Abstract
Races of stem rust fungus pose a major threat to wheat production worldwide. We mapped adult plant resistance (APR) to Ug99 in 141 lines of a PBW343/Muu recombinant inbred lines (RILs) population by phenotyping them for three seasons at Njoro, Kenya in field trials and genotyping them with Diversity Arrays Technology (DArT) markers. Moderately susceptible parent PBW343 and APR parent Muu displayed mean stem rust severities of 66.6 and 5 %, respectively. The mean disease severity of RILs ranged from 1 to 100 %, with an average of 23.3 %. Variance components for stem rust severity were highly significant (p < 0.001) for RILs and seasons and the heritability (h 2) for the disease ranged between 0.78 and 0.89. Quantitative trait loci (QTL) analysis identified four consistent genomic regions on chromosomes 2BS, 3BS, 5BL, and 7AS; three contributed by Muu (QSr.cim-2BS, QSr.cim-3BS and QSr.cim-7AS) and one (QSr.cim-5BL) derived from PBW343. RILs with flanking markers for these QTLs had significantly lower severities than those lacking the markers, and combinations of QTLs had an additive effect, significantly enhancing APR. The QTL identified on chromosome 3BS mapped to the matching region as the known APR gene Sr2. Four additional QTLs on chromosomes 1D, 3A, 4B, and 6A reduced disease severity significantly at least once in three seasons. Our results show a complex nature of APR to stem rust where Sr2 and other minor slow rusting resistance genes can confer a higher level of resistance when present together.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Ug99 race group of wheat stem rust fungus (Puccinia graminis Pers. f. sp. tritici Eriks. & E. Henn.) is documented as one of the foremost threats to wheat production and food security (Singh et al. 2011). The 1B/1R translocation carries resistance genes Lr26, Sr31, Yr9 and became widespread in wheat varieties and breeding materials developed at CIMMYT and various other countries during the 1980s and 1990s (Singh et al. 2008). The stem rust resistance gene Sr31 present on the translocation provided protection against all prevalent races of stem rust fungus until the appearance of the Ug99 race (Jin et al. 2008; Pretorius et al. 2010). Ug99 designated as TTKS on North American stem rust differential set, and some of its variants are the only known races that have virulence for Sr31 (Singh et al. 2011). The same race was re-designated as TTKSK when characterized on a fifth differential host set (Jin et al. 2008). This race is not only virulent to most Sr genes of wheat origin but also to the alien gene Sr38 brought into wheat from T. ventricosum (Singh et al. 2011). This unique virulence of Ug99 renders most wheat varieties susceptible worldwide. In 2006 and 2007, variants of Ug99 virulent on Sr24 and Sr36, respectively, were detected in Kenya (Jin et al. 2008, 2009). The combined virulence to Sr31 and Sr24 (race PTKST) was also identified in South Africa from isolates collected in 2009 (Pretorius et al. 2010). The widespread susceptibility of wheat varieties and germplasm was highlighted when the original strain spread to Yemen and Sudan in 2006, and fears of a spread into Asia were confirmed when this race was detected in Iran in 2007 (Nazari et al. 2009). This has raised concern of a major epidemic that could cause significant losses in wheat growing countries on all continents, as most popular varieties currently grown are susceptible to the Ug99 race group.
One of the aims of the CIMMYT wheat breeding program is to develop new wheat germplasm with durable resistance to stem rust. Identification and transfer of new sources of race-specific resistance from various wheat relatives is also underway to enhance the diversity for resistance. The race-specific resistance genes are involved in host-pathogen recognition in which these genes trigger a hypersensitive response when challenged by pathogen races that carry the corresponding avirulence alleles. However, a simple mutation in the pathogen renders the resistance gene ineffective (Stakman 1915; Singh et al. 2011). In germplasm with multiple effective resistance genes, multiple mutations from avirulence to virulence must occur in the same spore to overcome the resistance genes in a single event, which is considered implausible.
