Abstract
A mapping population of 126 doubled haploid (DH) lines derived from a cross between the English winter wheat cultivars Spark and Rialto was evaluated for response to Puccinia graminis f. sp. tritici in the greenhouse and in artificially inoculated field plots at two locations over 3 years (2011, 2012 and 2013). Genetic analysis indicated the involvement of two seedling genes (Sr5 and Sr31, contributed by Rialto) and three adult plant resistance genes. QTL analyses of field data showed the involvement of three consistent effects QTL on chromosome arms 1BS (contributed by Rialto), and 3BS and chromosome 5A (contributed by Spark) in the observed resistance to stem rust. These QTLs explained average phenotypic variation of 78.5, 9.0 and 5.9 %, respectively. With the presence of virulence for Sr5 and absence of Sr31 virulence in the field, the QTL detected on 1BS (QSr.sun-1BS) was attributed to the major seedling resistance gene Sr31. The QTL located on chromosome arm 3BS (QSr.sun-3BS) was closely associated with SSR marker gwm1034, and the QTL detected on 5A (QSr.sun-5A) was closely linked with SSR marker gwm443. DH lines carrying the combination of QSr.sun-3BS and QSr.sun-5A exhibited lower stem rust responses indicating the additive effects of the two APR genes in reducing disease severity. The markers identified in this study can be useful in pyramiding these QTLs with other major or minor genes and marker assisted selection for stem rust resistance in wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat, one of the world’s most important staple food crops, faces several production challenges due to abiotic factors like drought and heat stress, and biotic factors such as pests and diseases. Stem rust, caused by Puccinia graminis f. sp. tritici (Pgt), is one of the most important diseases of wheat on a global basis. It causes severe devastation, capable of reducing an apparently healthy looking wheat crop to broken stems and shrivelled grains in <1 month (Singh et al. 2008b). In susceptible cultivars, yield loss due to stem rust can be as high as 100 % (Singh and Rajaram 2002).
Historically, stem rust has been a significant problem in the major wheat-growing regions of Africa, the Middle East, Asia, Australia and New Zealand (Saari and Prescott 1985). Stem rust has been controlled in many parts of the world through the deployment of resistance genes (Bariana et al. 2004; Singh et al. 2004; Park 2008) and the eradication of common barberry plants in North America (Roelfs et al. 1992). Australian wheat production has also been well protected from stem rust by the deployment of cultivars carrying multiple genes for resistance (Park 2007; Bansal et al. 2008). However, the detection of a stem rust race TTKSK (“Ug99”) in Uganda in 1998 (Pretorius et al. 2000), combined with rapid evolution of new virulences and migration to different countries within Africa [Kenya, Ethiopia, Sudan, Tanzania, South Africa and Yemen (Singh et al. 2006; Pretorius et al. 2010; Singh et al. 2011a; Hale et al. 2014) and Iran (Nazari et al. 2009)], has again raised the global threat posed by stem rust. The movement of members of the “Ug99” lineage has been predicted to follow the same migratory route as Yr9-virulence, which in the late 1980s is believed to have arisen from the Eastern Africa highlands and then spread to Asia (Singh et al. 2006).
The East African highlands are known as a hot spot for the evolution and survival of new wheat rust pathotypes. The narrow genetic basis of resistance to stem rust in countries like Ethiopia (Beteselassie et al. 2007) has also contributed to the vulnerability of available genetic resources to the pathogen population. Recent stem rust outbreaks have occurred in Ethiopia, causing yield losses up to 100 % in severely affected areas. These epidemics first occurred in the 2013/2014 growing season and continued in 2014/2015. The pathotype responsible, TKTTF, affected the widely grown cultivar Digalu, which was known to carry the stem rust resistance gene SrTmp (Hodson 2015).
Genetic resistance is considered the most effective and sustainable way of controlling stem rust, particularly for farmers in developing countries where the use of fungicides is often unaffordable. To date, 58 genes conferring resistance to Pgt have been designated in wheat (McIntosh et al. 2014). Most of these genes confer seedling resistance, which are easily manipulated in a breeding programme but frequently overcome by new pathogen pathotypes (Park 2008; Singh et al. 2009; Herrera-Foessel et al. 2011). On the other hand, adult plant resistance (APR) based on minor, slow-rusting genes are believed to offer more durable resistance to the rust diseases of wheat. Although such minor resistance genes do not confer adequate levels of resistance individually, good levels of protection can be achieved by combining 4–5 of them (Singh et al. 2000). While polygenic APR to rust in wheat is considered durable and race non-specific, examples exist of single APR genes such as Sr2/Yr30, Lr34/Yr18/Sr57, Lr46/Yr29/Sr58 and Lr67/Yr46/Sr55 that have also proved to be durable (William et al. 2003; Herrera-Foessel et al. 2011; Singh et al. 2012).
