Abstract
Aluminium (Al) tolerance in barley is conditioned by the Alp locus on the long arm of chromosome 4H, which is associated with Al-activated release of citrate from roots. We developed a high-resolution map of the Alp locus using 132 doubled haploid (DH) lines from a cross between Dayton (Al-tolerant) and Zhepi 2 (Al-sensitive) and 2,070 F2 individuals from a cross between Dayton and Gairdner (Al-sensitive). The Al-activated efflux of citrate from the root apices of Al-tolerant Dayton was 10-fold greater than from the Al-sensitive parents Zhepi 2 and Gairdner. A suite of markers (ABG715, Bmag353, GBM1071, GWM165, HvMATE and HvGABP) exhibited complete linkage with the Alp locus in the DH population accounting 72% of the variation for Al tolerance evaluated as relative root elongation. These markers were used to map this genomic region in the Dayton/Gairdner population in more detail. Flanking markers HvGABP and ABG715 delineated the Alp locus to a 0.2 cM interval. Since the HvMATE marker was not polymorphic in the Dayton/Gairdner population we instead investigated the expression of the HvMATE gene. Relative expression of the HvMATE gene was 30-fold greater in Dayton than Gardiner. Furthermore, HvMATE expression in the F2:3 families tested, including all the informative recombinant lines identified between HvGABP and ABG715 was significantly correlated with Al tolerance and Al-activated citrate efflux. These results identify HvMATE, a gene encoding a multidrug and toxic compound extrusion protein, as a candidate controlling Al tolerance in barley.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminium (Al) toxicity is a major factor limiting crop production on acidic soils, which include about 40% of the arable land in the world (Kochian 1995). At low pH (<5), dissolution of Al-containing compounds is enhanced and the release of toxic Al cations (particularly Al3+) into soil solution can rapidly inhibit root growth. Barley (Hordeum vulgare L. 2n = 2x = 14) is one of the most Al-sensitive members of the Poaceae family (Mugwira et al. 1976). Most Al sensitive cultivars either die within a few weeks after germination on acidic soils or yield very poorly. Nevertheless, significant genetic variation for Al tolerance exists in the barley germplasm (Minella and Sorrells 1992; Reid et al. 1969) and this has been exploited in breeding programs to develop high-yielding varieties with greater tolerance to Al toxicity (Read and Oram 1995). The mechanism of tolerance, at least in some genotypes of barley, appears to rely on the Al-activated release of citrate from the roots (Ma et al. 2004). These citrate anions are thought to protect plants from Al toxicity in the same way that malate efflux protects wheat: by binding to the harmful Al3+ cations in the apoplasm and detoxifying them (Delhaize and Ryan 1995).
Dayton is one of the most Al-tolerant barley genotypes (Minella and Sorrells 1992) and a single locus (Alp) on the long arm of the chromosome 4H (4HL) conditions Al tolerance in populations derived from Dayton/Harlan Hybrid and Dayton/F6ant28B48-16 (Raman et al. 2003; Reid 1970; Tang et al. 2000). The same locus conditions Al tolerance in other populations including those generated from Yambla/WB229, Mimosa/WB229 (Raman et al. 2002), Harrington/Brindabella (Raman et al. 2001), Murasakimochi/Morex (Ma et al. 2004), Ohichi/F6ant28B48-16 (Raman et al. 2005a) and F6ant28B48-16/Honen (Wang et al. 2006). Minor gene effects for Al tolerance have also been suggested (Echart et al. 2002; Raman et al. 2005a; Reid 1970).
The SSR markers currently being used for marker-assisted selection (MAS) of Al tolerance in Australian barley breeding programs map at least 2.1 cM from Alp (Langridge and Barr 2003; Raman et al. 2003). Considering an average value of 4.4 Mb/cM (megabase/centimorgan ratios) calculated for the barley genome (Künzel et al. 2000), a genetic distance of 2.1 cM corresponds to a physical distance of 9.24 Mb. These markers are not ‘diagnostic’ for predicting Al tolerance and are of limited value for map-based cloning of the Alp gene. Furthermore, all the mapping studies conducted so far employed relatively small populations comprising 48–100 individuals (Raman et al. 2003; Tang et al. 2000). Therefore, a high-resolution map of the Alp genomic region is needed both for improving the efficiency of MAS and to identify candidate genes that encode for Al tolerance.