Adult plant resistance (APR) to stem rust based on multiple slow rusting, minor genes is another promising approach to achieve durable resistance. The resistance is readily detected in post-seedling growth stages and associated with non-race-specific resistance (Hare and McIntosh 1979; Knott 1982). Sources of quantitative APR in crop plants have proven to be durable, making it a promising breeding target for long-term stem rust resistance (Parlevliet 2002). However, the integration of this type of APR into adapted wheat germplasm is difficult because slow rusting is multi-genic (Knott 1982). Difficulties in phenotyping APR further obscure the breeding process because effective major genes interfere with the identification of APR. Marker-assisted selection (MAS) enables plant scientists to pyramid quantitative APR genes to enhance disease resistance durability (Varshney and Tuberosa 2007; William et al. 2007). Additionally MAS can also be used in pyramiding APR genes with major genes, which is almost impossible by conventional methods. Efforts are ongoing in developing wheat varieties resistant to Ug99, using molecular markers to pyramid several resistance genes, as well as APR slow rusting genes to reduce the probability of resistance breakdown by pathogen evolution (Leonard and Szabo 2005; Kuchel et al. 2007; Singh et al. 2011). Several sources of APR to Ug99 were identified in CIMMYT spring bread wheat germplasm (Njau et al. 2010) and mapping studies have identified genomic regions that contribute to APR (Bhavani et al. 2011; Yu et al. 2011).
The objectives of our study were: (1) to identify chromosomal regions associated with APR slow rusting QTLs in a spring bread wheat recombinant inbred line (RIL) population derived from the cross of moderately susceptible wheat ‘PBW343’ with APR wheat ‘Muu’, and (2) saturate the genomic regions on chromosome 5BL using SSR markers.
Materials and methods
Plant materials
A mapping population consisting of 141 RILs (F5 derived) was developed from a cross between moderately susceptible parent PBW343 and APR parent Muu. PBW343, a major variety in India, is a selection (GID2430154) from CIMMYT line ‘Attila’ with the pedigree ‘Nord Deprez/VG9144//Kalyansona/Bluebird/3/Yaco/4/Veery#5’, whilst Muu has pedigree ‘Pfau/Weaver//Kiritati’ (GID5090613). Muu was found to be susceptible at seedling stage but adult plants showed low disease severity to Ug99 during multiple years of field trials in Kenya (Njau et al. 2010). Njau et al. (2010) also postulated the presence of slow rusting gene Sr2 in Muu based on the presence of molecular marker Xgwm533 and the expression of pseudo-black chaff phenotype that are linked to it.
Stem rust evaluation
The parents, highly susceptible check ‘Cacuke’, and the RIL population were evaluated for stem rust severity at the Kenya Agricultural Research Institute (KARI), Njoro, during three crop seasons (main- and off-season 2010, and main-season 2011). The RILs and parents were grown in a completely randomized design with two replicates. Field plots consisted of two 1 m rows spaced 20 cm apart with 0.5 m pathway. Approximately 60–70 seeds were sown in each plot. The experimental block was surrounded by a spreader consisting of varieties differentially susceptible to the Sr24 virulent variant TTKST. Hills of spreaders were also planted in the middle of the pathway on one side of each plot to facilitate uniform disease build-up and spread. A suspension of freshly collected urediniospores suspended in distilled water was injected into the spreaders (1–3 plants/m) on at least two occasions just prior to booting, using a hypodermic syringe.
Disease responses in the field were assessed twice, first when the susceptible check variety Cacuke displayed about 50–60 % stem rust severity and a second time at the peak period of disease development, when Cacuke displayed 100 % stem rust during the mid-dough stage of plant growth. The percent disease severity on stem was assessed according to the modified Cobb Scale (Peterson et al. 1948). Second rating was considered for QTL analysis.
