Abstract
Crown rust, which is caused by Puccinia coronata f. sp. avenae, P. Syd. & Syd., is the most destructive disease of cultivated oats (Avena sativa L.) throughout the world. Resistance to the disease that is based on a single gene is often short-lived because of the extremely great genetic diversity of P. coronata, which suggests that there is a need to develop oat cultivars with several resistance genes. This study aimed to identify amplified fragment length polymorphism AFLP markers that are linked to the major resistance gene, Pc68, and to amplify the F6 genetic map from Pc68/5*Starter × UFRGS8. Seventy-eight markers with normal segregation were discovered and distributed in 12 linkage groups. The map covered 409.4 cM of the Avena sativa genome. Two AFLP markers were linked in repulsion to Pc68: U8PM22 and U8PM25, which flank the gene at 18.60 and 18.83 centiMorgans (cM), respectively. The marker U8PM25 is located in the linkage group 4_12 in the Kanota × Ogle reference oat population. These markers should be useful for transferring Pc68 to genotypes with good agronomic characteristics and for pyramiding crown rust resistance genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oats (Avena sativa L.) are considered an important option for grain production during the winter season in a no-till rotation with soybeans in sub-tropical environments. However, this environment is also favorable to growth of the main oat biotrophic pathogen, Puccinia coronata f. sp. avenae P. Syd. & Syd., which causes crown rust. This is the most destructive disease of cultivated oat, severely affecting the yield potential expression. In the Rio Grande do Sul state of Brazil, a grain weight loss of 50% was observed in susceptible oat cultivars because of this disease (Martinelli et al. 1994).
The most effective strategy to control diseases, particularly cereal rusts, is the development of resistant cultivars. More than 100 race-specific resistance genes to crown rust have been identified, of which 96 were defined as Pc, with the majority considered to be dominant genes (Zhu and Kaeppler 2003). However, because of its extreme genetic diversity, the P. coronata population has rapidly adapted to most major gene resistance in a short period of time (Leonard and Martinelli 2005). Pc68, on the other hand, which was introgressed in A. sativa from Avena sterilis L. (Wong et al. 1983), is considered to be one of the most effective genes against this disease and has been widely used in breeding programs across the United States and Canada (Chong 2000; Johnston et al. 2000).
Although virulence to Pc68 appeared in 2001, it was not until 2005 that cultivars with this gene were rusted in North America (McCallum et al. 2007). However, the frequency of virulence on the gene has been consistently low, particularly in parts of the world such as South America (Leonard and Martinelli 2005; Vieira et al. 2007). Therefore, Pc68 shows potential to be introgressed into the germplasm of A. sativa with good agronomic characteristics, especially if this is done in association with other strategies to improve the durability of resistance, such as gene pyramiding, which has been advocated for use in other pathosystems (Rubiales and Niks 2000). In this sense, the availability of molecular markers that facilitate an easy and reliable detection of the respective alleles is a prerequisite (Werner et al. 2005) since the use of the conventional rust-testing method based on differential reaction is difficult because of a lack of races with virulence combinations that are appropriate for testing for the presence of each gene independently (Chen et al. 2006). The presence of genes for complete resistance may mask the small effects of other genes during selection, so the availability of molecular markers is beneficial when combining genes for resistance (Rubiales and Niks 2000).
Molecular markers may also facilitate the cloning of the specific gene and as a result, the identification of the gene functions. On the other hand, because a number of crown rust resistance genes are found clustered in the oat genome (Chong et al. 1994), the development of molecular markers to one crown rust resistance gene may be useful in the study of other resistance genes located within the same cluster. The availability of genetic markers for oat is limited because the large hexaploid genome of oat (AACCDD) presents extensive gene duplication. This increases the difficulty in developing linkage maps probably because of the existence of numerous multigene families in homologous, homoeologous, and paralogous chromosomes, which causes the low level of polymorphism in any given population (Portyanko et al. 2001; Wight et al. 2003). Recently, the Kanota × Ogle reference map was improved in the density of coverage and the accuracy with which the linkage groups are represented by the addition of markers based on Diversity Array Technology (DArT) (Tinker et al. 2009). However, work is still needed to produce a stable consensus map for oat. Since 1990, efforts have been directed at developing genetic markers to the crown rust resistance genes in oat (Chen et al. 2006). Penner et al. (1993a) were the first to identify one molecular marker associated with crown rust resistance. A random amplified polymorphic DNA (RAPD) marker was obtained for Pc68 in a F3 derived from the lines ‘Makuru’*2//CAV4904/2*’SunII’ (with Pc68) and ‘Rodney O’ (without Pc68), and this marker was associated with repulsion in the gene. Other molecular markers such as a sequence characterized amplified region (SCAR) (Koeyer et al. 2000) and single nucleotide polymorphisms (SNPs) (Chen et al. 2006) for the Pc68 gene were identified, considering that Pc68 is clustered with Pg9, a resistance gene to stem rust (Chong et al. 1994). However, because the utility of any molecular marker is dependent on the polymorphism between the compared genotypes, it is unlikely that one single marker will be applicable across all oat germplasms, unless it differentiates the specific alleles that define the trait. Therefore, the availability of multiple independent markers is desired. Amplified fragment length polymorphism (AFLP) fingerprints can be used as a source of genetic markers to generate linkage maps or to identify markers linked to phenotypic traits (Vos et al. 1995). This technique provides the opportunity to map plants with large genomes and low polymorphic rates.