Comparative mapping studies using RFLP markers have revealed extensive synteny or co-linearity among the genomes of rice, wheat, barley, rye, oat, maize, and sorghum (Devos and Gale 2000). Wheat chromosome 4D, barley chromosome 4H, rye chromosome 4R and rice chromosome 3 possess regions syntenic with one another (Miftahudin et al. 2004; Namuth et al. 1994; Van Deynze et al. 1995) and all harbour loci for Al tolerance (Fontecha et al. 2007; Luo and Dvorak 1996; Miftahudin et al. 2004; Nguyen et al. 2003; Raman et al. 2005b; Riede and Anderson 1996). Of these loci only the Al-tolerance gene from wheat, TaALMT1, has been isolated to date (Sasaki et al. 2004). TaALMT1 encodes an Al-activated transporter of malate and Magalhaes (2006) suggested that the orthogolous loci in the other species also possess TaALMT-like genes. The synteny between the Alp locus in barley and other cereals can be assessed with markers from those species and the information can be used to develop new markers for high-resolution mapping.
In the present study, we exploited the synteny between barley, wheat, rye and rice to map the Alp genomic region in barley. From a high-resolution map of the Alp region constructed with recombinant lines selected from a large F2 population we identified a candidate gene designated as HvMATE controlling Al tolerance in barley.
Materials and methods
Genetic materials
A doubled haploid (DH) barley population consisting of 132 lines was constructed from a single F1 derived from a cross between Dayton (Al-tolerant) and Zhepi 2 (Al-sensitive) using pollen culture and was used as a low-resolution mapping population. For high-resolution mapping, 236 recombinants were selected from 2070 F2 individuals derived from the cross between Dayton (Al-tolerant) and Gairdner (Al-sensitive) using Bmag490 and Bmac310 markers flanking the Alp locus. The F2 recombinants were raised in the glasshouse and the F2 derived F3 seeds were subsequently used for Al tolerance testing.
Assessment of Al tolerance
DH lines and their parents were assessed for Al tolerance using a nutrient solution culture method. The composition of the nutrient solution is described by Raman et al. (2002). A spatial complete block design with two replicates was employed. Each replicate consisted of two aerated 40 L tubs containing circulated nutrient solution. The treatment tub contained 15 μM Al as AlK (SO4)2 (+Al) while Al was omitted from the control tub (−Al). The tubs were divided into replicate blocks of lines, each within an incomplete 9 × 16 array. Seedlings were allocated to positions within blocks using DiGGer (Coombes 2002) to give an efficient spatial design allowing for row and column effects and positive correlations between locations within a tub.
Sixteen seeds of uniform size from each of the parents and 132 DH lines were surface sterilized with 1% NaClO solution for 10 min and thoroughly rinsed. After 3 days of germination (2 days on moist filter paper in petri dishes in a refrigerator at 4°C and 1 day in nutrient solution at 25°C) and 1 day pre-growth in a 16/8 h (day/night) photoperiod, eight seedlings of each line were selected and the initial lengths of the longest roots were measured. Seedlings were then floated in the control and treatment solutions in polyurethane foam strips and the length of the longest root on each seedling were measured after a further 48 h. The predicted mean root elongation in control (RE − Al) and treatment (RE + Al) solutions were spatially analysed in Genstat using the restricted maximum likelihood method. The relative root elongation (RRE) was calculated as RRE (%) = (RE + Al)/(RE − Al) × 100. Roots of the DH lines of the Al-treated seedlings were stained with 0.1% erichrome cyanine R (ECR) after 3 days under Al stress (15 μM) and scored sensitive or tolerant as previously described (Wang et al. 2006).
For high-resolution mapping, 28 F3 individuals per F2 recombinant (F2:3) were assessed for Al tolerance using the same protocol (Wang et al. 2006). Parental lines were included as checks to ensure correct scoring.
Estimates of Al tolerance for the data in Fig. 6 were measured under different conditions. Seeds were germinated on moist filter paper for 2 days. The lengths of the longest root were recorded before the seedlings were divided between two 20 L tubs holding aerated nutrient solution containing (μM) 500 KNO3, 500 CaCl2, 500 NH4NO3, 150 MgSO4, 2 KH2PO4, 2 FeCl3, 11 H3BO3, 2 MnCl2, 0.4 ZnCl2, 0.2 CuCl2 (pH 4.5). One tub contained no added Al and the other tub contained 5 μM AlCl3. Net root elongation was measured after 4 days and RRE was calculated as described above.
Molecular marker analysis
Genomic DNA was isolated from young leaves of the Dayton/Zhepi 2 DH lines, from F2 individuals of the Dayton/Gairdner population and from bulk leaf samples of the F2:3 recombinant families. A framework map of the DH population constructed using 531 DArT, SSR and STS markers (Wenzl et al. 2006) were employed for mapping Al tolerance. The Alp locus was further saturated with markers (Table 1) previously mapped near the Al tolerance loci from chromosome 4H of barley (Künzel et al. 2000; Raman et al. 2002), from chromosome 4D of wheat (Raman et al. 2005b), and from chromosome 4R of rye (Miftahudin et al. 2004). Molecular analyses were performed following the recommended PCR profiles. Primers generating amplicons below 400 bp were tailed with the M13 sequence (Schuelke 2000). Capillary electrophoresis, data collection and analysis were performed according to Raman et al. (2005c). Primers generating amplicons over 400 bp were used as standard oligos (untailed) and PCR products were separated on 2% (w/v) agarose gels. Monomorphic amplicons generated using primer pairs of ABG715, MWG0948, MWG2134, MWG2268, and B1 markers were sequenced using the Applied Biosystems BigDyeTM Terminator method on AB 3730XL capillary sequencer (Australian Genome Research Facility, Brisbane, Australia) to detect the presence of single nucleotide polymorphisms (SNPs). Sequences were compared and analysed for restriction enzyme sites using BioEdit software (http://www.mbio.ncsu.edu). The digested products were resolved on 2% (w/v) agarose gels.