Statistical analysis
An analysis of variance for disease severity in the experiments was performed using PROC GLM of SAS 9.1 (SAS Institute 1994). Genotypes (RILs) were considered fixed factors while seasons (environments) were considered random factors. Mean percent disease severity was calculated for each RIL family. Pearson’s and Spearman’s rank correlation coefficients were calculated using PROC CORR of SAS 9.1 (SAS Institute 1994). Heritability on an entry mean basis was calculated using the mixed model analysis previously described by Holland et al. (2003).
Molecular analysis
DNA was extracted from lyophilized leaf tissue following the procedure described by Singh and Bowden (2011). A Nano-Drop ND1000 spectrophotometer (Thermo Fisher Scientific Inc, USA) was used for quantification of DNA samples. For Diversity Arrays Technology (DArT) genotyping, 500–1,000 ng of restriction grade DNA, suspended in TE with a final concentration of 50–100 ng/μL, were sent to Triticarte Pty. Ltd., Canberra, Australia (www.triticarte.com.au) for genome profiling (Wenzl et al. 2006; Neumann et al. 2010). Loci were scored as present (1) or absent (0). The overall call rate for the population was approximately 95 % and the Q (estimate of marker quality) value for most markers was above 80 %.
Parental screening for polymorphism
We saturated chromosome 5BL genomic region that carried an APR QTL with polymorphic SSR/EST-STS markers. This region was chosen due to its identification in other mapping populations also (Bhavani et al. 2011). For enrichment of this target region, 150 SSR markers from chromosome 5B were evaluated for polymorphism between parents (Pestsova et al. 2001; Röder et al. 1998; Song et al. 2005; Sourdille et al. 2004). Polymorphic DNA markers were genotyped on the entire PBW343/Muu RIL population to enrich the region and identify PCR-based markers close to the APR QTL. The population was also genotyped with marker Sr2G9 that is known to be closely linked to APR gene Sr2 (Mago et al. 2011).
Linkage mapping and QTL analysis
Inclusive composite interval mapping (ICIM) software was used for constructing linkage maps (Li et al. 2007). A PBW343/Muu linkage map was constructed using 465 informative DArT marker loci. Adult plant stem rust severity was incorporated into a DArT-based linkage map of the population. The effect of a marker and the LOD was determined by 1,000 permutation tests at p ≤ 0.01; this threshold corresponded to a highly conservative test for declaring the presence of QTL. ICIM tests both additive and epistasis effects, and it avoids the possible increase of sampling variance and the complicated background marker selection process that is a common problem in composite interval mapping (CIM; Li et al. 2007). For additive mapping, ICIM has increased detection power, reduced false detection rate, and less biased estimates of QTL effects compared to CIM (Li and Wang 2008).
Results
Phenotypic evaluation
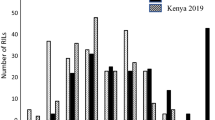
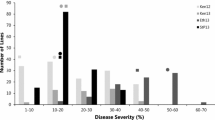
Adequate disease pressure was observed in all three crop seasons. Field evaluation revealed differences in stem rust rating between the parents and within the mapping population. The parents PBW343 and Muu differed significantly for stem rust responses (Fig. 1). The distribution of percent stem rust infection in the RIL population was continuous and skewed toward the lower percent infection across seasons. During 2010 main season the distribution appeared to be skewed toward resistance and somewhat bimodal, this could be due to the moderate effect of APR gene Sr2 derived from Muu. Sr2 is known to have an intermediate effect on APR (Singh et al. 2011). Resistance was hypothesized to be quantitatively inherited based on continuous distribution. The disease scores for the parents, RIL population means, population maximum and minimum for all seasons are given in Table 1. Mean stem rust severity of 5 % over seasons for the resistant parent Muu significantly differed from the susceptible mean stem rust severity of 66.6 % for moderately susceptible parent PBW343 (Table 1). The severity of RILs ranged from 1 to 100 % with a mean of 23.3 %. Variance components for stem rust severity were highly significant (p < 0.001) for RILs and seasons, however, the effects were non-significant for replication and interactions (Table 2). The heritability (h 2) for disease severity ranged from 0.78 to 0.89 across seasons. Percent infection among seasons was significantly (p < 0.001) correlated (Table 3). Correlation coefficients for disease severities ranged between 0.50 and 0.87 for three seasons indicative of a good resistance expression in all seasons. Results obtained from the correlation analysis indicated that data generated over the seasons and between replicates had significant relationships.
