Abstract
Cathepsin B is a cysteine protease lysosomal enzyme involved in several physiological functions. Overexpression of the enzyme enhances its proteolytic activity and causes the breakdown of amyloid precursor protein (APP) into neurotoxic amyloid β (Aβ), a characteristic hallmark of Alzheimer’s disease (AD). Therefore, inhibition of the enzyme is a crucial therapeutic aspect for treating the disease. Combined structure and ligand-based drug design strategies were employed in the current study to identify the novel potential cathepsin B inhibitors. Five different pharmacophore models were developed and used for the screening of the ZINC-15 database. The obtained hits were analyzed for the presence of duplicates, interfering PAINS moieties, and structural similarities based on Tanimoto’s coefficient. The molecular docking study was performed to screen hits with better target binding affinity. The top seven hits were selected and were further evaluated based on their predicted ADME properties. The resulting best hits, ZINC827855702, ZINC123282431, and ZINC95386847, were finally subjected to molecular dynamics simulation studies to determine the stability of the protein–ligand complex during the run. ZINC123282431 was obtained as the virtual lead compound for cathepsin B inhibition and may be a promising novel anti-Alzheimer agent.
Graphical abstract
The methodology utilized for the identification of novel cathepsin B inhibitors through combined structure and ligand-based drug design approach:

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD), described by Dr. Alois Alzheimer in a 51-year-old woman, is an age-related progressive neurodegenerative disorder and is the leading cause of dementia in elderly. It is often characterized by the extracellular deposition of amyloid-β (Aβ) plaques and intracellular accumulation of toxic neurofibrillary tangles (NFTs) of hyperphosphorylated tau protein [1, 2]. Several hypotheses have been put up to explain the development and progression of AD, including cholinergic deficiency, Aβ toxicity, tau protein hyperphosphorylation, synaptic dysfunction, glutamatergic excitotoxicity, vitamin B5 deficiency, oxidative stress, and neuroinflammation etc. [3]. About 0.1% of the cases of AD have roots with genetics through presence of either the APOE 4 allele, TREM2, or the mutation in any of the three genes encoding Presenilin 1 and 2, or the amyloid precursor protein (APP) [4]. Currently, only few drugs have been approved by the FDA for managing AD including acetylcholinesterase inhibitors (Donepezil, Rivastigmine, and Galantamine), NMDAR antagonist (Memantine), and recently approved monoclonal antibodies (Aducanumab and Lecanemab) [5]. However, only the symptomatic alleviation is offered by the drugs. Therefore, there is a dire need to develop disease-modifying therapies, which necessitates discovery and identification of new druggable AD targets and corresponding drugs [6].
Cathepsins are proteases essential for normal physiological functions. These are classified into several subtypes including A, B, C, D, E, F, G, H, K, L, O, S, V, W, and Z (Table S1) [7, 8]. Among all the cathepsins, cathepsin B has a considerable role in cognitive impairment associated with AD [9]. It is abundantly expressed in the CNS regions of AD patients and is produced from the rough endoplasmic reticulum (RER) as an inactive preprocathepsin B enzyme containing 339 amino acids. Cleavage of 17 amino acids off the N-terminal forms procathepsin B that is further transformed into the active cathepsin B form by the successive proteolytic action of different enzymes in the Golgi bodies (Figure S1) [10]. Studies suggests that cathepsin B enzyme has crucial role in producing the highly toxic pGlu-Aβ oligomers and subsequent neuroinflammation through activation of IL-1β, IL-18, NF-kB, NLRP3 inflammasome, caspase-dependent cleavage of antiapoptotic protein (Bcl-2), or the microglial release (Figure S2) [11,12,13]. Therefore, its inhibition is a considerable therapeutic approach for treating AD.
Cathepsin B inhibitors of both natural and synthetic origin are reported in the literature (Figure S3). One of the earliest identified inhibitors, i.e., E-64 (L-trans-epoxysuccinylleucylamido(4-guanidino)butane), was obtained from Aspergillus japonicus [10]. A series of stereoisomeric analogues of E-64 were designed and synthesized by Schaschke et al. and (2S, 3S) conformation was identified to be remarkably favored against cathepsin B [14]. Dipeptidyl nitriles (Greenspan et al.), peptidyl aryl vinyl sulfones (Mendieta et al.), peptidyl epoxides (Albeck et al.), fluoromethyl ketones (Rasnick et al.), and epoxysuccinates are the different classes of synthetic inhibitors identified from previous studies on the enzyme [15,16,17,18,19,20]. Sosic et al. synthesized novel derivatives of nitroxoline against cathepsin B enzyme by replacing positions 5, 7, and 8 of parent nuclei with different groups in order to evaluate their effect on endopeptidase and exopeptidase activity [21]. Most of the reported cathepsin B inhibitors bind irreversibly at the molecular level. The potent cathepsin B inhibitor, with activity in the nanomolar range, is also an epoxysuccinyl peptide CA-074 (N-(L-3-trans propylcarbamoyloxirane-2-carbonyl)-L-isoleucyl-L-proline). Studies have reported that it is highly selective to cathepsin B for binding over other cathepsins, including L, H, and S from the same family [22].