Development of markers using the synteny between the barley and rice genomes
To identify the genomic region in rice syntenic with barley Alp, the sequence of the co-segregating marker ABG715 identified from low-resolution map (see Results) was queried in BLASTn searches against the rice non-redundant database in NCBI (http://www.ncbi.nlm.nih.gov/). ABG715 identified the rice BAC clone OJ1193C08 and the sequences on this clone and two adjacent BAC clones OSJNBb0085F02 and OSJNBa0090D11 (http://www.tigr.org/tigr-scripts/e2k1/irgsp_orderedBAC.spl?db=osa1&chr=3) were used as queries in further BLASTn searches of the barley EST database in NCBI. Barley EST sequences identified this way were used to design EST-specific primers using primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The presence of repeats of dinucleotide tri-, tetra-, penta- and hexanucleotide motifs were searched and targeted for primer design as described previously (Raman et al. 2006).
Low-resolution mapping of the Alp locus
All the marker data from the DH mapping population were analysed to test the goodness of fit using Chi-square tests (p = 0.05). These tests were also applied to determine the goodness of fit of segregation patterns to Mendelian ratios for phenotypes scored with ECR staining. All the marker and phenotypic data were imported into Map Manager QTX version b20 (Manly et al. 2001) and used for linkage analysis using the Kosambi (1944) mapping function. Measurements on RE-Al, RE + Al and RRE were used for QTL analysis with Map Manager using the framework map of the DH population from Dayton/Zhepi2 (Wenzl et al. 2006). “Suggestive”, “significant”, and “highly significant” critical likelihood ratio statistics (LRS) were established by 1,000 permutation tests at p = 0.05 (Churchill and Doerge 1994) and used to declare QTLs.
High-resolution mapping of the Alp locus
Two markers (Bmag490 and Bmac310) flanking the Alp locus in the DH population from Dayton/Zhepi2 were used to select 236 recombinants from the 2070 F2 individuals derived from a Dayton/Gairdner. The recombinant lines were selfed and their F2:3 progeny were assessed for Al tolerance using ECR staining. Any markers co-segregating with Alp in the low-resolution DH population that were polymorphic in the Dayton/Gairdner population were used to construct a high-resolution map among the recombinants. The genotypic data were also integrated into the Map Manager and map distance was calculated using Kosambi (1944) function.
Citrate efflux
Seeds were germinated on moist filter paper for 2 days and transferred to the same nutrient solution described above for Fig. 6. After 4 days growth under laboratory lighting three replicates of ten root apices (3–4 mm) were excised from six to eight plants per line. The tissue was transferred to 5 ml sterilised glass vials containing 1 mL 0.2 mM CaCl2, pH 4.3). The tubes were sealed with ParafilmTM and placed on a shaker (∼60 rpm) for 40–60 min. The vials were removed from the shaker, the tissue rinsed twice with 1 mL of the same CaCl2 solution and then 1 mL of treatment solution (25 μM AlCl3 and 0.2 mM CaCl2, pH 4.3) was added to each vial and returned to the shaker for 4 h. The solution was collected, dried down in a rotary evaporator and made up with 100 μL assay solution. The assay solution was prepared by mixing 1 mL H2O, 120 μL 1 M TrisCl (pH 8.0), 10 μL NADH solution (prepared by dissolving 20 mg NADH and 20 mg NaHCO3 in 2 mL H2O), 1 μL malate dehydrogenase (10 mg/mL, Sigma-Aldrich), 1 μL lactate dehydrogenase (5 mg/mL, Sigma-Aldrich) and 12 μL 0.1 M sodium isocitrate. The dissolved sample was transferred to a micro-cuvette and a stable absorbance at 340 nm was obtained before and after adding 2 μL citrate lyase (∼0.2 units, Sigma-Aldrich). The disappearance of NADH is proportional to the citrate present and the initial citrate content in each sample was calculated from a standard curve.