The objectives of this study were to identify AFLP markers associated with the Pc68 gene using a F6 recombinant inbred line (RIL) population derived from crossing the genotypes Pc68/5*Starter and UFRGS 8, as well as to expand the genetic map of this population.
Materials and methods
Plant material and phenotypic screening
Approximately 130 RILs at the F5:6 generation were developed by single-seed descent (SSD) from the cross between the Pc68/5*Starter line, which contains the Pc68 resistance gene, and the cultivar UFRGS8, which does not contain the gene. The oat breeding line, Pc68/5*Starter, was developed at the University of Minnesota, in the USA. And the UFRGS8 (OA338 × X2682-1) line is a variety developed by the oat-breeding program of the Federal University of Rio Grande do Sul (UFRGS), in Brazil.
Phenotypic data were collected from seedlings. The parent genotypes and the F5:6 generation RILs were sowed in pots that were kept in a greenhouse. At an age of 12 days, the seedlings were sprayed with a solution of Tween® 0.01% and immediately afterwards inoculated with spores of the Pca race SQPT, by the shaking of infected, susceptible leaves over them. After the spores were deposited, the seedlings were sprayed again with a solution of Tween® 0.01% and kept in the dark for 20 h at 22 ± 2°C. The reaction to the pathogen was assessed 15 days after inoculation according to the method proposed by Chong et al. (2000), in which the infection type (IT) is characterized using a scale ranging from 0 to 4, where 0 represents the absence of uredia or other macroscopic infection symptoms, 1 represents the presence of small uredia surrounded by chlorosis or necrosis, 2 represents the presence of small- to medium-sized uredia surrounded by chlorosis, 3 represents the presence of medium-sized uredia in a chlorotic area, and 4 represents the presence of large uredia without chlorosis or necrosis. Plants showing responses of 0, 1, or 2 were considered to be resistant to the pathogen, and those with responses of 3 or 4 were considered to be susceptible. To determine the inheritance of this population, the ratio of the number of resistant to susceptible plants was evaluated by testing the goodness of χ2 with Yates’ continuity correction (Zar 2009). For mapping purposes, 92 lines were selected, of which 46 were characterized as resistant lines, and 46 were characterized as susceptible lines.
DNA extraction and AFLP analysis
The total genomic DNA was extracted from samples of approximately 0.1 g of young leaf tissue using the cetyl trimethylammonium bromide (CTAB) method (Roy et al. 1992). DNA quantification was performed with a spectrophotometer (Spectronic Genesys™), and the final DNA concentration of the AFLP reactions was 250 ng/μl.
Bulked segregant analysis (BSA) was performed to identify polymorphisms that are putatively linked to Pc68 based on the presence of differential alleles in the parents and the bulks. Each bulk was established by using equal amounts of DNA from ten resistant and ten susceptible lines (Michelmore et al. 1991).
AFLP analysis was conducted as described by Vos et al. (1995) with modifications. A total of 250 ng of genomic DNA was digested with MseI (Promega) and PstI (Gibco BRL). MseI and PstI adapters were ligated to the restriction fragments and diluted 1:10. Pre-selective amplification was performed using the primers with one additional nucleotide (MseI + C and PstI + A), and selective amplification was performed using the primers with three additional nucleotides. The PCR products were separated using 5% denaturing polyacrylamide gels (19:1 acrylamide:bis-acrylamide), at a constant power of 80 W for approximately 2 h and 30 min, accompanied with the DNA Ladder 123 pb marker (Invitrogen). DNA bands were visualized by silver staining according to the Silver Sequence™ (Promega Corporation) protocol (1996). Two independent readers performed visual scoring of the bands, and if divergence in the score was found, that marker was not used.
