University in 2010 (T2) and 2011 (T3). The seedlings were transplanted at 30 days after sowing. A completely randomized block design was employed with two replicates. Each plot consisted of 16 rice plants with 0.2 m between plants and 0.3 m between plots. When the rice plants grown under natural conditions reached maturity and the grains ripened, they were harvested and the seeds were separated from the vegetative parts by hand. The unfilled and filled grains were then taken apart, independently counted, and weighed. Yield parameters were scored for 12 rice plants per plot because the 2 rice plants with maximum and minimum scores per plant were excluded. The following agronomic traits were scored: total plant biomass fresh weight (TPW; g), culm fresh weight (CW; g), root fresh weight (RW; g), panicle number per hill (NP), spikelet number per panicle (NSP), grain filling rate (FR; %), total grain weight (TGW; g), and 1,000 grain weight (TOGW; g).
Statistical analysis
All experiments were carried out at least in triplicate and the results are expressed as the mean and standard deviation (SD). Relative data are presented relative to the segregative WT plants, which were defined as 100 %. The results for the phenotypes are the mean representative of seven independent TG rice plants under identical conditions.
Results and discussion
Development of transgenic rice plants expressing OsDHAR1
To assess transgenic (TG) rice plants expressing OsDHAR1 encoding a cytosolic DHAR, 19-independent TG events produced at the (T0) generation. Thereafter, the maize ubiquitin promoter and the nos terminator were harbored within each TG plant (Fig. 1a), because the primary (T0 generation) TG rice lines were screened in the presence of DL-phosphinothricin encoded by the bar gene. Seeds of the next generation (T1 generation) were separately harvested from each independent line, after which 16 plants per line were grown in a paddy field during farming season and their progeny (T2 seeds) were harvested. Among the (T2) TG seedlings, 10 independent homozygous transgenic lines were selected, and then 7 independent homozygous TG rice lines harboring OsDHAR1 (OX-1 to OX-7) were used for subsequent experiments. A single band was detected in 16 TG rice plants (1–16) per independent TG lines grown under paddy field conditions (2011) (Fig. 1b), whereas no bands were detected in segregative wild-type (WT) rice plants (Fig. 1c). To examine whether OsDHAR1 was constitutively expressed under the regulation of the ubiquitin promoter at transcriptional and translational levels, semi-quantitative RT-PCR and immunoblotting analyses were performed. OsDHAR1 expression levels were higher in seven independent TG rice plants than in WT rice plants, even though a signal in WT rice plants was also detected (Fig. 1d, e). OsDHAR1 incorporated into the genome of seven-independent homologous TG rice lines is constitutively expressed under the control of the ubiquitin promoter, and that this ubiquitin promoter, generally available and active in all or most cell types of monocotyledonous plants (McElroy et al. 1990; McElroya and Brettellb 1994), is useful in molecular breeding of crop plants such as rice.
Development of Ubi:OsDHAR1 TG rice plants. a A schematic diagram of the construct bearing OsDHAR1. Ubip, maize ubiquitin promoter; OsDHAR1, O. sativa cytosolic dehydroascorbate reductase gene; Tnos, Agrobacterium tumefaciens nopaline synthase gene. b (T3) generation (2011) phenotype analysis of homozygous TG rice (OX-1 to OX-7) and segregative WT rice plants at 60 days after rice planting under paddy field conditions. c PCR genotyping using genomic DNA. OsDHAR1 integration was performed by PCR. Numbers (1–16) represent independent TG plants per line. C, segregative WT rice plants. d Analysis using semi-quantitative RT-PCR. Semi-quantitative RT-PCR was carried out with leaf tissue of seven-independent TG lines (OX-1 to OX-7) and control WT rice plants. The PCR amplicon of the tubulin (Tub) gene was used as a housekeeping control. e Expression analysis of OsDHAR1 at the translational level using western blot. Crude protein extracts were isolated from leaf tissue of seedlings at 60 days after transplantation, separated on 12 % SDS-PAGE, and then visualized through the immunoblotting process. The tubulin (Tub) protein was used as a loading control
Comparative analysis of the photosynthetic ability and antioxidant enzyme activity of OsDHAR1-expressing TG plants under paddy field conditions
To elucidate the mechanism by which OsDHAR1 enables the adaptation of TG rice plants to environmental conditions, AsA and redox homeostasis, photosynthetic capacity, and antioxidant enzyme activity were investigated. Rice leaf tissues were sampled from seedlings grown for 60 days in the paddy fields (2011; Fig. 1b). First, chlorophyll-fluorescence was evaluated because the photochemical yield (Fv/Fm) of photosystem II is the most commonly used approach to determine the effects of stress or adaptation response to environmental conditions (Redillas et al. 2012). Leaf flats of TG and WT rice plants were treated with deionized distilled water and Fv/Fm values were measured at 12 h intervals for 72 h. The initial Fv/Fm values were approximately 0.8. Over time, the Fv/Fm values of TG plants were higher than those of WT plants, even though the values decreased over time in both WT and TG (OX-1 to OX-7) rice plants. The value of OX-1 rice plants was the lowest of TG rice plants (Fig. 2a). DHAR with AsA recycling activity affected the level of photosynthetic activity during leaf development and consequently influenced the rate of plant growth and leaf aging (Wang et al. 2010).