Quantitative mapping of APR to stem rust
Of the 1,355 DArT markers used for genotyping the RIL population, 465 were found to be polymorphic. Twenty-eight linkage groups were assigned to wheat chromosomes using published map locations of DArT markers as reference points. A DArT-based linkage map for the population, covering a total of 3,277 cM (1,322 cM for A genome, 1,680 cM for B genome, and 275 cM for D genome), was developed and used to identify the chromosome locations of QTL for stem rust resistance. Undetected QTL could exist on chromosomes with sparse marker coverage. ICIM analysis after integration of the rust data into the genetic map revealed eight significant QTL for stem rust resistance in total, however, only four were consistent over more than one seasons (Table 4). The consistent QTL (more than one environment) were located on chromosomes 2BS, 3BS, 5BL, and 7AS and designated as QSr.cim-2BS, QSr.cim-3BS, QSr.cim-5BL, and QSr.cim-7AS, respectively. All QTL were contributed by Muu, except 5BL QTL (contributed by PBW343). When the mean disease severity scores for the three seasons were used for QTL analysis, all four consistent QTLs were significant (Table 4). The three additional QTL on 3A, 4B, and 6A (from Muu), and a QTL on 1D (from PBW343) had significant effect only once in three seasons. Further verification is needed by testing the population for additional years or by genotyping with additional markers in the QTL regions.
The adjusted total R 2 obtained from all QTL effects varied from 52.3 to 65.5 % over seasons and in combined analysis. The DArT markers XwPt-9230 and XwPt-744022, flanking QTL on chromosome 2BS, were significantly associated with resistance during the 2010 off- and main-seasons with R 2 values of 5.3 and 6.2 %, respectively (Fig. 2a; Table 4). The DArT marker was converted into PCR-based assay (STS744022-2B-L AGC AGG GCA GCA CAC TCT AT and STS744022-2B-R CAA GGA TCT CCA AGC TGC AT). The QTL located on chromosome arm 3BS (Fig. 2b; Table 4) was consistent over seasons and explained 32.7–38.2 % of the phenotypic variance with LOD scores ranging from 12 to 15. DArT markers XwPt-3609 and Sr2G9 were associated with this QTL (Fig. 2c; Table 4). The QTL QSr.cim-5BL, on chromosome 5BL (derived from PBW343), gave significant effects in main-season 2011 and off-season 2010, and explained up to 10.9 % variation for disease severity. QTL QSr.cim-7AS also had consistent effects against stem rust across the seasons. This QTL was flanked with DArT markers XwPt-8043 and XwPt-2371 and explained up to 15.0 % phenotypic variation for the disease (Fig. 2d; Table 4).
Quantitative trait loci (QTL) interval maps for wheat chromosome 2B (QSr.cim-2B) (a), 3BS (QSr.cim-3BS) (b), 5BL (QSr.cim-5BL) (c), and 7A (QSr.cim-7A) (d) associated with stem rust resistance in PBW343/Muu recombinant inbred lines. The logarithm of the odds (LOD) score is indicated on the y axis; dotted line represents the LOD scores and colors green, blue, black, and red indicate Sr-OS-2010, Sr-MS-2010, Sr-MS-2011 and Sr-combined, respectively. Markers selected as co-factors highlighted in blue
To enrich the QSr.cim-5BL region with additional markers, 150 PCR-based markers located on this chromosome were screened for polymorphism between parents. Forty-nine markers showed polymorphism between the parents and were genotyped on the entire PBW343/Muu RIL population and incorporated into the DArT map. Seventeen markers from the long arm of the chromosome showed linkages with the resistance QTL QSr.cim-5BL (Fig. 2c). The APR QTL was placed on the long arm of chromosome 5B between markers Xwms604 and XwPt5896, within a distance of 6.6 cM (Table 4; Fig. 2c).
