Abstract
Aluminum (Al) toxicity is a major constraint on crop production in acid soils around the world. Hexaploid oat (Avena sativa L.) possesses significant Al tolerance making it a good candidate for production in these environments. Genetic improvement for Al tolerance in oat has traditionally been achieved through conventional plant breeding and could be enhanced by marker-assisted selection. The objectives of this study were to develop a chromosome-anchored genetic map for an oat recombinant inbred population and to identify SNP markers linked to quantitative trait loci (QTL) affecting root growth response to Al. Three QTL on chromosomes 7C-17A, 13A, and 19A conferring Al tolerance were identified using primary root regrowth of recombinant inbred lines derived from the cross between UFRGS 17 (Al tolerant) and UFRGS 930598-6 (Al sensitive). Localization of each QTL onto the sequenced rice genome revealed the genetic region on chromosome 13A might be associated with a putative malate transporter locus (LOC_Os06g15779). Studies of root apex tissue indicated that exudation of malate was increased in the Al-tolerant parent UFRGS17 and not in the sensitive parent. Based on these data, the malate transporter might be a candidate gene responsible for one of the Al tolerance QTL identified in this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum (Al) is the third most prevalent metal in the earth’s crust and is predominately insoluble and non-toxic to plants at a soil pH above 5.5. At a pH below 5.5, Al becomes soluble, especially as the Al3+ cation, and becomes toxic to plants (Kobayashi et al. 2013; Merino-Gergichevic et al. 2010) by reducing cell wall, plasma membrane, and cytoskeleton stability and inhibiting cell division and elongation in root (Ryan et al. 2011; Sivaguru et al. 2000). Stunted roots result in reduced nutrient and water uptake, thereby restricting plant growth (Foy et al. 1978). Overall, Al toxicity in low pH soils has a major impact on crop production.
Surface liming is a common practice for ameliorating topsoil acidity in Al-laden soils. This approach is limited under no-tillage agriculture where lime is applied only to the soil surface. In this situation, acidic conditions will occur in the subsoil layers (Rossiello and Netto 2006) resulting in Al toxicity. Development of more tolerant crops to Al could provide a more comprehensive and economical alternative. In several crop species, genetic variability for Al tolerance has been exploited to develop Al-tolerant varieties and to study the function of the genes involved in the tolerance (Collins et al. 2008).
Two mechanisms have been reported for Al tolerance in plants including exclusion from the root apex by secretion of organic acids and cellular tolerance (Kochian et al. 2005). In wheat (Triticum aestivum L.), barley (Hordeum vulgare L.), and sorghum (Sorghum bicolor L. Moench), Al tolerance has been attributed to the action of a single dominant gene, coding proteins that participate in the transport of organic acid (Delhaize et al. 2007; Magalhães et al. 2007; Minella and Sorrells 1997). However, in the wheat cv. Chinese Spring, three quantitative trait loci (QTL) that enhanced root growth under Al stress were identified, suggesting that inheritance of Al tolerance is a more complex (Ma et al. 2006). Similar to Arabidopsis thaliana and maize (Zea mays L.), in rice (Oryza sativa L.) and soybean (Glycine max L.), a polygenic inheritance is suggested by the identification of nine and five Al tolerance QTL, respectively (Bianchi-Hall et al. 2000; Maron et al. 2010; Nguyen et al. 2001). Genetic studies suggest the involvement of one or two genes in response to Al tolerance in oat (Avena sativa L.) (Castilhos et al. 2011; Nava et al. 2006; Sanchez-Chacon et al. 2000; Wagner et al. 2001). Oat, an allohexaploid species, with 2n = 6x = 42 chromosomes, originated from the aggregation of three ancestral diploid genomes AA, CC, and DD (Rines et al. 2006). Oat is an important cereal crop worldwide with a majority of the production focused in temperate climates of Europe and North America (Murphy and Hoffman 1992). In subtropical environments, oat plays an important role for grain production during the winter season in no-tillage rotation with soybean. It is considered an Al-tolerant species.
The identification of DNA markers tightly linked to Al tolerance genes or QTL in mapping populations provides efficient tools for the application of marker-assisted selection (MAS) in breeding programs and may help identify the mechanism associated with tolerance (Silva-Navas et al. 2012; Wang et al. 2007). The availability of genetic markers for oat is limited, because of the difficulty in developing linkage maps due to the large size and complexity of the genome (Arumuganathan and Earle 1991; Portyanko et al. 2001; Wight et al. 2003). In addition, numerous chromosome rearrangements disrupted the expected colinearity among oat sub-genomes (Jellen et al. 1994). Recently, substantial effort was directed to the generation of high-resolution maps for oat. A significant contribution in oat molecular biology was delivered in 2011 with the development of first oat single nucleotide polymorphism (SNP) markers. This achievement was followed by the development of the first Illumina GoldenGate SNP array in 2013, which facilitated the first physically anchored map for hexaploid oat representing all 21 chromosomes (Oliver et al. 2013). The GoldenGate SNP array was upgraded to the Illumina Infinium array targeting 4975 loci across the oat genome (Tinker et al. 2014). The objectives of this study were to develop a chromosome-anchored genetic map of a Brazilian oat population using SNP markers and to identify markers linked to QTL that affect the response to Al tolerance in oat.
Materials and methods
Plant material
A genetic population of recombinant inbred lines (RILs) derived from the cross between UFRGS 17 (COR2/CTZ3/PENDEK/ME1563/76-29-76-23/75-28/CI833) and UFRGS 930598-6 (UFRGS15/UFRGS881920) was analyzed in this study. The population consisted of 162 F5-derived RILs developed by single-seed descent. This population was selected based on the differential response to Al toxicity of the parental lines (Nava et al. 2006; Oliveira et al. 2005). Parental lines UFRGS 17 (Al tolerant) and UFRGS 930598-6 (Al sensitive) were developed by the oat breeding program at the Federal University of Rio Grande do Sul (UFRGS), Brazil.