Photosynthetic capacity, antioxidant enzyme activity, and AsA and redox homeostasis. a Changes in the Fv/Fm value in TG (OX-1 to OX-7) and WT rice plants under paddy field conditions. Leaf flats were pre-incubated in darkness overnight, after which Fv/Fm was measured at 25 °C and at a light intensity of 150 μmol photons m−2 s−1. b Enzyme activity of DHAR, MDHAR, APX, and GR. Enzyme activity was measured using leaf tissue (0.2 g) from seedlings grown under paddy field conditions, and is represented as relative enzyme activity to the 100 % of enzyme activity in WT rice plants. The 100 % activity of each enzyme is described as μmol substrate change/min/g fresh weight. c Analysis of the AsA pool and AsA/DHA ratio. tAsA, total ascorbate contents; AsA, reduced AsA; DHA, oxidized AsA. d Redox state analysis. Hydroperoxide levels were measured from leaf tissue of seedlings grown for 60 days under paddy field conditions after rice planting. The relative levels were calculated to be 100 % of those in the WT rice plants. Data are expressed as the mean ± SD of at least three replications from three independent experiments. Bars labeled with asterisks show significant differences (P < 0.05) between TG and WT rice plants as determined by analysis of the variance
Antioxidant enzyme activity involved in the AsA-GSH cycle, as a major cell rescue system, was also measured under natural paddy field conditions. The enzyme activity of DHAR, MDHAR, APX, and GR in OsDHAR1-expressing TG rice plants (OX-1 to OX-7) increased by 2.0-, 1.4-, 1.6-, and 1.5-fold, respectively, when compared to WT rice plants, even though there was a difference among 16 TG plants in each independent rice line (Fig. 2b). In particular, DHAR activity increased significantly in TG plants as a result of epistatic OsDHAR1 expression, and the activity levels were similar to the expression levels of the transcripts and proteins. Many studies have shown that the overexpression of antioxidant genes, including those encoding DHAR, MDHAR, APX (Lee et al. 2007; Yin et al. 2010), GR, superoxide dismutase, catalase, and glutathione peroxidase (Pasqualini et al. 2001; Ahmad et al. 2010), enhances tolerance to abiotic stresses by co-inducing the expression of a wide range of antioxidant enzymes in TG Arabidopsis (Ushimaru et al. 2006; Yoshida et al. 2006), tobacco (Kwon et al. 2003; Eltayeb et al. 2006), potato (Eltayeb et al. 2010; Qin et al. 2011), and tomato (LI et al. 2012). Thus, our results indicate that OsDHAR1 expression in TG rice plants improves their photosynthetic capacity and total antioxidant ability through the co-activation of cell rescue systems under paddy field conditions.