There have not been many reports of cathepsin B inhibitors [23]. But it is crucial to find novel, potential, and diverse leads that are safe, suited for chemical optimization, and can be developed into therapeutic medicines to combat AD. Computational tools provide a significant direction in the early-stage drug design in lesser time and cost-effective manner. Homology modeling, virtual screening, pharmacophore mapping, molecular docking, molecular dynamics simulations, and QSAR modeling are the most common techniques employed in computer-aided drug design. Among these, virtual screening is pivotal in computational drug discovery [24]. Virtual screening uses 3D visuals that facilitate more straightforward modification and comprehensive insight. Millions of druggable ligands from diverse chemical spaces (ZINC, ChEMBL, Enamine, PubChem database, etc.) can be screened altogether against the pharmacological target to find potential leads either through structure-based or ligand-based approach. Typically, virtual ligand search is optimized to screen molecules based on known ligands with particular physicochemical or drug-like characteristics that comply with Veber’s or Lipinski’s rule of molecular properties that control the oral bioavailability of drug molecules. More than 70 marketed drugs have been discovered using computational technique, marking its marvelous role beyond contemporary drug discovery [25].
In the present study, we have employed combined structure and ligand-based drug design strategies, including pharmacophore-based virtual screening, molecular docking, and dynamics simulations studies for the identification of novel ligands against cathepsin B enzyme that can act as possible lead(s) for the management of AD. The pharmacophoric features were mapped to screen the ZINC-15 database, and the obtained hits were subjected to different filters. The molecular docking was performed to determine the potential hits having good binding affinity to the target protein. Finally, molecular dynamics simulation studies were carried out to determine the stability of the protein–ligand complex of best hits having admittable ADME properties.
Materials and methods
All the computational studies described in the study (Fig. 1) were performed using DELL Inspiron 3505 system supported with Windows 11 home single language, version 22H2, 8.00 GB RAM, 64-bit operating system, and inbuilt AMD Ryzen 3 dual-core 3250U processor.
Pharmacophore feature mapping and model generation
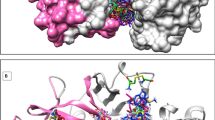
A pharmacophore model is an assembly of steric and electronic attributes required to enable the best supramolecular interactions with a particular biological target to modulate its biological response [26]. The co-crystallized ligand, CA-074 (propylamino-3-hydroxy-butan-1,4-dionyl)-isoleucyl-proline) in complex with cathepsin B protein (PDB ID: 1QDQ) was selected for pharmacophore modeling (Fig. 2). The protein file was downloaded in PDB file format from the protein data bank (https://www.rcsb.org/). The essential features of the ligand required for binding and inhibiting the target protein were identified using BIOVIA discovery studio visualizer software and available literature [27]. These were further mapped to construct eleven diversified pharmacophore models using the Pharmit search engine (https://pharmit.csb.pitt.edu/) (Table 1) [28]. The spatial coordinates of different features (hydrogen donor, hydrogen acceptor, and hydrophobic group) were used to construct the models summarized in Table S3.
Pharmacophore validation
The validation of pharmacophore models is essential to examine their potential to discriminate between sets of actives and decoys to accomplish the virtual screening objective. Here, the two most crucial validation parameters, viz. enrichment factor (E.F.) value, and Goodness of Hits (G.H.) score, were used for this purpose. The enrichment factor represents the actives present in a particular portion of the model-ordered database compared to a random sample [29]. A higher E.F. value for a model indicates its better ability to distinguish actives from inactives in a dataset. G.H. score helps to evaluate model selectivity and its effectiveness for database similarity search [30]. Its value ranges from 0 to 1, where one indicates the ideal model.