Phenotypic screening for Al tolerance
Parental lines and RILs from the cross UFRGS 17 × UFRGS 930598-6 were evaluated in hydroponic solutions under controlled conditions as described in previous study (Nava et al. 2006). In brief, the root regrowth after Al exposure of 159 RILs was evaluated in a complete randomized design. Each replicate consisted of a sample of about 10 seedlings per RIL. The hydroponic solutions consisted of an Al treatment and Al-free control. The Al-free solution contained 4 mM Ca(NO3)2·4H2O, 2 mM MgSO4·7H2O, 4 mM KNO3, 0.435 mM (NH4)2SO4, 0.5 mM KH2PO4, 2 µM MnSO4·1H2O, 0.3 µM CuSO4·5H2O, 0.8 µM ZnSO4·7H2O, 30 µM NaCl, 0.1 µM Na2MoO4·2H2O, and 10 µM H3BO3. The iron source was a high-performance iron chelate, comprised of 6 % of chelate iron and 92 % of an orthoisomer, with 0.9 µM of iron in the solution. The Al solution was identical to the Al-free solution with the exceptions of KCl in place of KH2PO4 to prevent Al precipitation and a tenfold reduction in the overall concentration. The Al source was aluminum sulfate (Al2(SO4)3·18H2O) at the final concentration of 740 µM.
Oat seedlings were first grown for 48 h in the hydroponic Al-free solution and then transferred to the aluminum solution for 48 h. After Al treatment, the seedlings were transferred back to the Al-free solution for an additional 72 h. Root growth reinitiated after removal from Al solution, and root regrowth of the primary root from each seedling was measured starting at the point of root thickening. The availability and activity of Al3+ in the solution was calculated using the software Visual MINTEQ 3.0 (Gustafsson 2010).
Aluminum localization in root apex across sections
Seeds from both parental lines with similar size were selected for Al localization studies. Lemma and palea tissues were manually removed, and the seeds were soaked for 1 min in a solution containing 0.75 % sodium hypochlorite and 0.05 % Tween 20. Seeds were washed five times with ddH2O, laid on germination paper, and incubated at 21 °C in the dark. After germination, seedlings with similar root size (2 mm) were transferred to a hydroponic solution containing either 0 or 500 µM Al. After 14 days, the roots were washed three times with ddH2O for 20 min and stained with hematoxylin 2 g L−1 and KIO3 0.2 g L−1 for 30 min. The excess of dye was removed by successive washes with ddH2O, and the roots were fixed in ethanol and stored in 50 % glycerol. Root tips were sectioned and analyzed using an optical microscope model Olympus BX41. Three independent biological replicates were used, each one containing 10 plants.
Aluminum and malate quantification
Seedlings from each parental line were transferred to small pots containing 40 mL of Al-free hydroponic solution and incubated for 5 days to allow root development. Seedlings were then transferred to either a new Al-free solution or a solution containing 500 µM Al. After 24 h, the hydroponic solutions were collected, frozen, lyophilized (L101, Liotop) to concentrate the solutes, and used to measure malate secreted by the plants with the l-malic acid enzymatic assay kit (Megazyme, Wicklow, Ireland). The roots and shoots from each plant were washed with ddH2O, separated, and macerated, and the amount of Al was measured by inductively coupled plasma mass spectrometry (ICP-MS). Three independent biological replicates were used, each one containing 20 plants.
Statistical analysis
The data compiled were submitted to analysis of variance (ANOVA), and the differences between the means were compared by t test at P ≤ 0.05.
DNA isolation and purification
Etiolated shoots of each parental line and RIL were harvested 7 days after planting, frozen in liquid nitrogen, and ground. DNA extraction was performed using a cetyl trimethyl ammonium bromide (CTAB)-based protocol as follows: Ground tissue was homogenized with 0.5 mL of extraction buffer containing 0.15 M sorbitol, 0.22 M Tris–HCl pH 8.0, 23 mM EDTA pH 8.0, 0.8 M NaCl, 0.8 % (w/v) CTAB, and 1 % (w/v) sarcosine, at 65 °C for 25 min. DNA was extracted with 0.3 mL of chloroform/isoamyl alcohol solution (24:1), precipitated with chilled 70 % isopropanol, washed with ethanol 70 % twice, dried overnight, and resuspended in TE pH 8.0. DNA was quantified using NanoDrop (ND 1000 Spectrophotometer) and diluted to 150 ng L−1.