Quantitative RT-PCR
HvMATE expression in root apices was assayed by quantitative real-time reverse transcriptase PCR (qRT-PCR) using procedures described previously (Delhaize et al. 2004; Raman et al. 2005b) except that the cDNA preparations were not passed through a spin-column but were diluted 100-fold and used directly in the assay. The forward primer was 5′-CGTTCGTGTTCGACGGCATC-3′ and the reverse primer was 5′-AGCTGATCCATCACTTCCGG-3′. Expression was quantified relative to the expression of the barley glyceraldehyde-3-phosphate dehydrogenase gene (Burton et al. 2004) using the forward primer 5′-GTGAGGCTGGTGCTGATTACG-3′ and reverse primer 5′-TGGTGCAGCTAGCATTTGAGAC-3′.
Results
Inheritance of Al tolerance
Aluminium tolerance was assessed in the 132 DH lines derived from Dayton/Zhepi2 using the ECR staining method and measurements of root growth. The roots of the Al-tolerant parent Dayton were unstained after Al treatment and had normal lateral roots, whereas roots of the Al-sensitive parent Zhepi2 were stained purple with stained nubs along the seminal roots where lateral roots would normally emerge (Fig. 1). Of the 132 DH lines tested, 64 were scored as Al-tolerant and 68 were scored as Al-sensitive which fits a 1:1, monogenic segregation ratio (χ2 = 0.12; P = 0.73). Quantitative measurement of RE + Al and RRE showed bimodal distributions consistent with the segregation pattern for a major gene (Fig. 2). Significant correlations were observed between RE + Al and RRE (r = 0.83), RE + Al and root staining (r = 0.82), and RRE and root staining (r = 0.89).
Level of Al tolerance as indicated by eriochrome cyanine R staining of roots after 3 days in 15 μM Al. (T tolerant plant; S sensitive plant; open right arrow Unstained root tips; open left arrow Lateral roots developed; right arrow Root tips heavily stained; left arrow heavily stained nubs along the seminal roots)
Frequency distribution for a control root elongation (RE − Al, mm); b root elongation under Al stress (RE + Al, mm); c relative root elongation (RRE, %). RE-Al, RE + Al and RRE of sensitive parent ‘Zhepi 2’ was 41.18 mm, 14.98 mm and 36.38% respectively; whereas the tolerant parent Dayton was 38.11 mm, 36.06 mm and 94.62% respectively
Parental polymorphism survey
Barley 4H markers
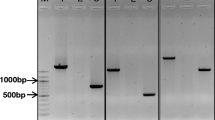
Several SSR markers tested were found to be polymorphic (13/16) between Dayton and Zhepi2. Among the STS markers (Table 1), only MWG2135, displayed a fragment length polymorphism. The amplicons of ABG715, MWG0948, MWG2134, and MWG2268 were monomorphic but subsequent sequencing revealed SNPs between Dayton and Zhepi 2 (Table 2) for all these except MWG2268, which allowed cleaved amplified polymorphic (CAPS) markers to be developed (Fig. 3).
CAPS analysis of markers ABG715, MWG0948 and MWG2134 resolved on 2% agarose gel (M-pGEM DNA marker, PT-tolerant parent, PS-sensitive parent). a Undigested PCR products of ABG715, b PCR products of ABG715 digested with restriction enzyme ApoI, c Undigested PCR products of MWG0948, d PCR products of MWG0948 digested with restriction enzyme AseI, e Undigested PCR products of MWG2134, f PCR products of MWG2134 digested with restriction enzyme AlwI
Wheat 4D markers
Among 15 wheat SSR markers tested (Table 1), only GWM165 and GWM608 amplified fragments in the parental lines of the DH population. Marker GWM165 was polymorphic between the parents and amplified a fragment of 238 bp in Zhepi 2 and 260 bp in Dayton. Marker GWM608 generated a 128 bp fragment, which was not polymorphic between the parents. The CAPS marker specific to TaALMT1, the gene conditioning Al tolerance in wheat (Sasaki et al. 2004), was not polymorphic in parental lines.
Rye 4R markers
The STS markers B1 and B4 linked to an Al tolerance locus in rye, Alt3 (Miftahudin et al. 2004), were tested for their linkage with the Alp locus in barley. The B1 marker amplified an expected fragment of approximately 500 bp in both the parental lines. The amplicons were sequenced and showed a 100% match with a barley sucrose transporter 1 gene (SUT1; AJ272309) but no SNPs were detected between Dayton and Zhepi 2. Marker B4 generated four bands in Zhepi 2 and two bands in Dayton between 222–676 bp allowing B4 to be used for molecular mapping.
Markers developed from the syntenic region of rice
The genomic region of Alp of barley was syntenic to three rice BAC clones OSJNBb0085F02, OJ1193C08 and OSJNBa0090D11 assembled on chromosome 3. Genes annotated on these BACs were used to identify potential orthologs in barley. Fourteen primer pairs were designed from the barley ESTs identified and used to generate markers. Some of these markers as well as the alignment of other barley markers to rice BAC clones are shown in Table 3. Two markers generated in this way, HvGABP and HvMATE were polymorphic in the DH population. HvGABP was also polymorphic for the Dayton/Gairdner population as a CAPS marker (Fig. 4) but the sequences of HvMATE amplicons were not polymorphic between the Dayton and Gairdner parental lines.