Both the bulk and parental samples were subjected to 60 combinations of the AFLP primers. The primer combinations that gave stable and consistent polymorphism between the bulks and the parents were used for the analysis of the 92 individual plants from the F5:6 generation to determine the associations between the AFLP markers and the resistance gene, Pc68. The markers were named according to the additional 3’ selective base pairs that were used on the primers and were complementary to the PstI and MstI sides, followed by the number of base pairs of the fragment.
Linkage map construction
The polymorphisms that were identified between the resistant and susceptible bulks were scored for the 92 plants. To the polymorphic candidate markers that were identified, we added 34 markers that were previously identified by Locatelli et al. (2006) in a study of flowering time in the same population. Chi-squared tests were performed on each locus for goodness-of-fit to the expected 1:1 segregation ratio. Markers that deviated significantly (P < 0.05) from the expected segregation ratio were excluded for linkage analysis and map construction.
The GQMOL program (Cruz and Schuster 2004) was used to assess the strength of the linkage between the resistance locus and the polymorphic bands. A logarithm of the odds (LOD) score of 3.0 was established to denote significant linkage, and the Kosambi’s mapping function was used to convert the recombination fractions into centiMorgans (cM). The maps were constructed with the GQMOL software.
Results and discussion
Phenotypic analysis
Cultivar UFRGS 8 presented an IT of 3, indicating that it is susceptible to the SQPT race of Pca, but the genotype PC68/5*Starter was immune to this race with an IT of 0. The inbred lines exhibited responses with IT values ranging from 0 to 3. Of those inbred lines, 62 lines were resistant to this race, and 64 lines were susceptible; this proportion was equal to the expected distribution for one gene (Table 1), indicating that resistance to the SQPT race is solely due to the Pc68 gene in this population. Similarly, Wong et al. (1983) and Koeyer et al. (2000), using other pathogen races and other segregating populations, observed the same inheritance model for this source of resistance. Resistance due to another gene is highly unlikely, because the Starter cultivar is susceptible to most races tested in the United States, and the Pc68/5*Starter genotype was resistant to all races of Pca tested for several years at diverse locations (Leonard 2002, 2003).
Bulk segregation analysis
The majority of the loci that were analyzed by the BSA method were monomorphic and therefore not able to distinguish between the two bulks. Of the 60 primer combinations that were analyzed, only 9 (15%) showed polymorphism between the bulks. Using these 9 combinations, 80 molecular markers were identified, giving an average of 9 polymorphic bands per primer combination (Table 2).
Despite the phenotypic diversity observed in the field between the parental genotypes, a small number of polymorphic fragments were expected. This phenomenon is a common characteristic of species with large genomes. Similar data were obtained by Yu and Herrmann (2006) when they mapped one resistance gene that was introgressed from Avena macrostachya in A. sativa. Using the BSA method, of the 256 AFLP primer combinations, only 21 (approximately 8%) showed polymorphisms between the contrasting genotypes. Li et al. (2000) analyzed 20 oat cultivars using simple sequence repeats (SSR) markers and obtained polymorphic markers in only 36% of them. Bryan et al. (1999), sequenced several DNA fragments from wheat, another hexaploid crop, and found a very low level of sequence variation among the genotypes. They suggested that the low genetic polymorphism found in crops such as oat and wheat is due to the evolution of these species. Several events of duplication of the repetitive regions probably caused the large genome size.
In studies like this one, the BSA method is efficient at detecting polymorphisms among inbred lines for one trait and avoids wasting time and resources. The combination of the BSA method with the AFLP technique is particularly useful because AFLP is a powerful tool that can tag genes in large genomes, especially when there is no previous knowledge about the gene location or SSR saturated map. The AFLP technique is more efficient than the restriction-fragment length polymorphism (RFLP) or RAPD techniques in finding polymorphisms and results in acquisition of more information than if another kind of molecular marker had been used.