Molecular markers play a significant role in the evaluation and identification of both major and minor genes (Suenaga et al. 2003; Collard and Mackill 2008). The development of molecular markers that are closely linked to target traits is important in expediting reliable selection and pyramiding of different genes in wheat breeding. This makes the breeding programme and selection process faster and more cost efficient (Kaur et al. 2008). Genetic mapping has been used effectively in identifying regions in the wheat genome that confer resistance to Pgt (Haile and Röder 2013). Molecular markers are available for several stem rust resistance genes including Sr2 (Spielmeyer et al. 2003; Mago et al. 2011), Sr13 (Klindworth et al. 2007; Periyannan et al. 2014), Sr22 (Khan et al. 2005; Periyannan et al. 2011), Sr24 and Sr26 (Mago et al. 2005), Sr31 (Das et al. 2006), Sr32 (Mago et al. 2013), Sr33 (Periyannan et al. 2008), Sr35 (Zhang et al. 2010), Sr39 (Mago et al. 2009), Sr40 (Shuangye et al. 2009), Sr45 (Periyannan et al. 2014), Sr52 (Qi et al. 2011), Sr53 (Liu et al. 2011), Sr55 (Herrera-Foessel et al. 2014), Sr56 (Bansal et al. 2014) and Sr57 (Spielmeyer et al. 2005).
In field tests in Australia, the European winter wheat cultivars Spark and Rialto were moderately resistant and highly resistant, respectively, against predominant Australian pathotypes of Pgt. In the current study, we characterised the genetic basis of the stem rust resistance observed in both cultivars through integrated greenhouse and field studies.
Materials and methods
Plant material
A DH winter wheat mapping population was generated from a cross between Spark (Pedigree: Moulin/Tonic) and Rialto (Pedigree: Haven/Fresco). The population was developed at the John Innes Centre (Simmonds et al. 2014) and introduced to Australia by the Plant Breeding Institute Cobbitty (PBIC) in 2005. A total of 126 DH lines were used for greenhouse and field stem rust phenotyping and QTL mapping.
Greenhouse phenotyping
Seedling tests were conducted under controlled greenhouse conditions at PBIC. The mapping population and parents, along with 35 Australian stem rust differential lines (McIntosh et al. 1995), were evaluated for seedling response against three pathotypes of Pgt (98-1,2,3,5,6 [PBIC Accession Number 781219]; 343-1,2,3,5,6 [840837] (virulent to Sr5 and avirulent to Sr31); and 21-0 [540129] (avirulent to both Sr5 and Sr31)) maintained in liquid nitrogen in the PBIC rust collection. Eight to 10 seedlings per line were evaluated by growing in 9-cm plastic pots. The pots were fertilised with complete fertiliser (Aquasol®, Hortico Pty. Ltd., Revesby, NSW, Australia) at the rate of 25 g/10L water for 200 pots. After fertilising, seeds were sown and covered with potting mix. Pots were moved to microclimate rooms maintained at 15–20 °C until they were ready for inoculation. Methods of sowing, disease assessment and pathotype nomenclature are explained in McIntosh et al. (1995). Seedlings were inoculated with each pathotype at the 1–1.5 leaf stage by atomising urediniospores suspended in light mineral oil (Isopar™ L, Australasian Solvents and Chemicals Pty. Ltd., Springwood, Queensland, Australia @ 10 mg urediniospores per 10 ml oil per 200 pots) using a hydrocarbon propellant pressure pack. To avoid contamination, the spray equipment was washed in alcohol and rinsed in running tap water between successive inoculations. Inoculated plants were incubated and placed in water-filled steel trays covered with moistened polythene sheet that maintained 100 % relative humidity within a greenhouse room. Inoculated seedlings were incubated for 48 h at a temperature maintained between 20 and 25 °C. The incubated seedlings were then transferred to a temperature and irrigation-controlled greenhouse growth rooms at 24–26 °C until ready for disease assessment. Individual DH lines were classified as resistant or susceptible at seedling growth stages based on infection type scale described by McIntosh et al. (1995).
Field phenotyping
The parents and the DH lines were assessed in the field for stem rust severity at PBIC in 2011, 2012 and 2013. In 2013, all lines were sown at two sites, Horse Unit (HRU) and Lansdowne (LDN). PBI is located at 34°1′2.20″S, 150°40′3.79″E. HRU and LDN are located at 34°1′15.79″S, 150°39′32.37″E and 34°1′18.43″S, 150°39′44.73″E, respectively. Approximately 20–30 seeds were planted evenly in a 0.75-m row with a spreader strip comprising stem rust susceptible genotypes adjacent to each row. The two parental lines were included at the beginning, middle and end of each nursery as controls. Urediniospores of selected mixtures of relevant Australian Pgt pathotypes [98-1,2,3,5,6 (781219), 343-1,2,3,5,6 (840837), 34-2,12,13 (840552), 34-1,2,7 +Sr38 (010130) in 2011 and 98-1,2,3,5,6, 343-1,2,3,5,6, 34-2,12,13, 34-1,2,7 +Sr38 and 98-1,2,3,5,6,7 (030312) in 2012 and 2013] suspended in mineral oil (Isopar™ L) were misted over the spreader using an ultra-low-volume applicator (Microfit”, Micron sprayer Ltd. Bromyard, Herefordshire, UK) on clear afternoons when there was likelihood of overnight dew formation. The pathotypes used were virulent to Sr5 and avirulent to Sr31.