Discussion
Accurate phenotypic assessment of APR to stem rust is a prerequisite for QTL mapping. Therefore, high and uniform disease pressure in field trials is necessary for reliable phenotyping of APR. PBW343 showed significantly higher stem rust severity of 66.6 % than the 5 % average severity observed for Muu (Table 1). The PBW343/Muu RILs showed a continuous variation for stem rust severity from 1 to 100 % (Table 1; Fig. 1). This shows the quantitative nature of APR to stem rust in a mapping population. The transgressive segregation of RILs that were either more resistant or susceptible than parents indicated the presence of diverse APR QTL in both parents. In main-season 2010 and 2011, 12 and 2 RILs, respectively, showed significant transgressive segregation toward increased resistance and had an average disease severity lower than the parents. High heritability of resistance indicates reliability of the data over seasons.
Our study revealed four consistent (more than one environment) APR QTL for stem rust resistance, of which three (QSr.cim-2BS, QSr.cim-3BS, and QSr.cim-7AS) were contributed by Muu and one (QSr.cim-5BL) by PBW343. Previously, the only known locus involved in stem rust quantitative APR was Sr2, located on chromosome 3BS (Mago et al. 2011; McIntosh 1988). An APR slow rusting QTL (QSr.cim-2B) on 2B with significant effect on rust severity had a LOD score above 3.0 and explained 3.2–6.2 % of the resistance expressed in the PBW343/Muu population with an additive effect. Kolmer et al. (2011) also reported APR on 2BL in Thatcher, though DArT marker XwPt5044 was associated with significantly lower stem rust severity. Yu et al. (2011) reported DArT markers XwPt7750, XwPt8460, and XwPt7200 from chromosome 2B to be significantly associated with Ug99 stem rust resistance in CIMMYT spring wheat germplasm through association analysis. DArT marker XwPt8460 was 18 cM away from the SSR marker Xwmc175 linked to Sr9a. A likely new race-specific resistance gene effective to Ug99 in a similar location was reported by Hiebert et al. (2010) in the RL6071/Webster population. However, the APR QTL in Muu must be different from Sr9 locus and the race-specific gene identified by Hiebert et al. (2010) as it does not confer race-specific resistance in seedlings. The close linkage of QSr.cim-3BS with the APR gene Sr2, as identified using the marker Sr2G9 and the presence of pseudo black chaff phenotype in Muu, suggest that Sr2 and QSr.cim-3BS are identical (Mago et al. 2011; McIntosh et al. 2003; Hare and McIntosh 1979; Spielmeyer et al. 2003). The resistance QTL QSr.cim-5BL was contributed by the moderately susceptible parent PBW343 and explained 8.6–10.9 % of phenotypic variance. A QTL for stem rust resistance on chromosome arm 5BL in ‘Arina’ was reported by Bansal et al. (2008). However, our study reports a more precise location and flanking markers for the 5BL QTL can be used for further validation in other populations. The QTL QSr.cim-7AS, not reported earlier, contributed 15 % of the resistance expressed in the population in 2011 (Table 4). Over three seasons, its average contribution to the phenotype was 7.5 %, with an additive effect of 3.6. APR genes underlying QSr.cim-2BS, QSr.cim-5BL, and QSr.cim-7AS represent important additions to the sources of APR to the Ug99 race group. The consistent detection of these QTL across experiments and their significant contribution toward reducing stem rust severity suggested that these QTL are distinct APR slow rusting genes.