A dataset of 510 molecules comprising ten actives and 500 decoys for cathepsin B protein was used to validate the pharmacophore models using the Pharmit web server. Active compounds analogous of CA-074 against cathepsin B protein were retrieved from available literature, and corresponding decoys were generated for benchmarking using the “DUD-E decoys” web server (https://dude.docking.org/) [31]. The E.F. value and G.H. score for each pharmacophore model were calculated using Eqs. (1) and (2).
where D corresponds to the total number of molecules in the dataset, Ht denotes the total number of hits obtained, Ha indicates the total number of active hits, and A represents the total actives in the database. The pharmacophore models with higher E.F. values and G.H. scores were further chosen for virtual screening.
Pharmacophore-based virtual screening
ZINC-15 database was utilized for performing virtual screening against five validated pharmacophore models using the Pharmit search engine. The obtained hits against each model were further reduced using two subsequent primary and secondary filters. Primary filtering was performed by limiting the maximum number of hits per conformation and molecule to one during the virtual screening process. A cut-off value of 2.0 mRMSD (root mean square deviation) was set as the secondary filter within the parameters section of the Pharmit web server to obtain only the hits that had mRMSD value in this range.
Duplicates removal
The duplicate ligands obtained during virtual screening were eliminated using the “descriptor calculation” feature of Open Babel GUI 2.4.1 software. InChI key (International Chemical Identifier) descriptor was generated for every hit molecule based on the SMILES string pattern (simplified molecular input line entry system) and further screened to remove duplicates [32, 33].
Pan-assay interference compounds (PAINS) and Brenk filter
Compounds possessing certain functional groups or substructures, including alkylidene, fused tetrahydroquinoline, rhodanine, phenolic Mannich base, catechol, quinone, enone, isothiazolone, and epoxide, fall under the category of PAINS moieties [34]. These often give false positive results in a biochemical assay owing to the interfering non-specific interaction with the target protein through different mechanisms. These are similar to the structural alerts of the BRENK filter [35]. Therefore, it is required to remove these from the high throughput screening results of the database. An integrated workflow based on the KNIME analytics platform (version 4.7.0) was prepared to screen out the PAINS and BRENK moieties (Fig. 3) [36]. The “SDF reader” node was used to draw in the compound’s library, which was further processed through the “RDKit molecule catalog filter” node to remove the interfering moieties containing PAINS and BRENK structural alerts. Finally, the “SDF writer” node exported the resulting screened molecules in SDF format.
Drug-likeliness filter
Lipinski’s rule of five is the most often used criterion for assessing the druggable potential of the obtained hits. The molecules that obey the rule with nil violations (molecular weight, clogP, number of hydrogen bond acceptors, and donors) are considered orally bioavailable [37]. To this attempt, a KNIME workflow (Fig. 3) employing different nodes such as SDF reader, Lipinski’s rule of five, row filter, duplicate row filter, and SDF writer, was designed to screen the molecules based on the drug-likeness criterion mentioned in Table S2 with zero violations. An additional “molecular properties filter” node was utilized to filter the resulting molecules based on their topological polar surface area (TPSA). The adjacent “row filter” node screened out the molecules with a cut-off TPSA value of 70 Å2, essential for molecules to cross the blood–brain barrier (BBB) [38].
Molecular structural diversity filter
The “RDKit diversity picker” node of KNIME analytics platform 4.7.0 was utilized to retain only the molecules that had high structural diversity. It uses the MaxMin algorithm to filter out the molecules in different rows based on the Tanimoto distance between the fingerprints generated from the respective molecules [39].
Protein and ligand preparation
Protein preparation and loop filling for missing residues
The X-ray crystal structure of cathepsin B protein in complex with CA-074 was downloaded in PDB format from the protein data bank using PDB ID: 1QDQ. This PDB was chosen because of its resolution of 2.18 Å, R-value of 0.152, and the co-crystallized ligand, CA-074 (IC50 = 44 nM), that has been highly explored for neurodegenerative diseases as reported in the literature (Fig. 4) [10, 22, 40, 41].
The protein structure was visualized using Discovery Studio. The water molecules, the co-crystallized ligand, and unnecessary ligand groups were removed from the protein structure. Finally, the molecule was carefully examined for missing amino acid residues using UCSF chimera (version 1.17.1) (https://www.cgl.ucsf.edu/chimera/download.html) [42]. Two amino acids, viz. glycine and arginine, at positions 48 and 49, were missing from the protein chain (Fig. 5). The amino acids were added at respective positions using the MODELLER module available in Chimera software [43]. After loop filling, the generated model was saved in PDB format and used for further studies.