Adult plant disease responses were assessed visually and scored based on disease severity and host response following the modified Cobb scale (Peterson et al. 1948). Disease scoring was performed at anthesis and repeated after 10 days. Coefficient of infection (CI) value was calculated by combining the disease severity and host response data, by multiplying severity by a constant for host response where R and TR, MR, MR-MS, MS, MSS, and S = 0.10, 0.25, 0.50, 0.75, 0.90 and 1.00, respectively. For example, a disease score of “60MS” was represented by CI = 60 × 0.75 = 45.
Statistical analysis and QTL mapping
Chi-squared (χ 2) analysis was used to evaluate the goodness of fit of observed to expected segregation ratios to estimate the number of genes conferring resistance. The program Map Manager QTX20 (Manly et al. 2001) was used to construct linkage maps. Inclusive composite interval mapping (ICIM) (Wang et al. 2007) was used to detect the QTLs. QTLs were considered significant at P < 0.05, with logarithm of odds (LOD) score determined by 1000 iterations for each trait (Doerge and Churchill 1996). Linkage groups were assigned to chromosomes on the basis of previously mapped and known genomic positions of the markers. Centimorgan (cM) values were calculated using the Kosambi (1943) mapping function. A total of 281 markers were used to localise the QTLs in the mapping population (Simmonds et al. 2014). QTL figures were drawn using MapChart V2.1 (Voorrips 2002). A one-way ANOVA was conducted, and the least significant difference (LSD) was used to verify the differences between the varieties or genotypes at P = 0.05.
Results
Parental response in the greenhouse and field
Under greenhouse conditions, seedlings of Rialto produced infection types (IT) “1+” to “2−” when tested with Pgt pathotypes 98-1,2,3,5,6 and 343-1,2,3,5,6 (Sr5-virulent and Sr31-avirulent). The ITs were similar to those produced by the Sr31 control genotype Mildress (Table 1). However, with pathotype 21-0 (avirulent to both Sr5 and Sr31), Rialto produced IT “0;” typical to that produced by the differential genotype Reliance carrying Sr5. On this basis, the seedling genes Sr5 and Sr31 were postulated in Rialto. In contrast, Spark showed IT “3+” against the three Pgt pathotypes and was therefore considered to lack effective seedling resistance to the pathotypes used (Table 1). When evaluated for adult plant stem rust response under field conditions, Rialto consistently exhibited high levels of resistance, while Spark was moderately susceptible across experiments/years to the Pgt pathotypes used (Table 1).
Progeny response at seedling stage
The DH population was also assessed in the greenhouse for response to stem rust. With a classification of IT “3” or less considered resistant, and greater than IT “3” as susceptible, the population segregated for a single dominant gene (χ 2 = 1.16, P > 0.281 and χ 2 = 0.97, P > 0.325), apparently Sr31, with the Sr5-virulent pathotypes 98-1,2,3,5,6 and 343-1,2,3,5,6 (Table 2). Three conspicuous ITs (“0;”, “2−” to “2-C” or “3+”) were observed in the population when tested against pathotype 21-0 (avirulent to Sr5): 86 lines (69.9 %) displayed a resistant reaction (“0;” or “2−” to “2-C”) and 37 (30.1 %) were susceptible (“3” to “3+”), while three lines did not germinate. The chi-squared analysis supported the involvement of two genes (χ 2 = 1.35, P > 0.246) from Rialto, consistent with the presence of seedling genes Sr5 and Sr31 (Table 2). The genotypes that showed IT “0;” were postulated to carry either Sr5 or Sr5 +Sr31, whereas those showing ITs “2−” to “2-C” were postulated to carry only Sr31.
Progeny response at adult plant stage
The parents and the DH lines were assessed for stem rust severity against Pgt pathotypes in the field in 2011, 2012 and 2013 (two locations). Consistent variation in stem rust infection was observed across the DH lines in all four experiments. Transgressive segregation was observed in some lines, which were either more resistant or more susceptible than the parents across all experiments. A total of 11 lines (8.73 %) that displayed high ITs (“3+”) to all three pathotypes at the seedling stage also showed high disease responses in the field (CI values of “70” to “90”), indicating a lack of seedling resistance and APR in these lines. Ten lines (7.94 %) that were susceptible to pathotypes 98-1,2,3,5,6 and 343-1,2,3,4,5,6, but resistant to pathotype 21-0 (postulated to carry Sr5), were also susceptible in the field because of the ineffectiveness of Sr5 against the field pathotypes. As expected, in the absence of pathotypes virulent to Sr31 in field, all DH lines that carried Sr31 (singly or in combination with Sr5) were highly resistant in the field (CI value of “0.1–7.5”). In this case, 57 (45.24 %) lines that showed resistance to pathotypes 98-1,2,3,5,6 and 343-1,2,3,4,5,6 at the seedling stage were also resistant in the field. However, 47 of the lines (37.30 %) that were susceptible to pathotypes 98-1,2,3,5,6 and 343-1,2,3,4,5,6 at the seedling stage were resistant to moderately susceptible at adult plant growth stages, indicating the presence of gene(s) conferring APR.