Marker segregation analysis
The markers that were obtained by Locatelli et al. (2006) were added to the 80 polymorphic markers that were identified in our BSA analysis, resulting in a total of 114 AFLP molecular markers, which were tested on the F6 inbred line population. The Mendelian segregation analysis showed that 77 (67%) of the marker loci segregated as expected for the F6 inbred lines (1:1, α* = 0.05). The value of alpha was corrected by Bonferroni’s method, which is indicated for set data analysis. In this case, the genomic significance (α*) was used, which is defined as the probability of rejecting at least one true null hypothesis, with an α* value of 1 indicating 100% probability of not rejecting any true null hypothesis (Silva and Vencovsky 2002). To calculate it, the significance level, α, must be estimated in each test to produce the significance level for the full experiment, α*.
If the individual significance level had been used (α = 0.05) another 10 loci would have presented distorted ratios and would have been left out of the analysis. For this reason, global significance levels are recommended when using multiple tests to avoid this type I error.
Of those combinations that segregated as expected, P-AAA/M-CAG and P-AGA/M-CCG produced a greater number of segregating bands (Table 2). Other combinations such as P-ACG/M-CTA and P-AGA/M-CAG produced a smaller number of markers; however, no distortion was observed on any of the loci (Table 2).
In this study, we observed that 32% of the molecular markers had distorted ratios. Distortions have been found in AFLP loci in several species. The distortion ratio seems to be dependent on the species and genotypes that are used to generate the segregating population. For example, distortion ratios of 14% were observed in melon (Wang et al. 1997), 4 to 10% in kiwi (Testolin et al. 2001), and greater than 40% in Cryptomeria japonica (Nikaido et al. 1999).
Distortion may arise as a true biological phenomenon or may be caused by preferential fertilization, DNA breakage during extraction, amplification of other genome regions, or genotyping or gel analysis error (Tan et al. 2001). The biological causes of distortion involve physical closeness of the marker loci and the genes or regions involved either in gamete formation or genotype viability after fertilization (Vogl and Xu 2000). Moreover, segregation distortion may occur because of mistakes in the identification of the endonuclease restriction site. Among the phenomena that lead to partial digestion is the lack of restriction site recognition by the PstI enzyme due to methylation. The addition of a methyl group to a cytosine may alter the ability of the enzyme to recognize the restriction site and is associated with several biological processes, including transcriptional regulation and silencing (Cervera et al. 2002). Changes in the methylation pattern may occur as result of de novo methylation or passive demethylation during DNA replication (Matsuo et al. 1998). Some enzymes, including PstI, are sensitive to methylation, which may explain the unspecific bands that appeared in the inbred lines.
At least part of the segregation distortion observed in this study arises from the use of the enzyme PstI. This conclusion is based on two observations. First, unspecific bands were observed in the inbred lines but not in the resistant or the susceptible parent (Fig. 1). Secondly, when only the loci obtained by Locatelli et al. (2006) were evaluated, the distortion was reduced to 26%. For the 80 markers obtained in this work, the restriction enzyme was PstI; however, for the 34 markers obtained by these authors, the enzyme was EcoRI, which is insensitive to methylation.
Segment of a polyacrylamide gel, showing AFLP bands that were identified for both parental genotypes, (P1) Pc68/5*Starter (resistant) and (P2) UFRGS 8 (susceptible), and 18 inbred lines. Bold arrows show specific fragments of one genitor segregating in the population. The dotted arrow shows fragments that are unique to the population and are not present in the parental genotypes
The distorted ratios influence the association between the markers and the segregation analysis data for the phenotypic trait. Thus, it is recommended that they be excluded from the data during the linkage map development because they can affect the statistical tests that are used to establish the linkage, causing false positives.
Mapping analysis
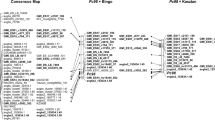
The linkage map was based on the analysis of 92 lines from the F5:6 inbred population that was derived from the cross between Pc68/5*Starter and UFRGS 8. The 77 molecular markers with an expected segregation ratio were used. Of the 77 molecular markers, 56 were grouped at 12 linkage groups (Fig. 2). One linkage group contained 30 markers. Eight linkage groups were formed by only two markers. The smallest distance between markers was 1.69 cM and the largest was 27.46 cM, both of which were found in the first linkage group. In the previously published map of this population, seven linkage groups were obtained, and as in this study, the first linkage group contained the largest number of markers. Eighteen molecular markers were not linked to any group.