Protein energy minimization
Energy minimization is essential to reduce atomic clashes in the structure of a molecule arising out of unfavorable torsion angles and bad contacts. Two distinct algorithms, viz., steepest descent and conjugate gradient, are widely used for this purpose. However, the conjugate gradient algorithm is preferred over the steepest descent in identifying local minima [44]. UCSF chimera (version 1.17.1) was used to perform energy minimization of the cathepsin B protein. Hydrogen atoms were added to the structure, and Gasteiger charges were applied, followed by a combination of 5000 steps of steepest descent and conjugate gradient integrator. The final energy-minimized structure obtained was saved in PDB format for docking purposes.
Ligands preparation and energy minimization
The hits obtained after pharmacophore-based virtual screening and subsequent filters were first drawn using ChemDraw 22.0.0 and then converted into the three-dimensional structure with the help of Chem3D software. Further, energy minimization for each ligand was performed by utilizing the MMFF94 force field. The resulting molecules were extracted in a folder and were saved in PDBQT format for further high throughput structure-based virtual screening.
Structure-based high throughput virtual screening and molecular docking studies
Grid generation and validation
For grid generation, amino acids (GLN23, TRP30, GLY73, GLY74, PRO76, HIS110, HIS111, ALA173, GLY198, HIS199, and TRP221) involved in the interaction with the co-crystallized ligand were identified by visualizing the protein–ligand complex in Discovery studio (Figure S5). Further, these amino acid residues were used to map the active site coordinates of cathepsin B. Grid box of size 74 × 48 × 48 and dimensions X: 9.391, Y: 19.262, and Z: − 7.337 was selected with a grid spacing of 0.375. To validate the docking protocol, we docked our co-crystallized ligand into the active site of cathepsin B protein using a grid box of the dimensions and size. Redocking was performed to calculate the RMSD value [45]. An RMSD value was found as of 2.652 Å, which is near to the acceptable limit (≤ 2 Å). Another method, which involves docking of actives and inactives (decoys) to the protein’s active site, was employed to validate the docking protocol [46]. Ten actives with corresponding 500 decoys (generated using the DUD-E decoys web server) were docked, and an ROC plot (receiver operating characteristic curve) was constructed using the StAR web server (http://melolab.org/star/home.php). The area under the curve (AUC) was 0.76, between the acceptable limit of 0.6 and 1.0 (Figure S6). Based on this value, it was assumed that the established grid parameters and the docking protocol would yield reproducible and reliable findings [45].
Docking
The ligands were docked to the binding site of the cathepsin B protein while following the validated docking protocol. Lamarckian algorithm was used to run the docking program, and the binding score in kcal/mol was calculated using the following Eq. (3):
Initially, the structure-based high throughput virtual screening was performed by docking the ligands using a fast-docking algorithm in open-access PyRx software. The ligands with binding energy less than − 6.5 kcal/mol (docking score of co-crystallized ligand, CA-074) were screened for the next docking phase.
AutoDock 4.2 was utilized to perform the subsequent two docking experiments, standard precision and extra precision. The parameters employed for docking the ligands in both stages are summarized in Table S4. The ligands with binding energy greater than − 7.0 kcal/mol were excluded after the standard precision docking stage, and the rest of the ligands were taken up for the next regressive phase, i.e., extra-precision docking. It was reported that double H-bonding with HIS110 and HIS111 of the protein is essential to inhibit the enzyme [27]. Therefore, the ligands that complied with the requirement were selected for further ADME studies.
In silico ADME parameters prediction
SwissADME, an open web server was used to calculate ADME properties for the selected ligands by giving SMILES code as the input (http://www.swissadme.ch/) [35]. Parameters such as gastrointestinal (GI) absorption, BBB permeability, p-glycoprotein substrate binding, and cytochrome P enzyme inhibition potential specific to ligand were considered for screening out best hits.
In silico toxicity prediction
Toxicity risk assessment for any drug candidate is a major concern to ensure its safety to the general population. Several FDA-approved drugs have been recalled from the market due to their toxicity symptoms. We performed the toxicity risk assessment for the selected hits, including human ether-a-go-go (hERG) inhibition, carcinogenicity, mutagenicity, and genotoxicity, using ADMETlab 2.0 web server (https://admetmesh.scbdd.com/) [47]. The hits that were found to be non-carcinogenic, non-mutagenic, and non-genotoxic were selected for further study.
Molecular dynamics (MD) simulations studies
Molecular dynamics (MD) simulations studies helps in analyzing the binding pattern and stability of ligand conformations within the binding pocket of the target protein molecule (receptor, enzyme, or lipidic membrane) in the simulated dynamic biological environment. The deep binding pockets, usually hidden and inaccessible to ligands for complex formation, can be quickly acclimatized through simulation studies. These studies are essential to determine the thermodynamic behavior and stability of the protein–ligand complex in the aqueous environment containing ions and electrolytes at physiological temperature (37 °C) and pressure [48]. Moreover, MD simulations helps to find out the actual movement of atoms and molecules over a given period of time [49].