AsA pool and redox homeostasis in OsDHAR1-expressing TG rice plants
AsA levels were examined to check whether the elevated activity of DHAR and MDHAR in OsDHAR-expressing TG rice plants affects AsA regeneration under environmental conditions. The total AsA (tAsA) and AsA contents of TG rice plants (OX-1 to OX-7) were approximately 1.11- and 1.32-fold higher than those of WT rice plants, respectively, while the DHA levels of TG rice plants were approximately 1.18-fold lower than those of WT rice plants. The increased AsA and decreased DHA levels of TG rice plants led to an approximately 1.58-fold increase in the AsA to DHA ratio as compared to WT rice plants (Fig. 2c). On the basis of the moderate increase of tAsA in TG rice plants, it is possible that tAsA accumulation can result from the AsA synthesis system. Nevertheless, the increased AsA levels in TG rice plants seemed to be due to AsA recycling through OsDHAR1 expression rather than AsA synthesis, since the plunge rate of tAsA was weak in TG rice plants as compared to that of AsA. The tAsA levels of OX-4 and OX-7 plants increased mildly (4.8 and 3.5 %, respectively) when compared to those of WT rice plants, whereas the AsA content of the same plants increased markedly (13.8 and 13.3 %, respectively; Fig. 2c). This result emphasizes the importance of AsA regeneration by OsDHAR1; if AsA is not recycled by DHAR, DHA is rapidly hydrolyzed into 2,3-diketogulonic acid and finally degraded into CO2, H2O2, and l-threarate (Horemansa et al. 2000; Qin et al. 2011). ROS produced by environmental stresses depletes the available AsA to unstable DHA, after which the produced H2O2 produces toxic-free radicals such as the hydroxyl radical, which cause damage to cellular components, e.g., proteins (Qin et al. 2011), lipids, and nucleic acids (Wang et al. 2010). The over-expression of OsDHAR1 in TG rice plants would increase the likelihood that DHA was converted to AsA before decaying, which would lead to an increase of AsA in TG rice plants. As stated above, the enhanced reduction of DHA from its degradation pathway might be the reason that the AsA concentration in OsDHAR1-expressing TG rice plants increased. The increase in AsA was accompanied by an increase in DHAR activity through OsDHAR1 overexpression, which suggests that the activity of endogenous DHAR is a limiting factor. This might enable rice plants to regulate their intracellular redox state, particularly in response to developmental process or to changes in environmental conditions. Some results pertinent to the correlation between the AsA pool and stress response have been reported in DHAR-expressing TG plants. For example, increased AsA content and AsA redox state (AsA/DHA ratio) confer stress tolerance to high salinity (Ushimaru et al. 2006) in TG tobacco plants over-expressing the human DHAR gene under optimum conditions (Kwon et al. 2003). Increased AsA content in Arabidopsis was accompanied by enhanced recycling that conferred tolerance to oxidative stress (Wang et al. 2010).
As stated above, OsDHAR1-expression in TG rice plants increased antioxidant enzyme activity (DHAR, MDHAR, APX, and GR) and AsA homeostasis under natural field conditions. On the basis of these results, we evaluated whether the activation of cell rescue systems affects the redox state under field conditions. The redox state was quantified by measuring hydroperoxide levels by using the FOX reagent. The hydroperoxide levels of TG rice plants (OX-1 to OX-7) were approximately 15.5 % lower than those of WT rice plants, even in the presence of a moderate difference in hydroperoxide levels among TG rice plants (Fig. 2d). Naturally, various abiotic stresses, including high salinity, UV radiation, ozone, drought, photoperiod, chilling, and high temperature lead to ROS production in plants. The presence of ROS leads to the elevation of antioxidant enzymes, such as DHAR, MDHAR, and APX, which are able to reduce the stress caused by the oxidizing conditions generated by ROS (Dinakar et al. 2010). TG plants expressing these enzymes decrease their cellular ROS levels by increasing AsA levels through enhanced AsA recycling, which leads to increased protection against paraquat, H2O2, ozone, and other abiotic stresses (Wang et al. 2010). Taken together, our results suggest that the expression of OsDHAR1 in TG rice plants improves AsA homeostasis by maintaining redox homeostasis under paddy field conditions and leads to improved adaptation under the same conditions.