The population showed the same pattern of segregation across all experiments, and chi-squared analysis supported the involvement of three genes (segregation ratio 7:1, Table 2). When chi-squared analysis was performed using 75 lines lacking Sr31, segregation within the population fitted a genetic ratio of 3:1, indicating the presence of two APR genes in the DH population.
QTL analysis
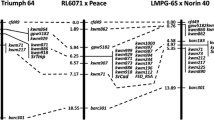
Inclusive composite interval mapping (ICIM) of field data with 281 markers revealed the involvement of resistance loci on chromosome arms 1BS and 3BS and chromosome 5A across all data sets. The QTL detected on chromosome arm 1BS, temporarily designated QSr.sun-1BS, was associated with the marker gwm18, where the major seedling resistance gene Sr31 was also located and contributed by Rialto. The QTL located on chromosome arm 3BS, designated QSr.sun-3BS, was closely associated with SSR marker gwm1034, and the QTL detected on chromosome 5A (QSr.sun-5A) was associated with SSR marker gwm443. The resistance effects of QTLs QSr.sun-3BS and QSr.sun-5A were both contributed by Spark.
QTL QSr.sun - 1BS (Sr31) explained 73–84 % of the phenotypic variation. The highest phenotypic variation (84 %) was explained in 2011 and in 2013HRU (LOD = 39.8), while the lowest phenotypic variation (73 %) was explained in 2012 (LOD = 29.78) (Table 3). The phenotypic variation explained by the QTL QSr.sun-3BS varied from 6 % to 12 %. The largest APR variance was explained in 2013LDN (12 %) with an LOD value 3.5, whereas the lowest was in 2012 explaining phenotypic variance of 6 % with the LOD value 1.59 at the gwm1034 locus (Table 3; Fig. 1a). The chromosome 3BS comprised a total of 20 loci covering a total distance of 122.9 cM with an average distance between markers of 6.15 cM. The QTL QSr.sun-5A (LOD value of 1.09–1.80) explained 4.43–7.30 % of the phenotypic variation with gwm443 being the closest marker (Table 3; Fig. 1b). This QTL had the largest effect on stem rust resistance (7.30 %) in 2013HRU and lowest (4.43 %) in 2012.
The effect of different QTL combinations on stem rust response of non-Sr31 carrying DH lines
The resistance conferred by each QTL alone and in combination was assessed by predicting the genotype of each DH line for the different QTL combinations based on the presence/absence of the most closely linked flanking markers. Lines that carried QSr.sun-3BS only produced comparatively lower disease responses (35.75 %) than those with QTL QSr.sun-5A, while DH lines carrying the two genes in combination showed lower stem rust responses (20.6 %) than lines with no gene or a single gene (Table 4). The mean stem rust severity of 20 lines (54.1 %) with no QTL was higher than lines with any QTL, whether present singly or in combination. Further analysis was performed using one-way ANOVA to see whether there were statistically significant differences in the effects of the QTLs and their combination. The results indicated that there was statistical difference between groups of all means except those of QTL QSr.sun-3BS and QSr.sun-5A.
Discussion
The study supported the presence of Sr5 and Sr31, two major seedling stem rust resistance genes, in Rialto. In contrast, Spark did not carry seedling resistance genes effective against the pathotypes used in this study. Previously, Singh et al. (2008a) also reported the presence of Sr5 and Sr31 in Rialto. Gene Sr5 is often cited as the “immunity” gene; while the infection type conferred by this gene is usually very low (“0” to “;”), it gives relatively higher infection types when transferred to susceptible genetic backgrounds (Luig and Rajaram 1972). Virulence to Sr5 occurs in many geographical areas and hence the gene is usually not effective in the field (Luig 1983; Vanegas et al. 2008). Linkage analysis based on the phenotypic data generated against Pgt pathotype 21-0 mapped Sr5 on chromosome arm 6DS associated with DArT marker wPt-3879 (data not shown). Previously, this gene was also mapped to the same region with the flanking markers barc183 and wPt-3879 (Prins et al. 2011). The resistance gene Sr31 is located in the 1BL/1RS translocation and has contributed significantly to protecting wheat crops from stem rust. It is very common in European, Chinese, USA and Indian wheat varieties (McIntosh et al. 1995) and has been deployed extensively worldwide, and remained effective and was considered durable until the detection of Pgt pathotype TTKSK (“Ug99”) in Uganda, which carries virulence for this gene (Pretorius et al. 2000; Das et al. 2006; Singh et al. 2008c). However, this gene is still used effectively in some wheat-growing countries where Sr31 virulence does not occur. Although effective in Australia, Sr31 has not been used widely in Australia due to the association of the rye introgression with a problem of dough stickiness (McIntosh 1988).