KASPar-based SNP analysis
Single nucleotide polymorphism markers (SNPs) from the Illumina GoldenGate Oligonucleotide Assay (Oliver et al. 2013) were used for genotyping the UFRGS 17/UFRGS 930598-6 mapping population based on KASPar technology (LGC, UK). A total of 960 contigs containing SNP loci were selected based on the polymorphism across the original oat diversity panel. KASPar SNP assays were then designed using the primer designer software PrimerPicker (KBioscience Ltd., Hoddesdon, UK) with default parameters. SNP genotyping was performed on the Fluidigm (Fluidigm Corp., South San Francisco, CA) nanofluidic 96.96 dynamic array (Wang et al. 2009) using competitive allele-specific PCR specified by the KASPar genotyping chemistry (KBioscience Ltd., Hoddesdon, UK). For genotyping on the 96.96 dynamic array chip, a 5-μL sample mix, consisting of 2.25 μL genomic DNA (20 ng/μL), 2.5 μL of 2× KASP reagent mix (KBioscience Ltd.), and 0.25 μL of 20× GT sample loading reagent, was prepared for each DNA sample. Similarly, a 4-μL 10× KASP assay, containing 0.56 μL of the KASP assay primer mix (allele-specific primers 12 μM, common reverse primer 30 μM), 2 μL of 2× assay loading reagent, and 1.44 μL DNase-free water, was prepared for each SNP assay. The assay mix and sample mix were then loaded onto a 96.96 dynamic array chip, mixed, and subjected to polymerase chain reaction (PCR) using an IFC Controller HX and FC1 thermal cycler (Fluidigm Corp., South San Francisco, CA) according to the manufacture’s protocols. PCR consisted of an initial denaturing step (70 °C—30 min; 25 °C—10 min), a hot-start Taq polymerase activation step (94 °C—15 min), followed by a touchdown amplification protocol as follows: 10 cycles of 94 °C for 20 s, 65 °C for 1 min (decreasing 0.8 °C per cycle), 26 cycles of 94 °C for 20 s, 57 °C for 1 min, hold at 20 °C for 30 s. Endpoint fluorescent images of the chip were acquired on an EP-1 imager (Fluidigm Corp., South San Francisco, CA) and the data analyzed with Fluidigm SNP Genotyping Analysis Software.
Polymorphisms were scored across 153 of the 162 RILs. Chi-square tests were performed on each locus for goodness of fit to the expected 1:1 segregation ratio. Markers that deviated significantly (P < 0.05) from the expected segregation ratio were excluded for linkage analysis and map construction.
Linkage mapping
A linkage map was developed for the mapping population using MapMaker/EXP version 3.0 software (Lander et al. 1987). Mapping information was available for 115 polymorphic markers that were known to belong to 15 of the 21 chromosomes of the oat genome, from the oat consensus map (Oliver et al. 2013). Markers were anchored to their corresponding chromosome, according to the reference map, and the software command compare was used to calculate all possible marker order within each group and the maximum likelihood of the map. After the calculation of optimized distances between markers, 11 of those 115 markers were removed because they were causing distortion. Linkage groups were recalculated with 30 unanchored markers using LOD 3, and the Kosambi’s mapping function was used to convert the recombination frequency into centimorgans (cM), and twelve of those were integrated into the map, totalizing 116 integrated markers in the map. Chromosomes 11A, 14D, 17A-7C, and 19A contained gaps and were broken into two linkage groups each to prevent distortion in the QTL analysis due to the large interval between markers. The final map was visualized using Mapchart 2.2 (Voorrips 2002).
QTL analysis
Quantitative trait loci (QTL) associated with primary root regrowth were detected using the single marker analysis and the simplified composite interval mapping procedure (sCIM) in MQTL (Tinker and Mather 1995). Stepwise regression with a threshold value of P = 0.05 was used to select cofactors for sCIM. Parameters for analysis were an interval of 0.5 cM and an experiment-wise Type 1 error rate of 5 %. In order to identify significant QTL, an experiment-wide false-positive rate was established by running 10,000 random permutations, for details see Churchill and Doerge (1994). The test statistic was then converted to a LOD score by multiplying with 0.22 (Tinker and Mather 1995). QTL intervals were assigned to the area within one LOD score from the QTL peak.
Results
Phenotypic screening for Al tolerance
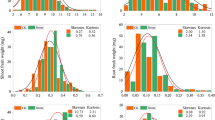
Twenty-eight percent of the Al in our experiments was in the Al3+ form (52.8 µM). The parental lines displayed very diverse response to Al. Roots of UFRGS 17 (tolerant) were significantly longer (t test, P ≤ 0.05) than those of UFRGS 930598-6 (sensitive). The mean root regrowth of UFRGS 17 was 22.9 mm, the minimum regrowth observed was 16 mm, and the maximum regrowth observed was 26 mm, whereas UFRGS 930598-6 displayed mean root regrowth of 2.6 mm, the minimum regrowth observed was 1.2 mm, and the maximum regrowth observed was 4.0 mm. The primary root regrowth distribution from the UFRGS 17/UFRGS 930598-6 mapping population varied from 1.2 to 48 mm (Fig. 1) with a mean and standard deviation of 10.6 and 9.5 mm, respectively. The heritability of primary root regrowth after Al exposure for this inbred population was 0.90.
UFRGS 930598-6 accumulates more aluminum in root apex than UFRGS 17
The concentration of Al was higher (t test, P ≤ 0.05) in the roots of UFRGS 930598-6 and UFRGS 17 (1001.63 and 616.25 µg g−1, respectively) than in the shoots (209.07 and 113.00 µg g−1, respectively), after 24 h of Al exposure. Less Al was observed by hematoxylin stain in the UFRGS 17 root apex than in the UFRGS 930598-6 apex after 14 days of exposure to 500 µM Al. External portions of roots were colored by oxidation of the hematoxylin–Al3+ complexes except the meristem region of UFRGS 17 roots (Supplementary Figure S1a). The analysis of sequential cross sections (1 mm from the root tip) showed difference in the Al localization between the parental lines. In UFRGS 17, the Al was restricted to outer cells, while it was observed throughout the cortex tissue in UFRGS 930598-6 (Supplementary Figure S1b).
UFRGS 17 exudes malate in response to aluminum
UFRGS 17 roots exuded significantly higher (t test, P < 0.001) amounts of malate after 24 h of exposure to Al than UFRGS 930598-6. UFRGS 17 exuded 49.33 ± 9.45 nmol/plant of malate in response to Al, whereas no measurable malate amounts were exuded by UFRGS 930598-6. Measurable malate exudation was not detected in the Al-free control for either parental line even in the absence of phosphorus.