Estimation of grain yield and biomass in TG rice plants under paddy field conditions
We identified that OsDHAR1 overexpression in TG rice plants improves photosynthetic capacity, antioxidant enzyme activity, AsA homeostasis, and redox homeostasis under paddy field conditions. Based on these results, we investigated whether OsDHAR1 overexpression affects grain yield and biomass under the same conditions. To perform this evaluation, seven independent homozygous TG rice lines (OX-1 to OX-7) and control non-transgenic WT rice plants were transplanted into a paddy field and cultivated to maturity during two seasons [2010 (T2) and 2011 (T3)] for which there were significant differences in mean temperature, total precipitation, and total sunshine duration within farming periods (May–October; Fig. 3a). Phenotypic and agronomic traits were then evaluated. The rice yield parameters analyzed were as follows: TPW, CW, RW, number of panicles per hill (NP), number of spikelets per panicle (NSP), FR, TGW, and TOGW. At the middle reproductive stage, OsDHAR1-expressing TG rice plants (OX1, OX-2, and OX-3) displayed a better phenotype than WT rice plants (Fig. 3b). In addition, the total length of the TG rice plants was less than that of the WT rice plants, whereas the number of panicles of the TG rice plants was higher than that of the WT rice plants during vegetative growth (data not shown) and the harvest stages after flowering (Fig. 3c). As shown in Fig. 3d, most of the yield parameters, with the exception of 1,000 GW (TOGW), was increased in OsDHAR1-expressing TG rice plants (OX1 to OX-7) when compared with the WT rice plants. This increase was pronounced in total fresh biomass weight (TPW), TGW, and panicle number per hill (NP), which were 21, 25, and 27 % higher, respectively, than those of the WT rice plants. Increased mean values of TPW, CW, RW, NP, NSP, FR, TGW, and TOGW in TG rice plants were 18.9 and 18.7, 15.0 and 16.4, 11.7 and 11.5, 19.7 and 19.3, 14.4 and 14.0, 14.3 and 14.0, 19.0 and 18.2, and 2.3 and 6.1 % in 2010 and 2011, respectively, as compared to the corresponding values in WT rice plants (Fig. 3d).
Comparison of agronomic traits between TG and WT rice plants under normal field conditions. a Monthly meteorological data during the farming season in 2010 (solid) and 2011 (open). Red circle, mean temperature (°C); olive upward triangle, maximum temperature (°C); green downward triangle, minimum temperature (°C); blue square, total sunshine duration (h); magenta diamond, total precipitation (mm). b Phenotype analysis at the middle reproductive stage in both TG (OX-1, OX-2, and OX-3) and WT rice plants in 2011. c Phenotype at the late reproductive stage in both TG (OX-1 to OX-7) and WT rice plants in 2010 (upper panel) and 2011 (lower panel). d Agronomic traits of TG rice plants (OX-1 to OX-7) and WT rice plants grown under paddy field conditions for two cultivating seasons (2010–2011). Agronomic traits of 12 independent homozygous (T2, 2010; upper panel) and (T3, 2011; lower panel) plants for each TG line (OX-1 to OX-7) together with control WT rice plants were plotted using Origin 8.0. Each data point represents the percentage of the mean values with WT rice plants assigned a reference value of 100 %. TPW total biomass weight; CW culm weight; RW root weight; NP number of panicles per hill; NSP number of spikelets per panicle; FR filling rate; TGW total grain weight; TOGW 1,000 grain weight. Data are expressed as the mean ± SD of at least three replications from three independent experiments. Bars labeled with asterisks show significant differences (P < 0.05) between TG and WT rice plants as determined by ANOVA
The correlation between gene expression and stress response has been previously reported. For example, accumulation of trehalose increased tolerance to drought and salt stress in rice plants (Redillas et al. 2012). However, these studies were limited to tolerance only at the vegetative stage, and the physiological mechanisms under natural field conditions and at the reproductive stage remained elusive (Jeong et al. 2012). Very valuably, the overexpression of the OsDHAR1 transgene significantly enhanced environmental adaptation at the reproductive stage of growth via enlarged total fresh biomass including root sand shoots (16–21 % in 2010 and 15–21 % in 2011), with a concomitant increase (15–25 % in 2010 and 12–24 % in 2011) in rice grain yield. Recently, similar results have been reported or TG rice. The grain yield of TG rice plants expressing OsNAC5 (N AM, A TAF, and C UC) under the regulation of either the root-specific (RCC3) or constitutive (GOS2) promoter was increased by 9–23 and 9–26 % under normal conditions, respectively (Jeong et al. 2012). Under the same promoter, OsNAC9 