The top hits retrieved after regress in silico toxicity studies were subjected to MD simulations studies for 100 ns timescale, performed using Desmond module of Schrodinger Maestro suite 2021. Protein–ligand complexes were processed using Desmond’s protein preparation wizard window, followed by generating an orthorhombic box surrounded with TIP3P (transferable intermolecular potential with three points) water molecules to solvate the protein–ligand complexes. Further, 0.15 M NaCl was added to neutralize the excess charge present in the complex. The pressure and temperature throughout the MD run were set to 1.013 bar and 310 K, respectively, to mimic the typical physiological environment while maintaining a constant number of atoms. The energy of the system was minimized for 2000 iterations on a convergence threshold of 1 kcal/mol/Å using OPLS3 (optimized potentials for liquid simulations) force field and subjected to MD simulations long run for 100 ns [48]. Finally, the MD trajectory obtained was evaluated using Desmond’s simulations interactions diagram window. The obtained RMSD, RMSF, and radius of gyration along with the solvent-accessible surface area for each protein–ligand complex were analyzed.
RMSD analysis
The root mean square deviation (RMSD) value determines and compares the structural conformational changes in the protein backbone from its initial to final pose. The deviations generated during the simulation process can be utilized to evaluate the protein’s stability concerning its conformation. Lesser deviation in the RMSD corresponds to better protein stability [49]. The RMSD value for the C-alpha backbone and ligand was calculated for 100 ns simulations run using Eq. (4) to determine the stability of the system corresponding to each generated frame (x).
where N is the total number of atoms in the atom selection, tref is the reference time for the first frame (reference frame) taken at time t = 0, tx denotes the recording time for any frame x, and r’ indicates the position of the selected atoms after superimposing frame x on the reference frame.
RMSF analysis
The root mean square fluctuation (RMSF) is utilized to assess the stiffness, flexibility, and regional variations in the protein chain, which gives the binding pattern of the ligand to the target [50]. The RMSF value for the cathepsin B enzyme following binding with the selected ligands were obtained from the trajectories recorded throughout the MD simulations run. This parameter is calculated using the following Eq. (5):
where T indicates trajectory time, tref denotes the reference time, ri represents the ith residue position, and riʹ is the atomic position of the ith residue obtained after overlaying on the reference. The angle brackets in the equation denote the average taken for the square distance calculated for atoms of protein’s residue. RMSF plot was constructed for each complex to illustrate the residues that underwent significant modifications during the MD simulations run. The peaks in a typical RMSF plot represent the most oscillating residues [51]. Proteins with a more significant number of flexible domains have higher RMSF values [52].
Radius of gyration (rGyr) analysis
Another critical parameter for assessing the size, structural compactness, and protein flexibility under the physiological environment is the radius of gyration [53]. It compares protein structure with the experimentally monitored hydrodynamic radius as a function of time. The radius of gyration was calculated to quantify structural changes in each complex utilizing the data generated from the trajectories obtained after the MD simulations run. The following formula is used to calculate the value of rGyr (Eq. 6) [54].
where ri and rcenter display atom coordinates and center of mass, respectively. N denotes the number of atoms in the protein. During the simulation, a protein structure with lower rGyr values is said to be more stiff [52].
Solvent accessible surface area (SASA) analysis
The SASA for any biomolecule, expressed in Å2, is the surface area the solvent molecules can access through Van der Waals contact. It is determined by imagining the molecule surface as a collection of dots and tracing the accessible surface area with a probe, typically a sphere. The quantitative analysis of the protein SASA is essential to illustrate complexes’ folding and unfolding patterns [55]. Protein molecules with lower SASA values are comparatively more rigid and compact. We calculated the SASA value for each of our protein–ligand complexes subjected to MD simulations using a spherical probe having Van der Waals radii of 1.4 Å.
Results and discussion
Pharmacophore model validation and virtual screening
Pharmacophoric validation was performed by calculating Enrichment factor and Goodness of hits score (Table 2) for each model in order to measure their capability to distinguish between active and decoy datasets. The dataset comprising 510 molecules, including both actives and inactives (decoys), was screened through individual models. The pharmacophore models 2, 3, 4, 9, and 10 (Fig. 6) rendered good results and were selected for virtual screening of ZINC library.
The different features of pharmacophore models were treated as inclusive spheres and used in mapping the ligands from the ZINC library against the co-crystallized ligand, CA-074. The hits so obtained were further refined using primary and secondary filters. The results of the virtual screening are summarized in Table 3. In total, 16,355 hits were obtained against queried features within mRMSD value of 2.0 Å.