The low level of polymorphism that was detected limited the ability to obtain a greater number of markers. The map covered 409.4 cM (14%) of the total Avena sativa genome, which is estimated to be 2900 cM (O’Donoughue et al. 1995). However, this represents a significant expansion compared to the first map of this population (Locatelli et al. 2006), which covered only 4% of the total genome of the species.
Of the 21 linkage groups expected for A. sativa, only 12 were obtained, and the marker distribution among these linkage groups was not uniform. For example, one linkage group contained 39% of the markers. This kind of concentration is commonly observed when only one type of molecular marker is used, and the number of markers is small. Ideally, the map must be saturated with markers. One alternative is to test other markers, such as SSR. Unfortunately, during the development of this study, none of the SSR markers that were tested showed polymorphism between the bulks (data not shown). Efforts have been made to increase the number of SSR markers, but currently, only a small number have been published, and only a subset of those is polymorphic for any given population. The new DArT markers could be an option in future work (Tinker et al. 2009). However, efforts to use these markers in oat have not yet produced a consensus map, in which all linkage groups are assigned to the expected 21 oat chromosomes.
AFLP markers have been associated with resistance genes in several species, including wheat (Weng and Lazar 2002; Randhawa et al. 2009), tomato (Giovanni et al. 2004; Golas et al. 2010), maize (Duble et al. 2003; Chen et al. 2004; Liu et al. 2009), canola (Kaur et al. 2009), and radish (Kamei et al. 2010). In the present study, the AFLP molecular markers U8PM22 and U8PM25, developed by Locatelli et al., were associated with the Pc68 gene. Both of these markers, from the linkage group 10, were linked in repulsion at 18.60 and 18.83 cM, respectively (Fig. 2). The Pc68 locus is flanked by those two markers in the susceptible lines. In spite of the large distance, these markers may be useful for marker-assisted selection. Brogin (2005) obtained high selection efficiency (91%) for a resistance gene to Asian soybean rust using a SSR marker that is 24.4 cM from the gene. Also, the use of markers flanking the gene from both sides is beneficial because unilateral markers are easily lost by a single crossing-over event, but a double event would be required to lose two markers.
The previously described SCAR and SNP molecular markers for Pc68 were developed based on RAPD (Orr et al. 1999) and RFLP (O’Donoughue et al. 1995) markers, which were initially obtained for the Pg9 gene (Koeyer et al. 2000; Chen et al. 2006). The use of Pg9 to select for Pc68 must be done with caution because neither the distance between the genes nor the cluster stability in different oat populations is known. Although the SNP identified by Chen et al. (2006) was also associated in repulsion to the Pc68 gene, it was developed in a manner that requires sequencing of the marker region to identify each genotype, a task that remains too expensive for the majority of oat breeding programs.
Linkage in repulsion is not an ideal situation because the markers are present in the absence of the trait. However, using RAPD markers, Haley (1994) found that repulsion phase linkage provided greater selection efficiency than coupling phase linkage. There are examples in the literature, in which the first markers that were identified were in repulsion and the genes were cloned with conversion to other markers (Renganayaki et al. 2002; Barzen et al. 2004). Curiously, all of the markers that have been identified to be associated with the Pc68 gene were linked in repulsion (Penner et al. 1993b; Orr et al. 1999). There is no explanation for such a coincidence. The Pc68 gene was introgressed in oat from another species, possibly as a cluster, but no significant difference is expected in the sequence pattern between the species. Some genomes are richer in specific bases. For example the virus SV40 genome is rich in the bases thymine and adenine, which affects the number of fragments that are expected when a restriction enzyme is used. This kind of difference could explain the absence of direct linkage. However, this is not the case for A. sterilis, and considering that different types of molecular markers were tested (based on different restriction sites and sequences), this coincidence remains unexplained.
Use of a wide array of resistance mechanisms in modern germplasms has been indicated to increase durability (Rubiales and Niks 2000). Combination of genes that cause relatively small effects on pathogen development and are race-non-specific with race-specific genes could allow for the development of cultivars with long-lasting resistance. Molecular markers, such those identified here, may overcome the difficulties in selection when genes for complete resistance mask the small effects of the other genes.