expression increased the rice grain yield by 13–18 and 13–32 % in TG rice (RCC3:OsNAC9 and GOS2:OsNAC9, respectively) under normal field conditions (Redillas et al. 2012). OsNAC10 overexpression in TG rice under the control of the RCC3 promoter increased grain yield by 5–14 % under normal field conditions, while under the GOS2 promoter the yield in transgenic plants remained similar to that of controls under the same conditions (Jeong et al. 2010). In addition, LOS5 and ZAT10 overexpression in TG rice under the control of the constitutive Actin1 and stress-inducible HVA22 promoter showed significantly higher yield per plant following high spikelet fertility than control WT rice plant under field conditions (Xiao et al. 2009). In contrast, the grain yield in transcription factor AP37- and AP39-expressing TG rice plants under the control of the constitutive rice CC1 promoter exhibited no significant difference or was reduced by 23–43 % under normal conditions as compared to control WT rice plants (Oh et al. 2009). On the other hand, some candidate genes were identified in follow-up analyses of associated loci for grain-related traits. For example, Bh4 (controlling seed hull color), DEP1 (controlling grain number), GW2 and qSW5 (controlling grain weight and grain width), GS3 (controlling grain length), and Waxy (controlling amylase content), ALK (controlling starch gelatinization temperature) were highly expressed at the stages of grain filling or panicle development (Huang et al. 2012).
These yield components are sensitive to water stress, UV radiation, and light/dark cycle at different stages of plant growth, such as anther dehiscence and panicle exertion (Jeong et al. 2012). Despite the apparent climate differences between 2010 and 2011, there were no differences in agronomic traits, including grain yield and biomass, for OsDHAR1-expressing TG rice plants under the control of the Ubi promoter. This indicates that OsDHAR1 plays a very important role in adaptation to natural field conditions during the farming season. Further, our results suggest that OsDHAR1 is a good candidate gene for the development of a rice plant tolerant to environmental conditions because the grain yield of OsDHAR1-expressing TG rice plants is similar to or better than that of the TG rice plants reported thus far.
Molecular modeling of the OsDHAR protein
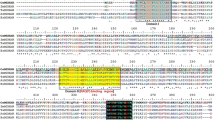
To examine the molecular properties of OsDHAR, multiple sequence alignment (MSA) was performed on OsDHAR by using the known HsDHAR, AtDHAR, GmDHAR, ZmDHAR, and CsDHAR, sequences. Pairwise alignment of OsDHAR with other DHARs was conducted using the NCBI BLAST software, and the conserved redox-active site was observed at Cys20 of OsDHAR (red box) as well as the conserved GSH-binding region (Lys8, Lys59, and Val60; gray box), the GSH-binding motif (Pro21 to Gln24, and Asp73 to Ser74; cyan box), the glutathione-S-transferase (GST) N-terminal domain (blue box), the C-terminal domain interface (Ala150 to His 160; yellow box), and GST C-terminal domain (green box; Fig. 4a). The predicted molecular weight of the OsDHAR enzyme is approximately 23.5 kDa. It has been reported that DHAR shows a loss of thiol transferase activity in the absence or mutagenesis of thiol (redox active site Cys20) (Harrop et al. 2001). OsDHAR showed 30, 70, 63, 61, and 70 % identity, and 51, 80, 81, 78, and 84 % similarity to HsDHAR, AtDHAR, GmDHAR, ZmDHAR, and CsDHAR, respectively. This is represented as a phylogenic tree in Fig. 4b. These results indicate that the molecular structure of the OsDHAR protein contains an active site and conserved domains. Since the 3D structure of O. sativa DHAR has not been elucidated, we introduced the 3D structure of a human DHAR isoform (CLIC1; accession no. AAD26137.1), which was 30 % homologous to OsDHAR. The 3D structure of the DHAR protein from O. sativa was constructed by homology modeling based on the PDB file obtained from the ESyPred3D Web Server 1.0 using the UCSF Chimera software. This structure consisted of amino acids 2-213 (Gly2-Ala213) out of a total of 213 amino acids (Fig. 4c). The active site and conserved domains of the OsDHAR model fit the human DHAR template well. In mammalian cells, DHAR (HsDHAR) protein has glutaredoxin (Saaranen et al. 2010; Wells et al. 1990, 1995) and protein disulfide isomerase (Girardini et al. 2002; Ken et al. 2009; Wells et al. 1990). The high sequence and structural homology of OsDHAR should indicate the ability of this protein to catalyze the glutathione-dependent DHA reduction and perform a wide range of biochemical reactions, such as the maintenance of a normal cellular thiol/disulfide ratio, redox regulation, and the regeneration of oxidatively damaged proteins.