Duplicates removal
The hits obtained through pharmacophore-based virtual screening were filtered for the presence of duplicates. InChI key descriptor was calculated using SMILES string as the input for each ligand. The screening rendered 15,105 molecules out of 16,355 total hits by eliminating 1250 duplicates.
Molecular properties filtering using KNIME workflow
The retrieved hits were analyzed based on their molecular properties such as PAINS, BRENK, drug-likeliness (Lipinski’s rule of five) and structural features using the KNIME workflow (Fig. 3). The “RDKit molecule catalog filter” node available in the KNIME analytics platform was used to screen out the hits containing PAINS and BRENK alerts. This node compares and matches the ligands against the SMARTS (SMILES arbitrary target specification) pattern generated using identified PAINS fragments or moieties. The ligands that match the SMARTS pattern are flagged as PAINS or BRENK molecules and are excluded from the dataset. A total of 10,686 hits were retrieved after PAINS and BRENK filtering.
Further, the screening of hits based on the drug-likeness criterion was carried out by determining the values of their molecular weight, clogP, number of hydrogen bond donors, number of hydrogen acceptors, number of heavy atoms, and topological polar surface area (TPSA) using same KNIME workflow in continuous manner. The hits with zero violations to the Lipinski rule of five and a TPSA value below 70 Å2 (CNS active) were selected. Out of 10,686 hits, only 8821 molecules aligned with the Lipinski rule of five. Further, only 946 hits with TPSA < 70 Å2 were retrieved out of 8821 molecules. The obtained hits can be regarded as CNS active. The drug-likeness results for the top hits are summarized in Table 4.
Finally, the ligands so obtained were subjected to the “RDKit diversity picker” node in the same KNIME workflow. It classified the molecules based on the Tanimoto coefficient calculated by aligning the molecules against their corresponding fingerprints row-wise. 250 out of 946 hits were retained and taken for the next stage, i.e., molecular docking.
Structure-based high throughput virtual screening and molecular docking studies
High throughput structure-based virtual screening was carried out on 250 hits by docking them against the cathepsin B protein using a fast-docking program in PyRx software. 78 hits with binding energy less than − 6.5 kcal/mol were selected from HTVS results. Further, AutoDock 4.2 software, a more regressive and robust docking software, was used to perform two-step molecular docking for the retained hits. The first docking step yielded 30 hits with a binding score of less than − 7.0 kcal/mol. The amino acid interactions and the binding score for the obtained hits (step 1) are summarized in Table S5. During the second docking step, the ligands that showed double hydrogen bonding with amino acids HIS110 and HIS111 of the occluding loop were selected apart from other essential interactions. Out of 30 hits available at step 1, only seven hits showed the requisite type of interactions with the target protein as illustrated in Table 5.
There are two principal lobes (S1ʹ and S2ʹ) that constitute the active site of the enzyme with their two further subdivision sites (P1, P2, P1ʹ, and P2ʹ) as described in Figure S4. S1ʹ lobe is constituted by amino acids VAL176, LEU181, MET196, HIS199, and TRP221 while S2ʹ lobe is constituted by amino acids GLN23, GLY24, HIS110, HIS111, GLY121, and TRP221. Double hydrogen bonding with amino acids HIS110 and HIS111 is essential for inhibiting the enzyme at S2ʹ lobe. Additionally, there is one hydrophobic pocket formed by backbone chain amino acids (PRO76, ALA173, and ALA200) for stabilizing the binding of interacting ligand. The selected ligands displayed favorable amino acids interactions similar to the reference compound, CA-074, for inhibiting the target enzyme as illustrated through their 2D and 3D interactions diagram (Figs. 7 and 8).