The marker U8PM25 was also polymorphic in the Kanota × Ogle reference oat population and was located at the linkage group KO 4_12 (Locatelli et al. 2006). Previous studies using quantitative trait loci (QTL) analysis (Bush and Wise 1996) and comparative mapping (O’Donoughue et al. 1995; Wight et al. 2003) had assigned other crown rust resistance genes to linkage group 4 in the reference oat map. A receptor-like kinase gene (LrK10) was also mapped to this region (Cheng et al. 2002). In addition, this linkage group was located by comparative mapping of the gene for resistance to stem rust, Pg9, (O’Donoughue et al. 1996). Pg9 has been shown to be linked to Pc68 (Chong et al. 1994). Furthermore, three putative loci associated with partial resistance to crown rust were located on the linkage group OT-11, which appears to be homologous to KO 4 (Jackson et al. 2006, 2007). It is clear that KO 4_12 is important for rust resistance, and efforts should be made to increase the number of markers in this region. The AFLP molecular markers U8PM22 and U8PM25 should prove useful for further study of this cluster of resistance genes.
References
Barzen E, Stahl R, Fuchs E, Borchardt DC, Salamini F (2004) Development of coupling-repulsion phase SCAR markers diagnostic for the sugar beet Rr1 allele conferring resistance to rhizomania. Mol Breed 3:231–238
Brogin RL (2005) Mapeamento de genes de resistência à ferrugem e de QTLs envolvidos à Septoriose em soja. Dissertation. Universidade de São Paulo, Piracicaba, 93 p
Bryan GJ, Stephenson P, Collins A, Kirby J, Smith JB, Gale MD (1999) Low levels of DNA sequence variation among adapted genotypes of hexaploid wheat. Theor Appl Genet 99:192–198
Bush AL, Wise RP (1996) Crown rust resistance loci on linkage groups 4 and 13 in cultivated oat. J Hered 87:427–432
Cervera MT, Ruiz-Garcia L, Martinez-Zapater JM (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol Genet Genomics 268:543–552
Chen CX, Wang ZL, Yang DE, Ye CJ, Zhao YB, Jin DM, Weng ML, Wang B (2004) Molecular tagging and genetic mapping of the disease resistance gene RppQ to southern corn rust. Theor Appl Genet 108:945–950
Chen G, Chong J, Gray M, Prashar S, Procunier JD (2006) Identification of single-nucleotide polymorphisms linked to resistance gene Pc68 to crown rust in cultivated oat. Can J Plant Pathol 28:214–222
Cheng DW, Armstrong KC, Tinker N, Wight C, He S, Lybaert A, Fedak G, Molnar SJ (2002) Genetic and physical mapping of Lrk10-like receptor kinase sequences in hexaploid oat (Avena sativa L.). Genome 45:100–109
Chong J (2000) Incidence and virulence of Puccinia coronata in Canada from 1996 to 1998. Can J Plant Pathol 22:99–109
Chong J, Howes NK, Brown PD, Harder DE (1994) Identification of the stem rust resistance gene Pg9 and its association with crown rust resistance and endosperm proteins in ‘Dumont’ oat. Genome 37:440–447
Chong J, Leonard KJ, Salmeron JJ (2000) A North American system of nomenclature for Puccinia coronata f.sp. avenae. Plant Dis 84:580–585
Cruz CD, Schuster I (2004) GQMOL: aplicativo computacional para análise de dados moleculares e de suas associações com caracteres quantitativos [Online]. Available at http://www.ufv.br/dbg/gqmol/gqmol.htm
Duble CM, Quint M, Melchinger AE (2003) Saturation of two chromosome regions conferring resistance to SCMV with SSR and AFLP markers by targeted BSA. Theor Appl Genet 106:485–493
Giovanni C, Dell’Orco P, Bruno A, Ciccarese F, Lotti C, Ricciardi L (2004) Identification of PCR-based markers (RAPD, AFLP) linked to a novel powdery mildew resistance gene (ol-2) in tomato. Plant Sci 166:41–48
Golas TM, Sikkem A, Gros J, Feron RMC, Van den Berg RG, Van der Weerden GM, Mariani C, Allefs J (2010) Identification of a resistance gene Rpi-dlc1 to Phytophthora infestans in European accessions of Solanum dulcamara. Theor Appl Genet 120:797–808
Haley SD (1994) Selection for monogenic pest resistance traits with coupling and repulsion-phase RAPD markers. Crop Sci 34:1061–1066