Low-resolution mapping of the Alp locus
A Chi-square test revealed that most of the polymorphic markers showed a 1:1 segregation ratio in the DH population except for HVM67, EBmac701, and B4 markers (results not shown). The six markers ABG715, GBM1071, GWM165, Bmag353, HvGABP and HvMATE showed complete linkage with the Alp locus (Table 4; Fig. 5a). Interestingly, marker B4, which is tightly linked to the Alt3 locus (4R) in rye (Miftahudin et al. 2004), did not map to 4H in barley but to chromosome 2H. The locus was flanked with DArT marker bPB-2219 and Bmac93 that mapped within 1.1 cM (see linkage map of Dayton/Zhepi 2. Additional file 1, http://www.biomedcentral.com/1471-2164/7/206/additional/, Wenzl et al. 2006).
Genetic linkage map of the Alp locus. a A low-density map generated in a DH population from Dayton/Zhepi 2. Vertical lines on the left side indicate the significantly associated genomic region for RRE and RE + Al. Solid bars indicate the peak locations. b A high-resolution map generated in 236 recombinant lines from Dayton/Gairdner. Genetic distance in centimorgans (cM) was calculated using the Kosambi (1944) function
QTL analysis with the Dayton/Zhepi 2 DH population revealed that a single genomic region on 4HL encompassing the Alp locus was significantly associated with RE + Al (LRS score: 119.9) and RRE (LRS score: 170.4). This region was flanked by markers Bmag490 and HVM68 and accounted for 59% of the genotypic variation in RE + Al and 72% of the variation in RRE (Fig. 5a). Dayton contributed favourable alleles both for both RE + Al and RRE as expected.
High-resolution mapping of the Alp locus
Among the 2070 F2 plants derived from Dayton/Gairdner, 236 recombinants were identified using two SSR markers flanking the Alp locus, Bmag490 and Bmac310 (Fig. 5a). Molecular analyses revealed that both markers exhibited a normal Mendelian segregation ratio as expected for a single gene. Where possible, the 236 recombinants were then genotyped with the other markers linked to Alp in the low-resolution DH population. These included the markers GWM165, ABG715, Bmag353 and HvGABP. The markers HvMATE and GBM1071 were monomorphic between Dayton and Gairdner. Two recombinants were identified between Alp and HvGABP and six were identified between Alp and ABG715 (Table 5). ABG715 mapped 0.15 cM distal to Alp and HvGABP mapped 0.05 cM proximal to Alp (Fig. 5b). Marker HvMATE was not polymorphic for the Dayton/Gairdner population and subsequent sequencing of the genomic sequence defined by the HvMATE EST did not identify any polymorphisms (Genbank #EF489298, EF588043).
Citrate efflux and HvMATE expression
Since the Al-tolerance of other barley genotypes appears to rely on Al-activated citrate efflux from roots (Ma et al. 2004) we investigated whether citrate efflux differed between the parental lines in the populations used here. Citrate efflux from the root apices of Dayton was 63 ± 4 pmol/apex/h compared to 2 ± 1 pmol/apex/h from Gairdner and 5 ± 0 pmol/apex/h from Zhepi 2. We then examined the HvMATE gene further because the rice/barley synteny indicates that the HvMATE and HvGABP genes are likely to be adjacent to one another. Furthermore, other MATE proteins are known to facilitate citrate efflux from Arabidopsis and sorghum cells (Durrett et al. 2006; Magalhaes et al. 2007).
Expression of the HvMATE gene was measured in the root apices of the parental lines using qRT-PCR. Relative expression of the HvMATE gene in the Al-tolerant Dayton was 30-fold higher than Gairdner and no expression could be detected in Zhepi2. We then investigated whether expression of the HvMATE gene correlated with Al tolerance and Al-activated citrate efflux in selected F2:3 families of the Dayton/Gairdner population including the eight informative recombinants identified with HvGABP and ABG715 markers (Table 5). Relative expression of HvMATE was significantly correlated with Al tolerance and Al-activated citrate efflux (Fig. 6). Of the 26 F2:3 families assayed, HvMATE expression and citrate efflux were greater in the F2:3 families from F2 individuals previously scored as homozygous tolerant than in the F2:3 families of F2 individuals scored as homozygous sensitive. F2:3 families from F2 individuals previously scored as being heterozygous for Al tolerance were generally intermediate for expression and citrate efflux. However, the variability of the expression measurements was too large to reliably differentiate between those F2:3 families that were homozygous for Al tolerance and those that were segregating for A1 tolerance. Nevertheless, HvMATE expression in the eight recombinant lines was consistent with the Alp alleles of these families predicted from the measurements of Al-tolerance and citrate efflux.