The ligand ZINC19663116 displayed conventional hydrogen bonding interactions with HIS110, CYS26, THR120, GLY121, and HIS111 (important amino acids for hydrogen bonding with S2ʹ subsite and occluding loop of the enzyme responsible for its inhibition), π-alkyl interactions with PRO118, HIS199 and TRP221 (hydrophobic core), and formed carbon-hydrogen bond with CYS119 and GLU122. ZINC123282431 produced conventional hydrogen bonding interactions with amino acids HIS110, HIS111, and GLY198 (essential for inhibition at S2ʹ lobe), and π-alkyl interactions with HIS199 at S1ʹ lobe. Apart from this, it also formed additional amino acids interactions with CYS29, PRO118, and CYS119. ZINC1390969 showed conventional hydrogen bonding interactions with amino acid residues HIS110 and HIS111 (essential for inhibiting the enzyme at S2ʹ lobe), van der Waals interaction with GLY27, π-alkyl interactions with CYS29 and CYS119, amide-π stacked interaction with CYS26, and π-cationic interactions with GLU122. Altogether, these interactions were favorable for inhibiting the enzyme. ZINC13565072 exhibited essential double H-bonding interactions with HIS110 and HIS111 (S2ʹ lobe), π-cationic interactions with GLU122, π-π stacked interactions with HIS199, π-alkyl interactions with VAL176, and even formed a carbon-hydrogen bond with CYS119. ZINC72392119 produced hydrogen bonding interactions with HIS110 and HIS111, π-π stacked interactions with HIS199, and π-alkyl interactions with VAL176 and CYS119. ZINC827855702 also displayed hydrogen bonding interactions with HIS110 and HIS111, additional π-π stacked interaction with HIS199 (S2ʹ lobe), π-sulfur interactions with CYS29, carbon-hydrogen bonding with THR120, and π-alkyl interactions with PRO118, CYS119, TRP221, VAL176, LEU181, and MET196. ZINC95386847 showed hydrogen bonding interactions with GLN23 (constituent of oxyanion hole of the cathepsin B enzyme), CYS29, HIS110, and HIS111 (S2ʹ lobe), van der Waals interaction with GLY197, carbon-hydrogen bonding with GLY197, π-π stacked interactions with MET196 and TRP221 (S1ʹ lobe), π-sulfur interaction with CYS119, and π-alkyl interactions with LEU181 and VAL176. Altogether, all the selected ligands displayed double hydrogen bonding with amino acids HIS110 and HIS111 of S2ʹ lobe (essential for inhibiting the enzyme) along with the other essential amino acids and had high binding energy compared to the reference molecule.
In silico ADME parameters prediction
The top hits retained after molecular docking were analyzed for their ADME properties using the SwissADME web server. For a drug to act on CNS, it must cross the BBB. All our compounds were found to cross BBB and had high gastrointestinal absorption, as illustrated by the results described in Table 6. Further, the hits were assessed for their affinity towards P-glycoprotein and cytochrome P450 enzymes. The P-glycoprotein is an essential ATP-binding cassette (ABC) transporter that restricts the entry or binding of foreign toxins or drugs to the cells via efflux mechanisms. Therefore, the drugs having an affinity towards the P-glycoprotein are extruded out, which in turn hampers their pharmacodynamic effect. From our dataset of seven compounds, only four compounds (highlighted in Table 6) were found to be non-substrate to P-glycoprotein.
Further, BOILED-egg analysis (Brain or intestinal estimated permeation) was performed for our selected ligands using the SwissADME web server. The BOILED-egg method uses a predictive algorithm to compute gastrointestinal absorption and BBB permeability of small molecules based on their lipophilicity and polarity. The results are represented through a graphical diagram shaped like a boiled egg, wherein the molecules are represented through small dots or points. The molecules falling in the white, elliptical region of the diagram have a high tendency to permeate the gastrointestinal tract passively. The egg yolk (yellow region) in the diagram represents the biochemical space for the molecules having a high probability of crossing the BBB. The molecules that are neither BBB permeant nor passively absorbed through the GI tract are placed in the grey region of the diagram. All compounds fell in the egg-yolk region, illustrating their capability to cross BBB. Three compounds were found substrate to P-glycoprotein marked blue in the diagram (Fig. 9). The compounds (2, 3, 6, and 7) symbolized in red (PGP-ve) were selected for further studies.
Toxicity risk assessment analysis
The toxicity risk assessment for the selected compounds was performed using the ADMETlab 2.0 web server. The AMES test for mutagenicity, hERG inhibition, acute oral toxicity, carcinogenicity, and genotoxicity were included. The ligands ZINC123282431, ZINC827855702, and ZINC95386847 showed zero carcinogenic, mutagenic, and genotoxic potential, as indicated through in silico data summarized in Table 7. Moreover, the ligands depicted acceptable value for acute oral toxicity.
Molecular dynamics (MD) simulations studies
The molecular dynamics simulations studies for the ligands ZINC123282431, ZINC827855702, and ZINC95386847 with cathepsin B protein were carried out to investigate their conformational stability over a time scale of 100 ns. The trajectories generated from MD simulations were used to calculate the values of RMSD, RMSF, radius of gyration, and solvent-accessible surface area (SASA) for each protein–ligand complex. Moreover, the binding behavior, protein–ligand contacts, and type of amino acid interactions involved between ligand and protein were examined throughout the simulations run.