KASPar SNP analysis and linkage map
A total of 960 SNP markers were evaluated in this study. From these, 145 were polymorphic among UFRGS 17/UFRGS 930598-6 RILs. Due to distortion in the Mendelian segregation analysis, 29 of the 145 polymorphic markers were removed. The 116 remaining SNP loci (80 %) were assembled into 19 linkage groups, which corresponded to 15 chromosomes on the physically anchored oat map (Table 1). Of the 15 corresponding groups, 1C, 3C, 4C, 5C, 6C, 7C-17A, 8A, 9D, 11A, 12D, 13A, 14D, 16A, 19A, and 20D were represented. Some linkage groups were very small, like in the linkage group 7C-17A, where the markers found GMI_ES15_444 and ES17_c18155_185 cosegregated with BA_grs_14655_201. The largest linkage group was chromosome 5C, which covered 180 cM and contained 22 markers (Fig. 2). Based on the marker location in the oat consensus map and to prevent distortion in the QTL analysis due to large interval between markers, two independent linkage groups were associated with each one of the chromosomes 7C-17A, 11A, 14D, and 19A. Overall, the linkage map assembled in this study covered 32, 62, and 28 % of the genomes A, C, and D, respectively, when compared to the oat consensus map (Table 1).
Locations and estimations of QTL
The statistical test established the threshold for QTL significance (LR) as 11.5 (LOD = 2.5). Three QTL were associated with Al tolerance based on the phenotypic variation in the root regrowth. The QTL were located on chromosomes 7C-17A, 13A, and 19A(2) (Fig. 2). Chromosome 7C-17A contained the QTL peak at the marker BA_grs_14655_201 with a test statistic (LR) of 16.9 (LOD = 3.72) and explaining 20.2 % of the phenotypic variation for root regrowth among assessed lines. The linkage group only consisted of three markers that were tightly linked (≤1.5 cM). A second QTL (LR of 15.7 or LOD = 3.45) explaining 17 % of variation was located between 0 and 4.1 cM on chromosome 13A with the nearest marker GMI_ES15_c2451_165. The third QTL was located between 12 and 53 cM on chromosome 19A (LR of 21.4 or LOD = 4.71) and explained 33.8 % of the phenotypic variation (Table 2). Since this linkage group had only three markers with a large distance from each other, the position of this QTL is less precise, and its peak was at the marker GMI_ES02_c435_145.
Discussion
In order to develop Al-tolerant oat varieties to cope with the negative effects caused by soil acidity, it is important to understand the genetic and molecular factors involved. Currently, the mechanism of Al tolerance in higher plants is only partially understood. In wheat, the tolerance is based on the expression of a malate transporter gene, ALMT1 (Sasaki et al. 2006). In barley, a citrate transporter gene HvAACT1 is required for tolerance (Zhou et al. 2013). A 1-kb DNA insertion upstream of the HvAACT1 gene in some Al-tolerant genotypes (Fujii et al. 2012) and a 21-bp indel have been suggested as causes of the difference between tolerant and sensitive barley genotypes (Bian et al. 2013). In oats, however, the genes related to Al tolerance have not yet been identified.
Al3+ activity in a hydroponic solution has efficiently discriminated tolerance in the parental lines UFRGS 17 and UFRGS 930598-6 as measured by root regrowth in this study. Root regrowth demonstrates the resistance of the root apical meristem to the toxic effects of Al. In fact, this trait is well correlated with plant Al tolerance in field; therefore, it represents an efficient and fast screening methodology (Nava et al. 2015). UFRGS 17 had significantly higher root regrowth than UFRGS 930598-6. This result was expected since Al tolerance in UFRGS 17 has been reported previously (Castilhos et al. 2011; Hervé et al. 2013; Nava et al. 2006). Earlier studies suggested that Al tolerance in UFRGS 17 may involve an external mechanism reducing the uptake of Al by the root apex as well as the induction of an antioxidant mechanism to cope with the oxidative stress caused by Al (Castilhos et al. 2011; Pereira et al. 2013).
Our results showed higher Al concentration in the roots than in the shoots for both parental lines. The root meristem region of UFRGS 17, however, seemed to be more protected from Al entrance. This was suggested by the reduced amount of Al in internal cells over UFRGS 930598-6 based on color formation from the hematein/Al complex. Possible Al detoxification mechanisms may include: (1) selective permeability of the plasmatic membrane to reduce Al uptake (Archambault et al. 1997), (2) proteins that are connected to Al secretion (Basu et al. 1999), (3) phosphate efflux (Huan-Xin et al. 2009; Pellet et al. 1996; Zheng et al. 2005), (4) secretion of phenolic compounds (Ofei-manu et al. 2001), and (5) organic acids (Liu et al. 2009), which chelate Al ions. Initially, citrate was considered the main organic acid exudate for Al tolerance (Zheng et al. 1998) by A. sativa roots; however, recent studies have shown only malate to increase its secretion in response to Al (Radmer et al. 2012). Our data showing malate exudation by Al in roots of UFRGS 17 and not UFRGS 930598-6 roots support this recent work.
The amount of malate secreted by UFRGS 17 roots was similar to those detected from other tolerant oat genotypes in response to Al (Radmer et al. 2012). Therefore, UFRGS 930598-6 may have a defective malate transporter mechanism or a disruption in the Al-response pathway. The presence of other concomitant mechanisms cannot be discarded. Sensitive oat genotypes sharing the same hydroponic solution with tolerant ones do not differ from those grown in vessels containing only sensitive plants. It suggests that the organic acid secreted in the solution by the tolerant genotype is not enough to reduce the amount of toxic Al ions in the whole solution. Therefore, either there is a mechanism other than organic acid secretion involved in the tolerance or a significant amount of organic acid is retained in the apoplast and/or in the mucilage of tolerant genotypes.