This study detected three consistent QTLs on chromosome arms 1BS, 3BS and chromosome 5A, all of which contributed field resistance and reduced stem rust severity significantly. Because Sr31 is effective against Australian Pgt pathotypes, the QTL detected on chromosome arm 1BS is most likely due to the major seedling resistance gene Sr31 or, less likely, a gene conferring APR. Previously, Letta et al. (2013) reported a highly significant QTL in tetraploid wheat associated with stem rust resistance near the centromere of chromosome 1B. The authors explained that in the case of hexaploid wheat, the markers identified in their study could tag either Sr14 in the centromeric region of 1B (McIntosh 1980) or Sr31 in the short arm of 1B (Zeller 1973). Through monosomic analysis, Sr14 was mapped on the long arm of chromosome 1B (McIntosh 1980). Letta et al. (2013) concluded that the QTL detected on chromosome 1B is likely Sr14 because this gene originated from tetraploid wheat, while Sr31 is present in hexaploid wheat germplasm only. In another study, a QTL associated with stem rust resistance was reported in a hexaploid spring wheat near the centromere of chromosome 1BS (Yu et al. 2011). Yu et al. (2012) also reported that some of the stem rust resistance genes identified in spring wheat were not found in winter wheat because they represent different gene pools. Thus, further testing and mapping of the common markers could help to clearly understand and determine whether the QTL QSr.sun-1BS and Sr31 are equivalent.
The second consistent QTL, QSr.sun-3BS, was located on chromosome arm 3BS where it mapped close to the SSR marker gwm1034. Previously, several stem rust QTLs have been reported on chromosome arm 3BS in different populations (Kaur et al. 2009; Haile et al. 2012; Njau et al. 2013; Singh et al. 2013a). However, in all of these studies, the authors stated that the QTL identified was likely Sr2. In a separate study (Getie 2015), the parents and a resistant bulk of ten lines carrying QSr.sun-3BS were screened with two Sr2 linked markers gwm533 (Spielmeyer et al. 2003) and csSr2 (Mago et al. 2011). Marker analysis based on these two markers indicated the absence of Sr2 in both parents and DH lines tested. Sr2 is also associated with the dark pigmentation trait pseudo-black chaff (PBC) (Hare and McIntosh 1979). In this study, PBC was not detected in either of the parents or any of the DH lines. Combined, these results indicate that the APR observed in Spark and in the population is highly unlikely to be controlled by Sr2. Studies of APR to leaf rust in the Spark/Rialto population (Getie 2015) found a QTL on the short arm of chromosome 3B at a genetic distance of 6.2 cM from the SSR marker gwm1034. However, the phenotypic data for leaf rust segregated independently of that for stem rust and therefore it is unlikely that the QTL located on chromosome arm 3BS for stem rust is associated with the gene identified in the same region as conferring APR to leaf rust. The third consistent QTL detected on chromosome 5A for APR to stem rust resistance was very minor and contributed by the moderately susceptible parent Spark. Although consistently detected, the LOD values of QSr.sun-5A were lower than the threshold. This could be either that the effect of the QTL is very minor or the presence of lower number of markers in this region. Use of more tightly linked markers could greatly improve the QTL mapping resolution and the LOD values. Stem rust QTLs in chromosome 5A were also reported in previous studies in different genetic backgrounds (Bhavani et al. 2011; Njau et al. 2013; Singh et al. 2013b; Letta et al. 2014). However, because of differences in the markers linked to these QTLs, it is difficult to determine whether QTL QSr.sun-5A detected in this study is same as these previously reported QTLs. Further work is therefore required to resolve the relationships between these loci.
Stem rust responses of lines carrying QSr.sun-3BS and QSr.sun-5A in combination were lower than in lines that carried either of the genes, clearly indicating the additive nature of the QTLs. This was further determined by one-way ANOVA that the combinations of the two QTLs were statistically significant from either of the QTLs based on LSD value obtained. Singh et al. (2011b) reported that cultivars carrying combinations of 4–5 APR genes, each with minor to intermediate effects, could provide a high level of resistance in the field. In view of this, there would be significant merit in combining both of these QTLs with other major or APR genes to stem rust resistance including Sr2/Yr30, Lr34/Yr18/Sr57/Pm38, Lr46/Yr29/Sr58/Pm39 and Lr67/Yr46/Sr55/Pm46. Minor genes with additive effects to the rust diseases are common in wheat germplasm (Johnson 1988; Singh et al. 2011b) and sufficient levels of APR are usually provided by several minor genes, each with a small to intermediate effect in reducing disease severity (Singh et al. 2008b). In order to provide adequate level of resistance in wheat varieties, the QTLs identified in this study can be useful in combination with other major or minor genes for developing germplasm with durable resistance to stem rust.