Alignment and predicted structure of OsDHAR. a Alignment between OsDHAR and 5 DHAR sequences. “asterisk” indicates that the residues in that column are identical in all sequences in the alignment, “colon” indicates the presence of conserved substitutions, and “dot” indicates the presence of semi conserved substitutions. Red box, active site; gray box, glutathione (GSH)-binding region; cyan box, GSH-binding motif; yellow box, C-terminal domain interface; blue box, glutathione-S-transferase (GST) N-terminal domain; green box, GST C-terminal domain. b Phylogenetic trees of OsDHAR and the 5 DHARs. c The predicted structure of the OsDHAR protein. The region of conserved active site and domains are depicted and boxed, and the N-terminal (Gly2) and C-terminal (Ala213) amino acids are indicated. The model was generated using the ESyPred3D Web Server 1.0 based on the structure of 1KOO_A (identity, 30 %) and visualized using Chimera software. Domain annotations of OsDHAR are represented on the basis of MSA and the predicted structure
In conclusion, the transgene OsDHAR1 was stably incorporated into the rice genome and effectively expressed under the control of the Ubi promoter. OsDHAR1 overexpression with AsA recycling, and glutaredoxin and protein disulfide isomerase activity improved AsA and redox homeostasis by co-activating antioxidant enzymes (MDHAR, APX, and GR) and enhancing photosynthetic capacity in TG rice plants under natural paddy field conditions, which led to increases in agronomic traits, including grain yield and biomass, through the high levels of yield parameters such as spikelet fertility when compared to WT rice plants.
Abbreviations
- APX:
-
Ascorbate peroxidase
- AsA:
-
Ascorbate
- AsA/DHA:
-
Ascorbate to dehydroascorbate ratio
- DHAR:
-
Dehydroascorbate reductase
- Fv/Fm :
-
Maximum quantum yield of PSII
- GR:
-
Glutathione reductase
- GSH:
-
Reduced glutathione
- MDHAR:
-
Monodehydroascorbate reductase
- Os:
-
Oryza sativa
- OX:
-
Overexpression
- ROS:
-
Reactive oxygen species
- TG:
-
Transgenic
References
Ahmad P, Jaleel CA, Salem MA, Nabi G, Sharma S (2010) Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit Rev Biotechnol 30:161–175
Boyer JS (1982) Plant productivity and environment. Science 218:443–448
Chen Z, Gallie DR (2005) Increasing tolerance to ozone by elevating foliar ascorbic acid confers greater protection against ozone than increasing avoidance. Plant Physiol 138:1673–1689
Chen Z, Young TE, Ling J, Chang SC, Gallie DR (2003) Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc Natl Acad of Sci USA 100:3525–3530
Deutsch JC (2000) Dehydroascorbic acid. J Chromatogr A 881:299–307
Dinakar C, Abhaypratap V, Yearla SR, Raghavendra AS, Padmasree K (2010) Importance of ROS and antioxidant system during the beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Planta 231:461–474
Eltayeb AE, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Morishima I, Shibahara T, Inanaga S, Tanaka K (2006) Enhanced tolerance to ozone and drought stresses in transgenic tobacco overexpressing dehydroascorbate reductase in cytosol. Physiol Plant 127:57–65
Eltayeb AE, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Shibahara T, Inanaga S, Tanaka K (2007) Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225:1255–1264
Eltayeb AE, Yamamoto S, Habora MEE, Yin L, Tsujimoto H, Tanaka K (2010) Transgenic potato overexpressing Arabidopsis cytosolic AtDHAR1 showed higher tolerance to herbicide, drought and salt stresses Breeding Sci 61:3–10
Foyer CH, Halliwell B (1976) The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta 133:21–25
Garcia V, Stevens R, Gil L, Gilbert L, Gest N, Petit J, Faurobert M, Maucourt M, Deborde C, Moing A, Poessel JL, Jacob D, Bouchet JP, Giraudel JL, Gouble B, Page D, Alhagdow M, Massot C, Gautier H, Lemaire-Chamley M, de Daruvar A, Rolin D, Usadel B, Lahaye M, Causse M, Baldet P, Rothan C (2009) An integrative genomics approach for deciphering the complex interactions between ascorbate metabolism and fruit growth and composition in tomato. C R Biol 332:1007–1021