Despite the phenotypic diversity observed between the parental lines as well as among the RILs, only a small number of polymorphic markers were found (15 %). This number is consistent with results from Oliver et al. (2013) where 16 % of the SNP loci interrogated by the Illumina GoldenGate assay were polymorphic across six different mapping populations on average. Low polymorphism is a common characteristic to species with large genomes (Kulcheski et al. 2010; Li et al. 2000; Yu and Herrmann 2006). It has been suggested that the low genetic polymorphism found in crops such as oat and wheat is due to the evolution of the species (Bryan et al. 1999). The polyploidy nature of the oat and wheat genomes could also be a major factor based on SNP assays binding to more than one genome. Illumina’s platform was developed based on Avena spp., and choosing genotypes with significant genetic distance increases the usefulness of the platform. However, it is expected to have lower efficiency for comparison between genotypes from the same species and developed using the same germplasm.
About 20 % of the polymorphic SNP markers exhibited distorted segregation ratio within the genetic population. Distortion may occur from several reasons, such as preferential fertilization, DNA breakage during extraction, amplification of other genome regions or analysis error, and occasionally, it is a true biological phenomenon (Tan et al. 2001). Biological reasons for distortion include physical nearness between markers and genes involved in either gamete formation or genotype viability after fertilization (Vogl and Xu 2000). Because distorted ratios impact the association between markers and the segregation data for the phenotypic characteristic, causing false positives, it is recommended their exclusion from the data before the linkage map development. Our results indicate that the use of KASPar assays in place of the Illumina GoldenGate for scoring oat SNP markers was feasible and efficient. KASPar assays have the advantage of being cheaper and therefore speed up the gain of information about different populations.
The genetic map covered 736 cM (one marker every 6.3 cM, on average). This map coverage represents about 37 % of the total Avena sativa genome, based on the 2000 cM estimated by the consensus map (Oliver et al. 2013) and 25 % considering previous estimated sizes (2932 cM) (O’Donoughue et al. 1995). Despite the 19 linkage groups formed, 15 chromosomes were represented in our genetic map, when comparing the markers/chromosome assignments in the oat consensus map. The physically anchored consensus map was generated using monosomic lines to define the marker location in the hexaploid oat chromosomes (Oliver et al. 2013). In the same study, Oliver et al. (2013) found unexpected smaller numbers of markers located on chromosomes 10D, 17A-7C, and 21D, which were not represented in our genetic map. Both studies, Oliver’s and ours, were based on markers derived from the same diversity panel. The lack of markers on 17A-7C may result from reciprocal translocation (Jellen and Beard 2000). Clearly, genome C was the best represented in our map. It covered 62 % of the genome C length regarding the oat consensus map size. The best coverage of genome C could be explained on the belief that the genomes A and D are very similar to each other. The new chip recently developed by Illumina containing 6000 SNP markers will be useful for increasing the map coverage (Tinker et al. 2014). It is foreseen that genotypes developed in a same location would carry similar genetic adaptation blocks, not only as a result of similar background but also because these blocks are essential for adaptation to the local environmental conditions, reducing polymorphism in those regions. A more drastic example is the oat genome D, which seems to have higher genetic conservation than the other two, indicated by a lower level of polymorphism and the prevalence of major oat disease resistance genes (Oliver et al. 2013). The “genome asymmetry” suggests that one genome may have played a more important role than the others during evolution and domestication (Peng et al. 2003).
Three QTL for Al tolerance were identified on three different chromosomes in the UFRGS 17/UFRGS 930598-6 mapping population explaining 71 % of the phenotypic variation. Previous studies support our results by indicating Al tolerance is conferred by one or two dominant genes in A. sativa (Castilhos et al. 2011; Nava et al. 2006; Oliveira et al. 2005; Wagner et al. 2001). It is important to point out that root regrowth is not a discrete trait and a large variation has been observed among genotypes and individuals from the same genotype (Hervé et al. 2013). Root regrowth may be influenced by genes related to root growth rate and their interaction with the environment, including the plant response to phosphate limitation. Aluminum interacts with phosphate forming a precipitate interfering with Al availability; therefore, to analyze Al tolerance in hydroponic solution, phosphate must be omitted. As a result, phosphate starvation responses may have been induced as well. A phosphate starvation response can include reduction in root cell division and elongation. In the diploid species A. strigosa, four QTL were found affecting Al tolerance (Wight et al. 2003). Moreover, the Al tolerance in wheat was also considered a monogenic trait, and more than one QTL has been found suggesting a more complex inheritance, due to one or two major loci and several minor interacting loci (Navakode et al. 2009).
BLAST was used to localize EST-based SNP loci from oat onto related species with sequenced genomes. SNP loci in two regions contained sequences similar to genes deposited in the NCBI, but none of them showed similarity to genes involved in Al tolerance in other species. GMI_ES15_c2451_165 and GMI_ES15_c12378_444 were similar to XM_003557226.1, an oxidase-like protein, and to XM_003568689.1, an ABA receptor PYL8-like from Brachypodium distachyon, respectively. The only oat SNP marker orthologous to rice was GMI_ES15_c2451_165 on oat chromosome 13A. This locus shared 87 % homology to the orthologous sequence LOC_Os06g01490 on rice chromosome 6.