RILs carrying a combination of QSr.cim-2BS, QSr.cim-3BS (or Sr2), QSr.cim-5BL, and QSr.cim-7AS, based on flanking markers, had lower stem rust responses, indicating the additive nature of these APR genes. APR genes with minor but additive effects on stem rust, stripe rust, and leaf rust are common in the wheat germplasm (Johnson 1988; William et al. 2006; Lillemo et al. 2008; Njau et al. 2010). Comparatively fewer studies have been conducted for APR to stem rust, compared to the other two wheat rusts. Wheat germplasm showing high to adequate APR levels usually has several minor genes, each with a small to intermediate effect in reducing disease severity (Knott 1988; Singh et al. 2011). The RIL population PBW343/Muu was segregated for the slow rusting APR gene Sr2, which interacted with other slow rusting QTL (QSr.cim-2BS, QSr.cim-5BL, and QSr.cim-7AS) identified in our study. We identified RILs that carried these QTL in various combinations and compared their stem rust responses with the contrasting counterparts. As expected, average stem rust severities of RILs with the resistance QTL were significantly lower than RILs without these QTL.
It is important to evaluate the contribution and interaction of slow rusting QTL with slow rusting APR gene Sr2. Yu et al. (2011) reported significant interactions of resistance loci, including Sr2 locus on 3BS, with other loci in the same and different chromosomes in spring wheat germplasm. Our results also revealed an additive interaction between Sr2 and 5BL QTL; RILs with these two QTL had lower disease severities compared to RILs that did not carry them or carried only one of them. PBW343, the moderately susceptible parent, had an average disease severity of 65 %, compared to 100 % for the susceptible check Cacuke and some of the RILs, suggesting that some alleles from PBW343 contributed to resistance in the RIL population. The QTL allele on 5BL of PBW343, with an increased phenotypic effect, in combination with the QTL alleles from Muu, resulted in transgressive RILs that had significantly lower disease severity of 1 % than the 5 % severity for the resistant parent Muu (Fig. 1). This can be explained by the fact that these few RILs combined QTL from Muu and PBW343. Similar observations were reported by Schnurbusch et al. (2003) where the susceptible parent ‘Forno’ contributed to resistance to Stagonospora glume blotch resistance. It has been reported that Sr2 contributes to APR through its interaction with other unknown minor genes (Singh et al. 2011). Interactions of Sr2 in our study support this hypothesis, though further specific validation is required to determine the contribution of each QTL.
The 5BL genomic region was saturated with SSR/EST-STS markers to develop PCR-based markers linked to QSr.cim-5BL to assist breeding for the resistance. Two closely linked molecular markers, Xwms604 and XwPt5896, were identified. The average phenotypic variation explained from QSr.cim-5B was 9.5 % and the QTL allele was contributed by PBW343. This is a very significant contribution from an APR slow rusting minor gene. Both PBW343 and Muu also carry pleiotropic slow rusting resistance gene Lr46/Yr29/Pm39 (unpublished results Sukhwinder-Singh), which confers slow rusting resistance to leaf rust, yellow rust, and powdery mildew. Recent studies have shown that this resistance gene also contributes to APR to stem rust (Bhavani et al. 2011; Yu et al. 2011), however, we could not determine the effect of Lr46/Yr29/Pm39 in the mapping population due to its homozygosity. Four additional minor QTL were also determined in single year analyses and the possibility of their role in conferring resistance cannot be ruled out because the mean severity of RILs lacking the four consistent QTL was only 48.8 %, lower than the mean severity for the moderately susceptible parent PBW343. These findings confirm previous reports that accumulating 4–5 minor APR slow rusting QTLs can reduce rust severity to negligible levels (Bhavani et al. 2011; Singh et al. 2008). ICIM has an advantage of detecting interactive QTLs where the phenotypic trait-enhancing alleles are contributed by the alternate parent in a bi-parental cross. The individual effects of such alleles contributing to minor QTL are usually undervalued, but combinations of such genes results in reduced disease severity. We conclude that slow rusting APR to stem rust is complex, however, high levels of resistance can be achieved by combining multiple minor genes.