Gay C, Collins J, Gebicki JM (1999) Hydroperoxide assay with the ferric-xylenol orange complex. Anal Biochem 273:149–155
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Gillespie KM, Ainsworth EA (2007) Measurement of reduced, oxidized and total ascorbate content in plants. Nat Protoc 2:871–874
Girardini J, Amirante A, Zemzoumi K, Serra E (2002) Characterization of an omega-class glutathione S-transferase from Schistosoma mansoni with glutaredoxin-like dehydroascorbate reductase and thiol transferase activities. Eur J Biochem 269:5512–5521
Harrop SJ, DeMaere MZ, Fairlie WD, Reztsova T, Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Jankova L, Warton K, Bauskin AR, Wu WM, Pankhurst S, Campbell TJ, Breit SN, Curmi PM (2001) Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution. J Biol Chem 276:44993–45000
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Horemansa N, Foyerb CH, Pottersa G, Asarda H (2000) Ascorbate function and associated transport systems in plants. Plant Physiol Biochem 38:531–540
Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, Li W, Guo Y, Deng L, Zhu C, Fan D, Lu Y, Weng Q, Liu K, Zhou T, Jing Y, Si L, Dong G, Huang T, Lu T, Feng Q, Qian Q, Li J, Han B (2012) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44:32–39
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK (2012) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J. doi:10.1111/pbi.12011
Kang HG, Jeon JS, Lee S, An G (1998) Identification of class B and class C floral organ identity genes from rice plants. Plant Mol Biol 38:1021–1029
Kausar R, Hossain Z, Makino T, Komatsu S (2012) Characterization of ascorbate peroxidase in soybean under flooding and drought stresses. Mol Biol Rep 39:10573–10579
Ken CF, Lin CY, Jiang YC, Wen L, Lin CT (2009) Cloning, expression, and characterization of an enzyme possessing both glutaredoxin and dehydroascorbate reductase activity from Taiwanofungus camphorata. J Agric Food Chem 57:10357–10362
Kerk NM, Feldman NJ (1995) A biochemical model for the initiation and maintenance of the quiescent center: implications for organization of root meristems. Development 121:2825–2833
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:25–34
Kim KY, Kwon SY, Lee HS, Hur Y, Bang JW, Kwak SS (2003) A novel oxidative stress-inducible peroxidase promoter from sweetpotato: molecular cloning and characterization in transgenic tobacco plants and cultured cells. Plant Mol Biol 51:831–838
Klimyuk VI, Carroll BJ, Thomas CM, Jones JD (1993) Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J 3:493–494
Kwon SY, Choi SM, Ahn YO, Lee HS, Lee HB, Park YM, Kwak SS (2003) Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J Plant Physiol 160:347–353
Lee YP, Kim SH, Bang JW, Lee HS, Kwak SS, Kwon SY (2007) Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Rep 26:591–598
Li Q, Li Y, Li C, Yu X (2012) Enhanced ascorbic acid accumulation through overexpression of dehydroascorbate reductase confers tolerance to methyl viologen and salt stresses in tomato. Czech J Genet Plant Breed 48:74–86
McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2:163–171
McElroya D, Brettellb RIS (1994) Foreign gene expression in transgenic cereals. Trends Biotechnol 12:62–68
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49:249–279
Noctor G, Veljovic-Jovanovic S, Foyer CH (2000) Peroxide processing in photosynthesis: antioxidant coupling and redox signalling. Philos Trans R Soc Lond B Biol Sci 355:1465–1475
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351
Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150:1368–1379