Currently, there is no expressive information about mechanisms, genes or even loci related to Al tolerance in oat. The only published work was on the diploid species A. strigosa and, as previously mentioned, identified four QTL associated with Al tolerance (Wight et al. 2003). Two of them mapped to homologous rice chromosomes 3, 5, 10, and 12 containing Al tolerance genes (Wight et al. 2003). Rice chromosome 3 shows microsynteny to Al tolerance important regions in wheat chromosome 4DL, where the gene TaALMT1 is located, as well as in barley chromosome 4H, HvMATE location, and rye chromosome 7RS, where ScALMT1 genes are situated (Collins et al. 2008; Raman et al. 2005; Wang et al. 2007). Comparative mapping allows the identification of orthologous regions affecting the trait across species and may help to identify the mechanism involved. Substantial macro-colinearity between oat and the model rice and B. dystachion genomes was identified recently (Oliver et al. 2013). The region of the QTL located at oat chromosome 7C-17A was collinear to the B. dystachyon chromosome 2, the region of the QTL at oat chromosome 13A was collinear to B. dystachyon chromosome 1 and rice chromosome 5, and finally, the one at oat chromosome 19A was collinear to B. dystachyon chromosome 2 and rice chromosome 1 (Oliver et al. 2013). At rice chromosome 1, a major QTL for Al tolerance exists and this region has extensive homology to chromosome 3 of Triticeae (Sorrells et al. 2003). Orthologous genes to the Al-related transcription factor, STOP1, were identified at B. dystachyon chromosome 2 and rice chromosome 1 (Genome sequencing and analysis of the model grass Brachypodium distachyon 2010; Project 2008; Consortium TRCS 2005). A MATE citrate transporter gene, OsFRDL4, conferring minor effects to Al tolerance was mapped to rice chromosome 1 (Yokosho et al. 2011). At rice chromosome 5, Nguyen et al. (2001) found a minor QTL for Al tolerance and the gene STAR2/ALS3 (ABC transporter) was identified (Huang et al. 2009). An orthologous of STAR2/ALS3 was found at B. dystachyon chromosome 2. Based on the parental lines differences in malate secretion in response to Al, we expected that at least one of the QTL would locate in a region collinear to rice chromosome 3. ALMT1 gene, the main malate transporter in the Triticeae, occurs in regions with high homology to rice chromosome 3 (Sorrells et al. 2003). Oat chromosome 19A also shows high homology to rice chromosome 3 (Oliver et al. 2013), however, not in the region of the identified QTL. Marker GMI_ES15_c2451_165 on oat chromosome 13A matched 87 % to orthologous sequence on the rice chromosome 6, where the gene LOC_Os06g01490 is located. This gene shows the same chromosome location as the gene LOC_Os06g15779, a putative malate transporter, and LOC_Os06g06440, an ABC transporter in rice genome. These gene families have been implicated in Al tolerance in other plant species; however, the distance between these genes in rice is large. LOC_Os06g01490 locates at chromosome 6 between 297 and 302 kbp, whereas LOC_Os06g06440 lies within 3001 and 3009 kbp and LOC_Os06g15779 within 8960 and 8964 kbp.
The identification of genetic markers near QTL for Al tolerance may facilitate the use of this tolerance in other crops. Introgression of Al resistance using marker-assisted backcrossing was obtained recently in barley (Soto-Cerda et al. 2013). There is a short distance between the markers associated with the QTL located at chromosomes 7C-17A and 13, which are candidates for further analysis. Furthermore, marker-assisted selection in oat has the potential to overcome the difficulties of conventional phenotypic selection, especially in environments that lack toxic levels of Al in the soil. The mapping may also help in the identification of the physiological mechanism involved in the Al tolerance, which is yet speculative in oats.
References
Archambault DJ, Zhang GC, Taylor GJ (1997) Spatial variation in the kinetics of aluminum (Al) uptake in roots of wheat (Triticum aestivum L.) exhibiting differential resistance to Al—evidence for metabolism-dependent exclusion of Al. J Plant Physiol 151:668–674
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–219
Basu U, Good AG, Taing-Aung T, Slaski J, Basu A, Briggs KG, Taylor GJ (1999) A 23 kDa protein root exudates polypeptide co-segregates with aluminium resistance in Triticum aestivum. Physiol Plant 106:53–61
Bian M et al (2013) Development of gene-specific markers for acid soil/aluminium tolerance in barley (Hordeum vulgare L.). Mol Breed 32:155–164. doi:10.1007/s11032-013-9859-3
Bianchi-Hall CM et al (2000) Aluminum tolerance associated with quantitative trait loci derived from soybean PI 416937 in hydroponics. Crop Sci 40:538–545. doi:10.2135/cropsci2000.402538x
Bryan GJ, Stephenson P, Collins A, Kirby J, Smith JB, Gale MD (1999) Low levels of DNA sequence variation among adapted genotypes of hexaploid wheat. Theor Appl Genet 99:192–198. doi:10.1007/s001220051224
Castilhos G, Farias JG, de Bernardi Schneider A, de Oliveira PH, Nicoloso FT, Chitolina Schetinger MR, Delatorre CA (2011) Aluminum-stress response in oat genotypes with monogenic tolerance. Environ Exp Bot 74:114–121. doi:10.1016/j.envexpbot.2011.05.007