References
Bansal UK, Bossolini E, Miah H, Keller B, Park RF, Bariana HS (2008) Genetic mapping of seedling and adult plant stem rust resistance in two European winter wheat cultivars. Euphytica 164:821–828
Bhavani S, Singh RP, Argillier O, Huerta-Espino J, Singh S, Njau P, Brun S, Lacam S, Desmouceaux N (2011) Mapping durable adult plant stem rust resistance to the race Ug99 group in six CIMMYT wheats. In McIntosh R (ed) Proceedings of borlaug global rust initiative, Technical Workshop, Saint Paul, Minnesota, USA, pp 43–53, 13–16 June 2011
Hare RA, McIntosh RA (1979) Genetics and cytogenetic studies of durable adult-plant resistance in ‘Hope’ and related cultivars to wheat rusts. Z Pflanzenzuchtung 83:350–367
Hiebert CW, Thomas JB, McCallum BD, Humphreys DG, DePauw RM, Hayden MJ (2010) An introgression of wheat chromosome 4DL in RL6077 (Thatcher*6/PI250413) confers adult plant resistance to stripe rust and leaf rust (Lr67). Theor Appl Genet 122:239–249
Holland JB, Nyquist WE, Cervantes-Martinez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Jin Y, Pretorius ZA, Singh RP, Fetch T (2008) Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis 92:923–926
Jin Y, Szabo L, Rouse M, Fetch T, Pretorius ZA, Wanyera R, Njau P (2009) Detection of virulence to resistance gene Sr36 within race TTKS lineage of Puccinia graminis f. sp. tritici. Plant Dis 93:367–370
Johnson R (1988) Durable resistance to yellow (stripe) rust in wheat and its implications in plant breeding. In: Simmonds NW, Rajaram S (eds) Breeding strategies for resistance to the rusts of wheat. CIMMYT, Mexico, pp 63–75
Knott DR (1982) Multigenic inheritance of stem rust resistance in wheat. Crop Sci 22:393–399
Knott DR (1988) Using polygenic resistance to breed for stem rust resistance in wheat. In: Simmonds NW, Rajaram S (eds) Breeding strategies for resistance to the rusts of wheat. CIMMYT, Mexico, pp 39–47
Kolmer JA, Garvin DF, Jin Y (2011) Expression of a Thatcher wheat adult plant stem rust resistance QTL on chromosome arm 2BL is enhanced by Lr34. Crop Sci 51:526–533
Kuchel H, Fox R, Reinheimer J, Mosionek L, Willey N, Bariana H, Jefferies S (2007) The successful application of a marker-assisted wheat breeding strategy. Mol Breed 20(4):295–308
Leonard KJ, Szabo LJ (2005) Stem rust of small grains and grasses caused by Puccinia graminis. Mol Plant Pathol 6(2):99–111
Li H, Wang J (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260
Li H, Ye GY, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:1–14
Lillemo M, Asalf B, Singh RP, Huerta-Espino J, Chen XM, He ZH, Bjørnstad Å (2008) The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor Appl Genet 116:1155–1166
Mago R, Brown-Guedira G, Dreisigacker S, Breen J, Jin Y, Singh R, Appels R, Lagudah ES, Ellis J, Spielmeyer W (2011) An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theor Appl Genet 122:735–744
McIntosh RA (1988) The role of specific genes in breeding for durable stem rust resistance in wheat and triticale. In: Simmonds NW, Rajaram S (eds) Breeding strategies for resistance to the rusts of wheat. CIMMYT, Mexico, pp 1–9
McIntosh RA, Yamazaki Y, Devos KM, Dubcovsky J, Rogers WJ, Appels R (2003) Catalogue of gene symbols for wheat. In: Pogna NE, Romano M, Pogna A, Galterio G (eds) Proceedings of the 10th international wheat genetics symposium, Paestum, Italy
Nazari K, Mafi M, Yahyaoui A, Singh RP, Park RF (2009) Detection of wheat stem rust (Puccinia graminis f. sp. tritici) race TTKSK (Ug99) in Iran. Plant Dis 93:317