Relationships between HvMATE expression and Al tolerance and Al-activated citrate efflux. Data represent the mean of values and standard errors (SE) for expression (n = 3), citrate efflux (n = 3 or 4) and Al tolerance (n = 5−8). To account for the accumulation of errors associated with deriving RRE the SEs were calculated as follows: SERRE = RRG [(SE x /x)2 + (SE y /y)2]1/2 where x and y represent the mean root length in the control and aluminum treatments. The symbols represent whether the F2:3 families tested were previously found to be segregating for Al tolerance when mapping Alp (filled circle) homozygous Al-tolerant, (open circle) segregating for Al tolerance, (inverted filled triangle) homozygous Al-sensitive. Also indicated are the parental lines Dayton (Al-tolerant) and Gairdner (Al-sensitive). The correlation coefficients are r = 0.738 (P < 0.001) for a and r = 0.85 (P < 0.001) for (b)
Discussion
A single Alp locus conditions Al tolerance
Aluminum tolerance in the barley lines was estimated staining the roots with ECR, RE + Al and RRE. Staining indicates whether exclusion of Al from the root tips is occurring (Ma et al. 2001), whereas measurements of root elongation could reflect a combination of mechanisms that affect tolerance. Statistical analysis revealed that all three assessments of tolerance were significantly correlated with one another. The segregation analysis for root staining patterns and frequency distributions of RE + Al and RRE revealed that a single locus conditions Al tolerance in the barley cultivar Dayton. A single-gene model for Al tolerance scored on the basis of hematoxylin staining (Tang et al. 2000), root re-growth (Raman et al. 2002, 2003), ratio of root to shoot fresh weight (Ma et al. 2004), and ECR staining (Wang et al. 2006) has been reported previously for different barley populations. Linkage and QTL analysis confirmed that the major locus Alp, located on the long arm of chromosome 4H encodes Al tolerance in the populations from Dayton/Zhepi 2 and Dayton/Gairdner. Our results are consistent with previous reports that identified loci linked to acid-soil tolerance and Al tolerance in barley (Ma et al. 2004; Minella and Sorrells 1997; Raman et al. 2002, 2003; Stølen and Andersen 1978; Wang et al. 2006).
Comparative and high resolution mapping of Alp
We showed that the wheat SSR marker (GWM165-4DL) was located 0.45 cM from the Alp locus on the long arm of 4H. This marker has been located 1.5 cM apart from BCD1117 in the cMap of wheat chromosome 4D (http://www.rye.pw.usda.gov). Tang et al. (2000) have reported that BCD1117 and CDO1395 markers flank the Alp locus (Fig. 6). CDO1395 also explained approximately 20% of the genetic variation for Al tolerance in a rice population (Nguyen et al. 2003). BCD1230 exhibited tight linkage with the Al tolerance locus Alt3 in rye (Miftahudin et al. 2004), and Alt BH in wheat (Riede and Anderson 1996) but was mapped 33 cM away from Alp locus in barley (Tang et al. 2000). Marker GWM165 tightly linked with Al tolerance in the present study was mapped about 12 cM apart from the Al tolerance locus in wheat (Raman et al. 2005b). Interestingly, rye marker B4, which is tightly linked with the Alt3 locus on 4R (Miftahudin et al. 2004), mapped to chromosome 2H in barley instead of the expected 4H. It is possible that multiple copies of B4 may exist in the barley genome, or there may be some conservation of genes between chromosomes 2H and 4R. Similarly, Gruber et al. (2006) identified an ALMT1 homolog in barley (HvALMT1), which mapped in the vicinity of the B4 locus on 2H in the DH population from Dayton/Zhepi 2. Recently Fontecha et al. (2007) reported that an ALMT1 homolog was located on barley 4H, which is consistent with the expected synteny between the wheat 4DL and barley 4HL. However this conclusion relied on PCR amplification and needs to be verified by Southern blot analysis.
The rice/barley synteny was exploited to develop additional markers for high-resolution mapping of the Alp locus. The locus was finally delineated to a 0.2 cM region in the Dayton/Gairdner population by the flanking markers HvGABP and ABG715 (Fig. 5b). The region of the rice genome found to be syntenic with the Alp locus spans approximately 120 kb on chromosome 3 (Fig. 7) and is flanked by Os03g11590 (ABG715) and Os03g11720 (HvGABP) (http//www.tigr.org/tdb/e2k1/osa1/). Within this region there are no homologs of TaALMT1, the Al tolerance gene of wheat. This result indicates that despite a similar chromosomal location for Al tolerance loci in wheat and barley, the genes are likely to encode different proteins and are therefore not orthologous.