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Collins NC, Shirley NJ, Saeed M, Pallotta M, Gustafson JP (2008) An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.). Genetics 179:669–682. doi:10.1534/genetics.107.083451
Consortium TRCS (2005) Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res 15:1284–1291. doi:10.1101/gr.3869505
Delhaize E, Gruber BD, Ryan PR (2007) The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett 581:2255–2262. doi:10.1016/j.febslet.2007.03.057
Foy CD, Chaney RL, White MC (1978) The physiology of metal toxicity in plants. Annu Rev Plant Physiol 29:511–566. doi:10.1146/annurev.pp.29.060178.002455
Fujii M et al (2012) Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 3:713. http://www.nature.com/ncomms/journal/v3/n3/suppinfo/ncomms1726_S1.html
Genome sequencing and analysis of the model grass Brachypodium distachyon (2010). Nature 463:763–768. http://www.nature.com/nature/journal/v463/n7282/suppinfo/nature08747_S1.html
Gustafsson JP (2010) Visual MINTEQ ver. 3.0
Hervé CB, Calai FA, Nava IC, Delatorre CA (2013) Tolerância ao alumínio tóxico em germoplasma brasileiro elite de aveia. Ciência Rural 43:1364–1370
Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21:655–667. doi:10.1105/tpc.108.064543
Huan-Xin J, Ning T, Jin-Gui Z, Yan L, Li-Song C (2009) Phosphorus alleviates aluminum-induced inhibition of growth and photosynthesis in Citrus grandis seedlings. Physiol Plant 137:298–311
Jellen E, Beard J (2000) Geographical distribution of a chromosome 7C and 17 intergenomic translocation in cultivated oat. Crop Sci 40:256–263
Jellen EN, Gill BS, Cox TS (1994) Genomic in situ hybridization differentiates between A/D- and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena). Genome 37:613–618. doi:10.1139/g94-087
Kobayashi Y et al (2013) Molecular and physiological analysis of Al3+ and H+ rhizotoxicities at moderately acidic conditions. Plant Physiol 163:180–192. doi:10.1104/pp.113.222893
Kochian LV, Piñeros MA, Hoekenga OA (2005) Physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274:175–195
Kulcheski FR, Graichen FAS, Martinelli JA, Locatelli AB, Federizzi LC, Delatorre CA (2010) Molecular mapping of Pc68, a crown rust resistance gene in Avena sativa. Euphytica 175:423–432
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li CD, Rossnagel BG, Scoles GJ (2000) The development of oat microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor Appl Genet 101:1259–1268. doi:10.1007/s001220051605
Liu J, Magalhães JV, Shaff J, Kochian LV (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. doi:10.1111/j.1365-313X.2008.03696.x
Ma H-X, Bai G-H, Lu W-Z (2006) Quantitative trait loci for aluminum resistance in wheat cultivar Chinese Spring. Plant Soil 283:239–249. doi:10.1007/s11104-006-0008-1
Magalhães JV et al (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminium tolerance in sorghum. Nat Genet 39:1156–1161
Maron LG et al (2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J 61:728–740. doi:10.1111/j.1365-313X.2009.04103.x
Merino-Gergichevic C, Alberdi M, Ivanov AG, Reyes-Diaz M (2010) Al+3–Ca+2 interaction in plants growing in acid soils: Al-phytotoxicity response to calcareous amendments. J Soil Sci Plant Nutr 10:217–243
Minella E, Sorrells ME (1997) Inheritance and chromosome location of Alp, a gene controlling aluminum tolerance in ‘Dayton’ barley. Plant Breed 116:465–469. doi:10.1111/j.1439-0523.1997.tb01032.x
Murphy JP, Hoffman LA (1992) The origin, history, and production of oat. In: Marshall HG, Sorrells ME (eds) Oat science and technology. ASA, CSSA Publishers, Madison, pp 1–28
Nava IC, Delatorre CA, Duarte I, Pacheco MT, Federizzi LC (2006) Inheritance of aluminum tolerance and its effects on grain yield and grain quality in oats (Avena sativa L.). Euphytica 148:353–358
Nava IC, Delatorre CA, Pacheco MT, Scheeren PL, Federizzi LC (2015) Aluminum tolerance of oat cultivars under hydroponic and acid soil conditions. Exp Agric. doi:10.1017/S0014479715000046
Navakode S, Weidner A, Lohwasser U, Röder MS, Börner A (2009) Molecular mapping of quantitative trait loci (QTLs) controlling aluminium tolerance in bread wheat. Euphytica 166:283–290. doi:10.1007/s10681-008-9845-8
Nguyen VT, Burow MD, Nguyen HT, Le BT, Le TD, Paterson AH (2001) Molecular mapping of genes conferring aluminum tolerance in rice (Oryza sativa L.). Theor Appl Genet 102:1002–1010. doi:10.1007/s001220000472
O’Donoughue LS et al (1995) A molecular linkage map of cultivated oat. Genome 38:368–380. doi:10.1139/g95-048
Ofei-manu P, Wagatsuma T, Ishikawa S, Tawaraya K (2001) The plasma membrane strength of the root-tip cells ans root phenolic compounds are correlated with Al tolerance in several common woody plants. Soil Sci Plant Nutr 47:359–375
Oliveira PH, Federizzi LC, Milach SCK, Gotuzzo C, Sawasato JT (2005) Inheritance in oat (Avena sativa L.) of tolerance to soil aluminum toxicity. Crop Breed Appl Biotechnol 5:125–133
Oliver RE et al (2013) SNP discovery and chromosome anchoring provide the first physically-anchored hexaploid oat map and reveal synteny with model species. PLoS ONE 8:e58068. doi:10.1371/journal.pone.0058068
Pellet DM, Papernik LA, Kochian LV (1996) Multiple aluminum-resistance mechanisms in wheat: roles of root apical phosphate and malate exudation. Plant Physiol 112:591–597
Peng J, Ronin Y, Fahima T, Röder MS, Li Y, Nevo E, Korol A (2003) Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci 100:2489–2494. doi:10.1073/pnas.252763199