Owens S (2001) Salt of the earth. Genetic engineering may help to reclaim agricultural land lost due to salinisation. EMBO Rep 2:877–879
Pasqualini S, Batini P, Ederli L, Porceddu A, Piccioni C, De Marchis F, Antonielli M (2001) Effects of short-term ozone fumigation on tobacco plants: response of the scavenging system and expression of the glutathione reductase. Plant, Cell Environ 24:245–252
Qin A, Shi Q, Yu X (2011) Ascorbic acid contents in transgenic potato plants overexpressing two dehydroascorbate reductase genes. Mol Biol Rep 38:1557–1566
Redillas MC, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK (2012) The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J 10:792–805
Saaranen MJ, Karala AR, Lappi AK, Ruddock LW (2010) The role of dehydroascorbate in disulfide bond formation. Antioxid Redox Signal 12:15–25
Sakihama Y, Mano J, Sano S, Asada K, Yamasaki H (2000) Reduction of phenoxyl radicals mediated by monodehydroascorbate reductase. Biochem Biophys Res Commun 279:949–954
Shin SY, Kim IS, Kim YH, Park HM, Lee JY, Kang HG, Yoon HS (2008) Scavenging reactive oxygen species by rice dehydroascorbate reductase alleviates oxidative stresses in Escherichia coli. Mol Cells 26:616–620
Tang L, Kim MD, Yang KS, Kwon SY, Kim SH, Kim JS, Yun DJ, Kwak SS, Lee HS (2008) Enhanced tolerance of transgenic potato plants overexpressing nucleoside diphosphate kinase 2 against multiple environmental stresses. Transgenic Res 17:705–715
Urano J, Nakagawa T, Maki Y, Masumura T, Tanaka K, Murata N, Ushimaru T (2000) Molecular cloning and characterization of a rice dehydroascorbate reductase. FEBS Lett 466:107–111
Ushimaru T, Nakagawa T, Fujioka Y, Daicho K, Naito M, Yamauchi Y, Nonaka H, Amako K, Yamawaki K, Murata N (2006) Transgenic Arabidopsis plants expressing the rice dehydroascorbate reductase gene are resistant to salt stress. J Plant Physiol 163:1179–1184
Wang Z, Xiao Y, Chen W, Tang K, Zhang L (2010) Increased vitamin C content accompanied by an enhanced recycling pathway confers oxidative stress tolerance in Arabidopsis. J Integr Plant Biol 52:400–409
Wells WW, Xu DP, Yang YF, Rocque PA (1990) Mammalian thioltransferase (glutaredoxin) and protein disulfide isomerase have dehydroascorbate reductase activity. J Biol Chem 265:15361–15364
Wells WW, Rocque PA, Xu DP, Meyer EB, Charamella LJ, Dimitrov NV (1995) Ascorbic acid and cell survival of adriamycin resistant and sensitive MCF-7 breast tumor cells. Free Radic Biol Med 18:699–708
Xiao BZ, Chen X, Xiang CB, Tang N, Zhang QF, Xiong LZ (2009) Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol Plant 2:73–83
Yin L, Wang S, Eltayeb AE, Uddin MI, Yamamoto Y, Tsuji W, Takeuchi Y, Tanaka K (2010) Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 231:609–621
Yoshida S, Tamaoki M, Shikano T, Nakajima N, Ogawa D, Ioki M, Aono M, Kubo A, Kamada H, Inoue Y, Saji H (2006) Cytosolic dehydroascorbate reductase is important for ozone tolerance in Arabidopsis thaliana. Plant Cell Physiol 47:304–308
Zoua L, Lib H, Ouyanga B, Zhanga J, Yea Z (2006) Cloning and mapping of genes involved in tomato ascorbic acid biosynthesis and metabolism. Plant Sci 120:120–127
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ008115012013), Rural Development Administration, and funded by the Priority Research Centers Program (2011-0030748) through the National Research Foundation (NRF) of the Ministry of Education, Science and Technology, Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Y.-S. Kim and I. S. Kim contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, YS., Kim, IS., Bae, MJ. et al. Homologous expression of cytosolic dehydroascorbate reductase increases grain yield and biomass under paddy field conditions in transgenic rice (Oryza sativa L. japonica). Planta 237, 1613–1625 (2013). https://doi.org/10.1007/s00425-013-1862-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1862-8