Within the syntenic region of rice on BAC clone OSJNBa0090D11 a gene encoding a MATE stands out as a candidate homolog for the Al tolerance gene of barley. Members of the MATE family were recently shown to facilitate citrate efflux from Arabidopsis and sorghum cells (Durrett et al. 2007; Magalhaes et al. 2007) and it is conceivable that HvMATE encodes a protein responsible for the Al-activated efflux of citrate from Al-tolerant barley. Although the HvMATE marker was not polymorphic in the Dayton/Gairdner population, expression levels did differ between the parental genotypes and therefore could be used to score the F2:3 families. The finding that HvMATE expression was correlated with citrate efflux and Al tolerance identifies HvMATE as a candidate for the gene controlling Al tolerance in barley.
References
Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley. Quantitative analysis of transcript reveals two groups of co-expressed genes. Plant Physiol 134:224–236
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Coombes NE (2002) The reactive tabu search for efficient correlated experimental designs. PhD Thesis, John Moores University, Liverpool
Delhaize E, Ryan PR (1995) Aluminum toxicity and tolerance in plants. Plant Physiol 107:315–321
Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H (2004) Engineering high level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA 101:15 249–15 254
Devos KM, Gale MD (2000) Genome relationships: the grass model in current research. Plant Cell 12:637–646
Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol (in press, doi:10.1104/pp.107.097162
Echart CL, Barbosa-Neto JF, Garvin DF, Cavalli-Molina S (2002) Aluminum tolerance in barley: methods for screening and genetic analysis. Euphytica 126:309–313
Fontecha G, Silva-Navas J, Benito C, Mestres MA, Espino FJ, Hernández-Riquer MV, Gallego FJ (2007) Candidate gene identification of an aluminium-activated organic acid transporter gene at the Alt4 locus for aluminium tolerance in rye (Secale cereale L.). Theor Appl Genet 114:249–260
Gruber BD, Ryan PR, Richardson AE, Hebb DM, Raman H, Zhou M, Wang J, Howitt SM, Delhaize E (2006) The identification and characterisation of ALMT1 homologs in the Triticeae. In: Proceedings of 8th international congress of plant molecular biology. Adelaide, Australia., p 185
Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Künzel G, Korzun L, Meister A (2000) Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154:397–412
Langridge P, Barr AR (2003) Preface to “Better barley faster: the role of marker assisted selection”. Aust J Agric Res 54:I–IV
Liu ZW, Biyashev RM, Saghai-Maroof MA (1996) Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor Appl Genet 93:869–876
Luo M, Dvorak J (1996) Molecular mapping of an aluminium tolerance locus on chromosome 4D of Chinese Spring wheat. Euphytica 91:31–35
Ma JF, Nagao S, Sato K, Ito H, Furukawa J, Tekeda K (2004) Molecular mapping of a gene responsible for Al-activated secretion of citrate in barley. J Exp Bot 55:1335–1341
Ma JF, Ryan PR, Delhaize E (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6:273–278
Magalhaes JV (2006) Aluminum tolerance genes are conserved between monocots and dicots. Proc Natl Acad Sci USA 27:9749–9750
Magalhaes JV, Liu J, Guimarães CT, Lana UGP, Alves VMC, Wang Y-H, SchaVert RE, Hoekenga OA, ShaV JE, Piñeros MA, Klein PE, Coelho CM, Kochian LV (2007) A member of the multidrug and toxic compound extrusion ‘MATE’ family is a major gene that confers aluminum tolerance in sorghum. Nat Gen (in press)
Manly KF, Cudmore JRH, Meer JM (2001) Map manager QTX, cross platform software for genetic mapping. Mamm Genome 12:930–932
Miftahudin, Scoles GJ, Gustafson JP (2004) Development of PCR-based codominant markers flanking the Alt3 gene in rye. Genome 47:231–238
Miftahudin T, Chikmawati T, Ross K, Scoles GJ, Gustafson JP (2005) Targeting the aluminum tolerance gene Alt3 region in rye, using rice/rye micro-colinearity. Theor Appl Genet 110:906–913
Minella E, Sorrells ME (1992) Aluminum tolerance in barley: genetic relationships among genotypes of diverse origin. Crop Sci 32:593–598
Minella E, Sorrells ME (1997) Inheritance and chromosome location of Alp, a gene controlling aluminium tolerance in ‘Dayton’ barley. Plant Breed 116:465–469
Mugwira LM, Elgawhary SM, Patel KI (1976) Differential tolerances of Triticale, wheat, rye and barley to aluminum in nutrient solution. Agron J 68:782–787
Namuth DM, Lapitan NLV, Gill KS, Gill BS (1994) Comparative RFLP mapping of Hordeum vulgare and Triticum tauschii. Theor Appl Genet 89:865–872