Pereira LB et al (2013) Differential speed of activation in antioxidant system in three oat genotypes. J Inorg Biochem 128:202–207. doi:10.1016/j.jinorgbio.2013.07.025
Portyanko VA, Hoffman DL, Lee M, Holland JB (2001) A linkage map of hexaploid oat based on grass anchor DNA clones and its relationship to other oat maps. Genome 44:249–265
Project RA (2008) The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res 36:D1028–D1033. doi:10.1093/nar/gkm978
Radmer L, Tesfaye M, Somers D, Temple S, Vance C, Samac D (2012) Aluminum resistance mechanisms in oat (Avena sativa L.). Plant Soil 351:121–134. doi:10.1007/s11104-011-0937-1
Raman H et al (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48:781–791. doi:10.1139/g05-054
Rines H, Molnar S, Tinker N, Phillips R (2006) Oat. Genome mapping and molecular breeding in plants. Cereals Millets 1:211–242
Rossiello RO, Netto JJ (2006) Toxidez de alumínio em plantas: novos enfoques para um velho problema. In: Fernandes MS (ed) Nutrição mineral de plantas. Sociedade Brasileira de Ciência do Solo, Viçosa, pp 375–418
Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E (2011) The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot 62:9–20. doi:10.1093/jxb/erq272
Sanchez-Chacon CD, Federizzi LC, Milach SCK, Pacheco MT (2000) Viabilidade genética e herança da tolerância à toxicidade do aluminio em aveia. Pesqui Agropecu Bras 35:1797–1808
Sasaki T et al (2006) Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol 47:1343–1354. doi:10.1093/pcp/pcl002
Silva-Navas J, Benito C, Téllez-Robledo B, Abd El-Moneim D, Gallego FJ (2012) The ScAACT1 gene at the Q alt5 locus as a candidate for increased aluminum tolerance in rye (Secale cereale L.). Mol Breed 30:845–856. doi:10.1007/s11032-011-9668-5
Sivaguru M et al (2000) Aluminum-induced 1-3-B-D-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol 2:991–1006
Sorrells ME et al (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827. doi:10.1101/gr.1113003
Soto-Cerda B, Peñaloza E, Montenegro A, Rupayan A, Gallardo M, Salvo-Garrido H (2013) An efficient marker-assisted backcrossing strategy for enhancing barley (Hordeum vulgare L.) production under acidity and aluminium toxicity. Mol Breed 31:855–866. doi:10.1007/s11032-013-9839-7
Tan YD, Wan C, Zhu Y, Lu C, Xiang Z, Deng HW (2001) An amplified fragment length polymorphism map of the silkworm. Genetics 157:1277–1284
Tinker NA, Mather DE (1995) MQTL: software for simplified composite interval mapping of QTL in multiple environments. J Agric Genomics 1:1–4
Tinker NA et al (2014) A SNP genotyping array for hexaploid oat. Plant Genome. doi:10.3835/plantgenome2014.03.0010
Vogl C, Xu S (2000) Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics 155:1439–1447
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wagner CW, Milach SCK, Federizzi LC (2001) Genetic inheritance of aluminum tolerance in oat. Crop Breed Appl Biotechnol 1:22–26
Wang J et al (2007) High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 115:265–276. doi:10.1007/s00122-007-0562-9
Wang J et al (2009) High-throughput single nucleotide polymorphism genotyping using nanofluidic Dynamic Arrays. BMC Genomics 10:561
Wight CP et al (2003) A molecular marker map in ‘Kanota’ x ‘Ogle’ hexaploid oat (Avena spp.) enhanced by additional markers and a robust framework. Genome 46:28–47
Yokosho K, Yamaji N, Ma JF (2011) An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J 68:1061–1069. doi:10.1111/j.1365-313X.2011.04757.x
Yu J, Herrmann M (2006) Inheritance and mapping of a powdery mildew resistance gene introgressed from Avena macrostachya in cultivated oat. Theor Appl Genet 25:329–335
Zheng SJ, Ma JF, Matsumoto H (1998) Continuous secretion of organic acids is related to aluminium resistance during relatively long-term exposure to aluminium stress. Physiol Plant 103:209–214
Zheng SJ, Yang JL, He YF, Yu XH et al (2005) Immobilization of aluminum with phosphorus in roots is associated with high aluminum resistance in buckwheat. Plant Physiol 138:297–303
Zhou G, Delhaize E, Zhou M, Ryan PR (2013) The barley MATE gene, HvAACT1, increases citrate efflux and Al3+ tolerance when expressed in wheat and barley. Ann Bot 112:603–612. doi:10.1093/aob/mct135
Acknowledgments
The authors thank Dr. Eric Jellen and Dr. Jeff Maughan from Brigham Young University for the support during the SNP analysis; Dr. Marcelo T. Pacheco for the heritability analysis, and Mauricio D. Salomon for figure preparation. This study was supported by grants, scholarships, and fellowships from the Brazilian National Council for Scientific and Technological Development (CNPq), the Coordination for the Improvement of Higher Education Personnel (CAPES) from Brazil, The Prairie Oat Growers Association, and the US Department of Agriculture.
Conflict of interest
The authors declare that they have no conflict of interest and the experiments complied with the current laws of Brazil and USA, where they were performed.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schneider, A.B., Nava, I.C., Hervé, C.B. et al. Chromosome-anchored QTL conferring aluminum tolerance in hexaploid oat. Mol Breeding 35, 121 (2015). https://doi.org/10.1007/s11032-015-0315-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0315-4