References
Bansal UK, Bossolini E, Miah H, Keller B, Park RF, Bariana HS (2008) Genetic mapping of seedling and adult plant stem rust resistance in two European winter wheat cultivars. Euphytica 164:821–828
Bansal UK, Bariana HS, Wong D, Randhawa M, Wicker T, Hayden M, Keller B (2014) Molecular mapping of an adult plant stem rust resistance gene Sr56 in winter wheat cultivar Arina. Theor Appl Genet 127:1441–1448
Bariana H, Willey N, Venkata B, Lehmenseik A, Standen G, Lu M (2004) Breeding methodology to achieve durability for rust resistance in wheat. In: Black CK, Panozzo JF, Rebetzke GJ (eds) Proceedings of the 54th Australian cereal chemistry conference and 11th wheat breeders assembly’. Canberra, ACT. Wheat Breeding Society of Australia, pp 8–12
Beteselassie N, Fininsa C, Badebo A (2007) Sources of resistance to stem rust (Puccinia graminis f. sp. tritici) in Ethiopian tetraploid wheat accessions. Genet Resour Crop Evol 54:337–343
Bhavani S, Singh RP, Argillier O, Huerta-Espino J, Singh S, Njau P, Brun S, Lacam S, Desmouceaux N (2011) Mapping durable adult plant stem rust resistance to the race Ug99 group in six CIMMYT wheats. In: McIntosh R (ed) Proceedings of Borlaug global rust initiative technical workshop. Saint Paul, Minnesota, pp 43–53
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B 363(1491):557–572
Das BK, Saini A, Bhagwat SG, Jawali N (2006) Development of SCAR markers for identification of stem rust resistance gene Sr31 in the homozygous or heterozygous condition in bread wheat. Plant Breeding 125:544–549
Doerge RW, Churchill GA (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142:285–294
Getie B (2015) Identification, genetic studies and molecular characterisation of resistance to rust pathogens in wheat. PhD Thesis, The University of Sydney, NSW, Australia
Haile JK, Röder MS (2013) Status of genetic research for resistance to Ug99 race of Puccinia graminis f. sp. tritici: a review of current research and implications. Afr J Agric Res 8:6670–6680
Haile JK, Miloudi MN, Hammer K, Badebo A, Röder MS (2012) QTL mapping of resistance to race Ug99 of Puccinia graminis f. sp. tritici in durum wheat (Triticum durum Desf.). Mol Breed 30:1479–1493. doi:10.1007/s11032-012-9734-7
Hale I, Mamuya I, Singh D (2014) Sr31-virulent races (TTKSK, TTKST, and TTTSK) of the wheat stem rust pathogen Puccinia graminis f. sp. tritici are present in Tanzania. Mol Breed 34:871–881
Hare RA, McIntosh RA (1979) Genetic and cytogenetic studies of durable adult-plant resistances in Hope and related cultivars to wheat rusts. Zeitschrift fur Pflanzenzuchtung 83:350–367
Herrera-Foessel SA, Lagudah ES, Huerta-Espino J, Hayden MJ, Bariana HS, Singh D, Singh RP (2011) New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor Appl Genet 122:239–249
Herrera-Foessel SA, Singh R, Lillemo M, Huerta-Espino J, Bhavani S, Singh S, Lan C, Calvo-Salazar V, Lagudah ES (2014) Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor Appl Genet 127:781–789
Hodson D (2015) Ethiopia- update on localized stem rust epidemic. Rust tracker.org: a global wheat rust monitoring system. CIMMYT Wheat Atlas Country Page. http://rusttracker.cimmyt.org/?page_id=40. Accessed 10 Feb 2015
Johnson R (1988) Durable resistance to yellow (stripe) rust in wheat and its implications in plant breeding. In: Simmonds NW, Rajaram S (eds) Breeding strategies for resistance to the rusts of wheat. CIMMYT, Mexico, pp 63–75
Kaur N, Street K, Mackay M, Yahiaoui N, Keller B (2008) Molecular approaches for characterization and use of natural disease resistance in wheat. Eur J Plant Pathol 121:387–397
Kaur J, Bansal UK, Khanna R, Saini RG, Bariana HS (2009) Molecular mapping of stem rust resistance in HD2009/WL711 recombinant inbred line population. Int J Plant Breed 3:28–33
Khan R, Bariana H, Dholakia B, Naik S, Lagu M, Rathjen A, Bhavani S, Gupta V (2005) Molecular mapping of stem and leaf rust resistance in wheat. Theor Appl Genet 111:846–850
Klindworth DL, Miller JD, Jin Y, Xu SS (2007) Chromosomal locations of genes for stem rust resistance in monogenic lines derived from tetraploid wheat accession ST464. Crop Sci 47:1441–1450
Kosambi D (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Letta T, Maccaferri M, Badebo A, Ammar K, Ricci A, Crossa J, Tuberosa R (2013) Searching for novel sources of field resistance to Ug99 and Ethiopian stem rust races in durum wheat via association mapping. Theor Appl Genet 126:1237–1256
Letta T, Olivera P, Maccaferri M, Jin Y, Ammar K, Badebo A, Salvi S, Noli E, Crossa J, Tuberosa R (2014) Association mapping reveals novel stem rust resistance loci in durum wheat at the seedling stage. Plant Genome 7:1–13
Liu W, Rouse M, Friebe B, Jin Y, Gill B, Pumphrey MO (2011) Discovery and molecular mapping of a new gene conferring resistance to stem rust, Sr53, derived from Aegilops geniculata and characterization of spontaneous translocation stocks with reduced alien chromatin. Chromosome Res 19:669–682