Jackson EJ, Avant JB, Overturf KE, Bonman JM (2006) A quantitative assay of Puccinia coronata f. sp. avenae DNA in Avena sativa. Plant Dis 90:629–636
Jackson EJ, Obert DE, Menz M, Hu G, Bonman JM (2007) Qualitative and quantitative trait loci conditioning resistance to Puccinia coronata pathotypes NQMG and LGCG in the oat (Avena sativa L.) cultivars Ogle and TAM O-301. Theor Appl Genet. doi:10.1007/s00122-007-0687-x
Johnston MR, Carsten LD, Douglas L, Sands DC (2000) Epidemic development and virulence in 1995–1998 of Puccinia coronata, a potential biocontrol agent of wild oats on San Clemente Island. Biol Control 17:250–257
Kamei A, Tsuro M, Kubo N, Hayashi T, Wang N, Fujimura T, Hirai M (2010) QTL mapping of clubroot resistance in radish (Raphanus sativus L.). Theor Appl Genet 120:1021–1027
Kaur S, Cogan NOI, Ye G et al (2009) Genetic map construction and QTL mapping of resistance to blackleg (Leptosphaeria maculans) disease in Australian canola (Brassica napus L.) cultivars. Theor Appl Genet 120:71–83
Koeyer D, Orr W, Lybaert A, Deyl J, Chenier C, Tinker N, Mcelroy A, Chong J, Molnar S (2000) Scar markers linked to the Pc68 resistance allele are an effective tool for selection. In: Cross RJ (ed) International oats conference. New Zealand Institute for Crop and Food Research Limited, Christchurch, New Zealand, pp 137–140
Leonard KJ (2002) Oat lines with effective adult plant resistance to crown rust. Plant Dis 86:593–598
Leonard KJ (2003) Regional frequencies of virulence of oat crown rust in the United States from 1990 through 2000. Plant Dis 87:1301–1310
Leonard KJ, Martinelli JA (2005) Virulence of oat crown rust in Brazil and Uruguay. Plant Dis 89:802–808
Li CD, Rossnagel BG, Scoles GJ (2000) The development of oat microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor Appl Genet 101:1259–1268
Liu XH, Tan ZB, Rong TZ (2009) Molecular mapping of a major QTL conferring resistance to SCMV based on immortal RIL population in maize. Euphytica 167:229–235
Locatelli AB, Federizzi LC, Milach SCK, Wight CP, Molnar SJ, Chapados JT, Tinker NA (2006) Loci affecting flowering time in oat under short-day conditions. Genome 49:1528–1538
Martinelli JA, Federizzi LC, Bennedeti AC (1994) Redução do rendimento de grãos de aveia em função da severidade da ferrugem da folha. Summa Phytopathologica 20:116–118
Matsuo K, Silke S, Georgiev O, Marti P, Giovannini N, Rungger D (1998) An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J 17:1446–1453
McCallum BD, Fetch T, Chong J (2007) Cereal rust control in Canada. Aust J Agr Res 58:639–647
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis—a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Nat Acad Sci USA 88:9828–9832
Nikaido A, Yoshimaru H, Tsumura Y, Suyama Y, Murai M, Nagasaka K (1999) Segregation distortion for AFLP markers in Cryptomeria japonica. Genes Genetic Syst 74:55–59
O’Donoughue LS, Kianian SF, Rayapati PJ et al (1995) A molecular linkage map of cultivated oat. Genome 38:368–380
O’Donoughue LS, Chong J, Wight CP, Fedak G, Molnar SJ (1996) Localization of stem rust resistance genes and associated molecular markers in cultivates oat. Phytopathology 86:719–727
Orr W, Koeyer D, Chenier C, Tinker N, Molnar SJ (1999) SCAR markers for rust resistance genes Pc 68, Pg3, and Pg9 designed for marker assisted selection in oats. Plant & Animal Genome VII Conference, San Diego, CA, p 345
Penner GA, Chong J, Levesquelemay M, Molnar SJ, Fedak G (1993a) Identification of a Rapd marker linked to the oat stem rust gene-Pg3. Theoret Appl Genetics 85:702–705
Penner GA, Chong J, Wight CP, Molnar SJ, Fedak G (1993b) Identification of an Rapd marker for the crown rust resistance gene Pc68 in oats. Genome 36:818–820
Portyanko VA, Hoffman DL, Lee M, Holland JB (2001) A linkage map of hexaploid oat based on grass anchor DNA clones and its relationship to other oat maps. Genome 44:249–265