Neumann K, Kobiljski B, Denčić S, Varshney RK, Börner A (2010) Genome-wide association mapping: a case study in bread wheat (Triticum aestivum L.). Mol Breed 27(1):37–58
Njau PN, Jin Y, Huerta-Espino J, Keller B, Singh RP (2010) Identification and evaluation of sources of resistance to stem rust race Ug99 in wheat. Plant Dis 94:413–419
Parlevliet JE (2002) Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124:147–156
Pestsova E, Ganal MW, Röder MS (2001) Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43:689–697
Peterson RF, Campbell AB, Hannah AE (1948) A diagrammatic scale for estimating rust intensity of leaves and stem of cereals. Can J Res Sect C26:496–500
Pretorius ZA, Bender CM, Visser B, Terefe T (2010) First report of a Puccinia graminis f. sp. tritici race virulent to the Sr24 and Sr31 wheat stem rust resistance genes in South Africa. Plant Dis 94:784
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
SAS Institute (1994) The SAS system for Windows. Release 6.10 SAS Inst., Cary, NC
Schnurbusch T, Paillard S, Fossati D, Messmer M, Schachermayr G, Winzeler M, Keller B (2003) Detection of QTLs for Stagonospora glume blotch resistance in Swiss winter wheat. Theor Appl Genet 107:1226–1234
Singh S, Bowden RL (2011) Molecular mapping of adult-plant race-specific leaf rust resistance gene Lr12 in bread wheat. Mol Breed 28:137–142
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Njau P, Wanyera R, Herrera-Foessel SA, Ward RW (2008) Will stem rust destroy the world’s wheat crop? Adv Agron 98:271–309
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel SA, Singh PK, Singh S, Velu G (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat. Funct Integr Genomics 4:12–25
Spielmeyer W, Sharp PJ, Lagudah ES (2003) Identification and validation of markers linked to broad spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.). Crop Sci 43:333–336
Stakman EC (1915) Relation between Puccinia graminis and plants highly resistant to its attack. J Agr Res 4:193–299
Varshney RK, Tuberosa R (2007) Genomics-assisted crop improvement: an overview. In: Varshney RK, Tuberosa R (eds) Genomics assisted crop improvement, vol I., Genomics approaches and platformsSpringer, Dordrecht, pp 1–12
Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesna J, Cakir M, Poulsen D, Wang J, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A (2006) A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and phenotypic traits. BMC Genomics 7:206
William HM, Singh RP, Huerta-Espino J, Palacios G, Suenaga K (2006) Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49:977–990
William HM, Trethowan R, Crosby-Galvan EM (2007) Wheat breeding assisted by markers: CIMMYT’s experience. Euphytica 157:307–319
Yu L, Liu S, Anderson A, Singh RP, Jin Y, Dubcovsky J, Brown Guedira GL, Bhavani S, Morgounov A, He Z, Heurta-Espino J, Sorrells M (2011) Haplotype diversity of stem rust resistance loci in uncharacterized wheat lines. Mol Breed 26:667–680
Acknowledgments
We acknowledge financial resources from the Durable Rust Resistant Wheat Project led by Cornell University and supported by the Bill & Melinda Gates Foundation. Editing assistance received from Emma Quilligan is also highly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Ordon.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, S., Singh, R.P., Bhavani, S. et al. QTL mapping of slow-rusting, adult plant resistance to race Ug99 of stem rust fungus in PBW343/Muu RIL population. Theor Appl Genet 126, 1367–1375 (2013). https://doi.org/10.1007/s00122-013-2058-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2058-0