Nguyen BD, Brar DS, Bui BC, Nguyen TV, Pham LN, Nguyen HT (2003) Identification and mapping of QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., to indica rice (Oryza sativa L.). Theor Appl Genet 106:583–593
Raman H, Karakousis A, Moroni JS, Raman R, Read BJ, Garvin DF, Kochian LV, Sorrells ME (2003) Development and allele diversity of microsatellite markers linked to the aluminium tolerance gene Alp in barley. Aust J Agric Res 54:1315–1321
Raman H, Moroni JS, Sato K, Read BJ, Scott BJ (2002) Identification of AFLP and microsatellite markers linked with an aluminium tolerance gene in barley (Hordeum vulgare L.). Theor Appl Genet 105:458–464
Raman H, Moroni S, Raman R, Karakousis A, Read B, Sato K, Scott BJ (2001) A genomic region associated with aluminium tolerance in barley. In: Proceedings of the 10th Australian Barley Technical Symposium. Canberra, 16–20 September (http://www.regional.org.au/au/abts/2001/t3/raman.htm)
Raman H, Raman R, Wood R, Martin P (2006) Repetitive indel markers within the ALMT1 gene conditioning aluminium tolerance in wheat (Triticum aestivum L.). Mol Breed 18:171–183
Raman H, Wang JP, Read B, Zhou MX, Venkataganappa S, Moroni JS, O’ Bree B, Mendham N (2005a) Molecular mapping of resistance to aluminium toxicity in barley. In: Proceedings of Plant and Animal Genome XIII Conference, January 15–19, San Diego, pp154 (http://www.intl-pag.org/13/abstracts/ PAG13_P328.html)
Raman H, Zhang K, Cakir M, Appels R, Moroni J S, Maron LG, Kochian LV, Raman R, Imtiaz M, Drake-Brockman F, Garvin DF, Waters I, Martin P, Sasaki T, Yamamoto Y, Matsumoto H, Hebb DM, Delhaize E, Ryan PR (2005b) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48:781–791
Raman R, Raman H, Johnstone K, Lisle C, Smith A, Martin P, Allen H (2005c) Genetic and in silico comparative mapping of the polyphenol oxidase gene in bread wheat (Triticum aestivum L.). Funct Integr Genomics 5:185–200
Ramsay L, Macaulay M, Ivanissevich S. degli, MacLean K, Cardle L, Fuller J, Edwards KJ, Tuveson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, Waugh R (2000) A simple sequence repeat based linkage map of barley. Genetics 156:1997–2005
Read BJ, Oram RN (1995) Hordeum vulgare (Barley) cv. Brindabella. Aust J Exp Agric 35:425
Reid DA (1970) Genetic control of reaction to aluminum in winter barley. In: Proceedings of the 2nd International barley genetics symposium, Pullman, pp 409–413
Reid DA, Jones GD, Armiger WH, Foy CD, Koch EJ, Starling TM (1969) Differential aluminum tolerance of winter barley varieties and selections in associated greenhouse and field experiments. Agron J 161:218–222
Riede CR, Anderson JA (1996) Linkage of RFLP markers to an aluminum tolerance gene in wheat. Crop Sci 36:905–909
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37:645–653
Schuelke M (2000) An economic method for the fluorescent labelling of PCR fragments. Nat Biotech 18:233–234
Stolen O, Andersen S (1978) Inheritance of tolerance to low soil pH in barley. Hereditas 88:101–105
Tang Y, Sorrells ME, Kochian LV, Garvin DF (2000) Identification of RFLP markers linked to the barley aluminum tolerance gene Alp. Crop Sci 40:778–782
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Van Deynze AE, Nelson JC, Yglesias ES, Harrington SE, Braga DP, McCouch SR, Sorrells ME (1995) Comparative mapping in grasses. Wheat relationships. Mol Gen Genet 248:744–754
Wang JP, Raman H, Read B, Zhou M X, Mendham N, Venkatanagappa S (2006) Validation of an Alt locus for aluminium tolerance scored with eriochrome cyanine R staining method in barley cultivar Honen (Hordeum vulgare L.). Aust J Agric Res 57:113–118
Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesná J, Cakir M, Poulsen D, Wang J, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A (2006) A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics 7: 206 (http://www.biomedcentral. com/1471-2164/7/206)
Acknowledgments
The authors would like to thank the NSW Agricultural Genomic Centre, The Tasmanian Institute of Agricultural Research and the Grains Research and Development Corporation (GRDC) of Australia (UT8) for providing financial support for this project. GBM primer sequences were kindly supplied by Drs. Andreas Graner and Martin Ganal from Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Waugh.
Rights and permissions
About this article
Cite this article
Wang, J., Raman, H., Zhou, M. et al. High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 115, 265–276 (2007). https://doi.org/10.1007/s00122-007-0562-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0562-9