Randhawa HS, Popovic Z, Menzies J, Knox R, Fox S (2009) Genetics and identification of molecular markers linked to resistance to loose smut (Ustilago tritici) race T33 in durum wheat. Euphytica 169:151–157
Renganayaki K, Fritz AK, Sadasivam S et al (2002) Mapping and progress toward map-based cloning of Brown planthopper biotype-4 resistance gene introgressed from Oryza officinalis into cultivated Rice, O. sativa. Crop Sci 42:2112–2117
Roy A, Frascaria N, Mackay J, Bousquet J (1992) Segregating random amplified polymorphic DNAs (RAPDs) in Betula alleghaniensis. Theor Appl Genet 85:173–180
Rubiales D, Niks RE (2000) Combination of mechanisms of resistance to rust fungi as a strategy to increase durability. Options Mediterraneennes 40:333–339
Silva HD, Vencovsky R (2002) Testes múltiplos e associação de marcas associadas a QTL’s. Sci Agr 99:755–762
Tan YD, Wan CL, Zhu YF, Lu C, Xiang ZH, Deng HW (2001) An amplified fragment length polymorphism map of the silkworm. Genetics 157:1277–1284
Testolin R, Huang WG, Cipriani G, Retamales J (2001) Towards a linkage map in kiwifruit (Actinidia chinensis Planch.) based on microsatellites and satured with AFLP markers. Acta Horticultural 498:79–84
Tinker NA, Kilian A, Wight CP, Heller-Uszynska K, Wenzl P, Rines HW, Bjornstad A, Howarth CJ, Jannink J-L, Anderson JM, Rossnagel BG, Stuthman DD, Sorrells ME, Jackson EW, Tuvesson S, Kolb FL, Olsson O, Federizzi LC, Carson ML, Ohm HW, Molnar SJ, Scoles GJ, Eckstein PE, Bonman JM, Ceplitis A, Langdon T (2009) New DArT markers for oat provide enhanced map coverage and global germplasm characterization. BMC Genomics 10:39. doi:10.1186/1471-2164-10-39
Vieira EA, Carvalho FIF, Chaves MS, ACde Oliveira, Benin G, Hartwig I, Silva JAG, Bertan I, Martins AF, Martins LF (2007) Virulence variability of Puccinia coronata f. sp. avenae isolates collected in three counties from Rio Grande do Sul. Plant Dis 91:66–70
Vogl C, Xu SZ (2000) Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics 155:1439–1447
Vos P, Hogers R, Bleeker M et al (1995) Aflp—a new technique for DNA-fingerprinting. Nucleic Acids Res 23:4407–4414
Wang YH, Thomas CE, Dean RA (1997) A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) markers. Theor Appl Genet 95:791–798
Weng Y, Lazar MD (2002) Amplified fragment length polymorphism- and simple sequence repeat-based molecular tagging and mapping of greenbug resistance gene Gb3 in wheat. Plant Breed 121:218–223
Werner K, Friedt W, Ordon F (2005) Strategies for pyramiding resistance genes against the barley yellow mosaic virus complex (BaMMV, BaYMV, BaYMV-2). Mol Breed 16:45–55
Wight CP, Tinker NA, Kianian SF et al (2003) A molecular marker map in ‘Kanota’ × ‘Ogle’ hexaploid oat (Avena spp.) enhanced by additional markers and a robust framework. Genome 46:28–47
Wong LSL, Mckenzie RIH, Harder DE, Martens JW (1983) The Inheritance of Resistance to Puccinia-Coronata and of Floret Characters in Avena-Sterilis. Can J Genet Cytol 25:329–335
Yu J, Herrmann M (2006) Inheritance and mapping of a powdery mildew resistance gene introgressed from Avena macrostachya in cultivated oat. Theor Appl Genet 113:429–437
Zar JH (2009) Biostatistical analysis. Prentice Hall, New Jersey, USA
Zhu S, Kaeppler HF (2003) Identification of quantitative trait loci for resistance to crown rust in oat line MAM17-5. Crop Sci 43:358–366
Acknowledgment
The authors thank to the Brazilian Council for Scientific and Technological Development (CNPq) for the scholarship provided to the first author.
Author information
Authors and Affiliations
Corresponding author
Additional information
Part of the Master of Science dissertation of Franceli R. Kulcheski.
Rights and permissions
About this article
Cite this article
Kulcheski, F.R., Graichen, F.A.S., Martinelli, J.A. et al. Molecular mapping of Pc68, a crown rust resistance gene in Avena sativa . Euphytica 175, 423–432 (2010). https://doi.org/10.1007/s10681-010-0198-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0198-8