Luig NH (1983) A survey of virulence genes in wheat stem rust, Puccinia graminis f. sp. tritici. Advances in plant breeding, supplement 11 to journal of plant breeding. (Paul Parey: Berlin.). In: McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes
Luig N, Rajaram S (1972) The effect of temperature and genetic background on host gene expression and interaction to Puccinia graminis tritici. Phytopathology 62:1171–1174
Mago R, Bariana HS, Dundas IS, Spielmeyer W, Lawrence GJ, Pryor AJ, Ellis JG (2005) Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor Appl Genet 111:496–504
Mago R, Zhang P, Bariana HS, Verlin Dc, Bansal UK, Ellis JG, Dundas IS (2009) Development of wheat lines carrying stem rust resistance gene Sr39 with reduced Aegilops speltoides chromatin and a simple PCR marker for marker-assisted selection of the gene. Theor Appl Genet 119:1441–1450
Mago R, Brown-Guedira G, Dreisigacker S, Breen J, Jin Y, Singh R, Appels R, Lagudah ES, Ellis J, Spielmeyer W (2011) An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theor Appl Genet 122:735–744
Mago R, Verlin D, Zhang P, Bansal U, Bariana H, Jin Y, Ellis J, Hoxha S, Dundas I (2013) Development of wheat–Aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2S#1 chromosome. Theor Appl Genet 126:2943–2955
Manly KF, Cudmore RH Jr, Meer JM (2001) Map manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
McIntosh R (1980) Chromosome location and linkage studies involving the wheat stem rust resistance gene Sr14. Cereal Res Commun 8:315–320
McIntosh RA (1988) The role of specific genes in breeding for durable stem rust resistance in wheat and triticale. In: Simmonds NW, Rajaram S (eds) Breeding strategies for resistance to the rusts of wheat. CIMMYT, Mexico, pp 1–9
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. CSIRO, Canberra
McIntosh R, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xia XC, Azul B (2014) Catalogue of gene symbols for wheat: 2013–2014 supplement. http://maswheat.ucdavis.edu/CGSW/2013-2014_Supplement.pdf
Nazari K, Mafi M, Yahyaoui A, Singh R, Park R (2009) Detection of wheat stem rust (Puccinia graminis f. sp. tritici) race TTKSK (Ug99) in Iran. Plant Dis 93:317
Njau PN, Bhavani S, Huerta-Espino J, Keller B, Singh RP (2013) Identification of QTL associated with durable adult plant resistance to stem rust race Ug99 in wheat cultivar ‘Pavon 76’. Euphytica 190:33–44
Park RF (2007) Stem rust of wheat in Australia. Aust J Agric Res 58:558–566
Park RF (2008) Breeding cereals for rust resistance in Australia. Plant Pathol 57:591–602
Periyannan S, Bansal UK, Hayden MJ, Dvorak J, Lagudah ES, Bariana HS (2008) Identification of markers linked with stem rust resistance genes Sr33 and Sr45. In: Appels R, Eastwood R, Lagudah E, Langridge P, Mackay M, McIntyre L, Sharp P (eds) The 11th international wheat genetics symposium proceedings, Brisbane, Queensland, Australia
Periyannan S, Bansal UK, Bariana HS, Pumphrey M, Lagudah ES (2011) A robust molecular marker for the detection of shortened introgressed segment carrying the stem rust resistance gene Sr22 in common wheat. Theor Appl Genet 122:1–7
Periyannan S, Bansal U, Bariana HS, Deal K, Ming-C Luo, Dvorak J (2014) Identification of a robust molecular marker for the detection of the stem rust resistance gene Sr45 in common wheat. Theor Appl Genet 127:947–955
Peterson RF, Campbell AB, Hannah AE (1948) A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can J Res 26:496–500
Pretorius ZA, Singh RP, Wagoire WW, Payne TS (2000) Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis 84:203
Pretorius Z, Bender C, Visser B, Terefe T (2010) First report of a Puccinia graminis f. sp. tritici race virulent to the Sr24 and Sr31 wheat stem rust resistance genes in South Africa. Plant Dis 94:784
Prins R, Pretorius ZA, Bender CM, Lehmensiek A (2011) QTL mapping of stripe, leaf and stem rust resistance genes in a Kariega × Avocet S doubled haploid wheat population. Mol Breed 27:259–270
Qi LL, Pumphrey MO, Friebe B, Qian C, Bowden RL, Rouse MN, Jin Y, Gill BS (2011) A novel Robertsonian event leads to transfer of a stem rust resistance gene (Sr52) eff ective against race Ug99 from Dasypyrum villosum into wheat. Theor Appl Genet 123:159–167
Roelfs AP, Singh RP, Saari EE (1992) Rust diseases of wheat: concepts and methods of disease management. CIMMYT, Mexico, pp 1–81
Saari EE, Prescott JM (1985) World distribution in relation to economic losses. In: Roelfs AP, Bushnell WR (eds) The cereal rusts, Vol. II: Diseases, distribution, epidemiology and control. Academic Press, Orlando, pp 259–298
Shuangye W, Pumphrey M, Bai G (2009) Molecular mapping of stem-rust resistance gene Sr40 in wheat. Crop Sci 49:1681–1686
Simmonds J, Scott P, Leverington-Waite M, Turner AS, Brinton J, Korzun V, Snape J, Uauy C (2014) Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat (Triticum aestivum L.). BMC Plant Biol 14:191
Singh RP, Rajaram S (2002) Breeding for disease resistance in wheat. In: Curtis BC, Rajaram S, Gomez Macpherson H (eds) Bread wheat. FAO plant production and protection series (FAO), no. 30. FAO, Rome, Italy, pp 317–330
Singh R, Huerta-Espino J, Rajaram S, Barna B, Kiraly Z (2000) Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol Hung 35:133–139
Singh RP, William HM, Huerta-Espino J, Rosewarne G (2004) Wheat rust in Asia: meeting the challenges with old and new technologies. In: New directions for a diverse planet: proceedings of the 4th international crop science congress. September 26–October 1, 2004, Brisbane, Australia
Singh RP, Hodson DP, Jin Y, Huerta-Espino J, Kinyua MG, Wanyera R, Njau P, Ward RW (2006) Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour 54:1–13
Singh D, Park R, McIntosh R, Bariana H (2008a) Characterisation of stem rust and stripe rust seedling resistance genes in selected wheat cultivars from the United Kingdom. J Plant Pathol 90:553–562
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Njau P, Wanyera R, Herrera-Foessel SA, Bhavani S, Singh D, Singh PK (2008b) Global status of Ug99 spread and efforts to mitigate the threat. In: Singh GP, Prabhu KV, Singh AM (eds) Proceeding of international conference on wheat stem rust Ug99—a threat to food security. Indian Agricultural Research Institute, New Delhi, pp 1–85
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Njau P, Wanyera R, Herrera-Foessel SA, Ward RW (2008c) Will stem rust destroy the world’s wheat crop? Adv Agron 98:271–309
Singh D, Simmonds J, Park RF, Bariana HS, Snape JW (2009) Inheritance and QTL mapping of leaf rust resistance in the European winter wheat cultivar ‘Beaver’. Euphytica 169:253–261
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V (2011a) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481
Singh RP, Huerta-Espino J, Bhavani S, Herrera-Foessel SA, Singh D, Singh PK, Velu G, Mason RE, Jin Y, Njau P (2011b) Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 179:175–186
Singh R, Herrera-Foessel S, Huerta-Espino J, Bariana H, Bansal U, McCallum B, Hiebert C, Bhavani S, Singh S, Lan C, Lagudah E (2012) Lr34/Yr18/Sr57/Pm38/Bdv1/Ltn1 confers slow rusting, adult plant resistance to Puccinia graminis tritici. In: Chen WQ (ed) Proceedings of the 13th international cereal rusts and powdery mildews conference, Beijing, China
Singh A, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Campbell HL, Singh D, Bhavani S, Fetch T, Clarke F (2013a) Identification and mapping in spring wheat of genetic factors controlling stem rust resistance and the study of their epistatic interactions across multiple environments. Theor Appl Genet 126:1951–1964
Singh S, Singh RP, Bhavani S, Huerta-Espino J, Eugenio L-VE (2013b) QTL mapping of slow-rusting, adult plant resistance to race Ug99 of stem rust fungus in PBW343/Muu RIL population. Theor Appl Genet 126:1367–1375
Spielmeyer W, Sharp P, Lagudah E (2003) Identification and validation of markers linked to broad-spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.). Crop Sci 43:333–336
Spielmeyer W, McIntosh RA, Kolmer J, Lagudah ES (2005) Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor Appl Genet 111:731–735
Suenaga K, Singh RP, Huerta-Espino J, William HM (2003) Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology 93:881–890
Vanegas CDG, Garvin DF, Kolmer JA (2008) Genetics of stem rust resistance in the spring wheat cultivar Thatcher and the enhancement of stem rust resistance by Lr34. Euphytica 159:391–401
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang S, Basten C, Zeng Z (2007) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
William HM, Singh RP, Huerta-Espino J, Ortiz-Islas S, Hoisington D (2003) Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 93:153–159
Yu L-X, Lorenz A, Rutkoski J, Singh RP, Bhavani S, Huerta-Espino J, Sorrells ME (2011) Association mapping and gene-gene interaction for stem rust resistance in CIMMYT spring wheat germplasm. Theor Appl Genet 123:1257–1268
Yu L-X, Morgounov A, Wanyera R, Keser M, Singh SK, Sorrells M (2012) Identification of Ug99 stem rust resistance loci in winter wheat germplasm using genome-wide association analysis. Theor Appl Genet 125:749–758
Zeller FJ (1973) 1B/1R wheat-rye chromosome substitutions and translocations. In: Sears ER, Sears LMS (eds) Proceedings of the 4th international wheat genetics symposium. Agricultural Experiment Station, College of Agriculture, University of Missouri, Columbia, Missouri, USA, pp 209–221
Zhang W, Olson E, Saintenac C, Rouse M, Abate Z, Jin Y, Akhunov E, Pumphrey M, Dubcovsky J (2010) Genetic maps of stem rust resistance gene Sr35 in diploid and hexaploid wheat. Crop Sci 50:2464–2474
Acknowledgments
The Bill and Melinda Gates Foundation and the Grains Research Development Corporation, Australia, financially supported this study. We are thankful to Mr. Matthew Williams for providing technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Getie, B., Singh, D., Bansal, U. et al. Identification and mapping of resistance to stem rust in the European winter wheat cultivars Spark and Rialto. Mol Breeding 36, 114 (2016). https://doi.org/10.1007/s11032-016-0537-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0537-0