MD analysis of ZINC123282431 with cathepsin-B protein
The analysis of trajectories obtained from the MD simulations studies of the ZINC123282431 with cathepsin B protein depicted slight fluctuations in protein backbone (Cα) for the initial 20 ns time interval, which later became stable throughout the simulation run of 100 ns. The average RMSD value of the protein backbone chain for the whole run was 1.83 Å with a maximum deviation of 2.23 Å at 90 ns, which lies between the acceptable limit of 1–3 Å for small and globular proteins. Ligand was relatively stable during the run with an average RMSD value of 1.84 Å. The RMSF plot of the protein revealed more fluctuations at residues between the 48 and 52 positions, away from the positions of amino acids (HIS110 and HIS111) involved in hydrogen bonding with the ligand. Therefore, the protein complex was stable during the MD simulations run. On examining the interactions diagram, it was found that the ligand formed hydrogen bonding with amino acids HIS110, HIS111, GLY198, and TRP221 throughout the run. Hydrophobic contacts with amino acids CYS29, PRO118, and TRP221 were also established between the ligand and protein. The average values for the radius of gyration and solvent-accessible surface area for the protein–ligand complex during the run were also calculated using the data obtained from MD trajectories and found to be 4.31 Å and 178.76 Å2, respectively. Figure 10 summarizes the MD simulation results for the ligand ZINC123282431 with the protein complex.
MD simulations data of the ligand ZINC123282431 with protein cathepsin B enzyme for 100 ns: a RMSD plot of protein–ligand complex, b Protein (cα) RMSF plot, c Ligand RMSF plot, d 2D diagram of amino acids interactions with the ligand, e Bar diagram of protein–ligand contacts, where green, grey, and blue colors represent H-bonding, hydrophobic interactions, and water bridges, respectively, f radius of gyration (rGyr) in Å, and g solvent accessible surface area (SASA) in Å2
MD analysis of ZINC827855702 with cathepsin-B protein
The MD simulation data for the ligand ZINC827855702 revealed significant fluctuations in the protein backbone (Cα) during the run. The RMSD indicated an unstable trajectory of the Cα chain for the initial 10 ns, followed by steadiness for the next 35 ns. The protein backbone chain became stable throughout the run with an acceptable RMSD value of 2.04 Å. The ligand showed a maximum RMSD of 5.08 Å and the average RMSD value of 3.31 Å during the simulations. The RMSF plot depicted the stability of the protein–ligand complex as there were minuscule fluctuations at 100 to 115 positions in the protein chain. The radius of gyration for the protein complex was found to be 4.43 Å. The SASA was quantified as 172.35 Å2 utilizing the MD simulations data of the complex. The ligand formed hydrogen bonding with GLY24, HIS110, HIS111, CYS119, GLY121, GLU122, GLY197, and TRP221, while hydrophobic contacts were established with amino acids MET196, HIS199, and TRP221 of the protein chain during the entire simulation run of 100 ns timescale. Figure 11 summarizes the MD simulation data for the ligand ZINC827855702 with the protein complex.
MD simulations data of the ligand ZINC827855702 with protein cathepsin B enzyme for 100 ns: a RMSD plot of protein–ligand complex, b Protein (cα) RMSF plot, c Ligand RMSF plot, d 2D diagram of amino acids interactions with the ligand, e Bar diagram of protein–ligand contacts, where green, grey, and blue colors represent H-bonding, hydrophobic interactions, and water bridges, respectively, f radius of gyration (rGyr) in Å, and g solvent accessible surface area (SASA) in Å2
MD analysis of ZINC95386847 with cathepsin-B protein
The trajectory of the ligand ZINC95386847 with the protein showed high fluctuations for the initial 20 ns, which later became steady throughout the run. The average RMSD value for the protein backbone was calculated as 1.90 Å, which was under acceptable limits. The ligand in the protein complex showed a high RMSD value of 4.88 Å. Moreover, fluctuations in the protein chain at positions between 100 and 120 were seen through the RMSF plot. It signifies the instability of the protein–ligand complex during the simulations run. The values for the radius of gyration and SASA for the protein complex were calculated to be 4.01 Å and 145.43 Å2, respectively. Analysis of the interaction diagram depicted that the ligand formed frequent hydrogen bonding with amino acids GLN23, CYS29, HIS110, THR120, and TRP221 and intense hydrophobic interactions with amino acids CYS29, VAL176, PHE180, LEU181, MET196, HIS199, TRP221, and TRP225 of the protein. Figure 12 summarizes the MD simulation results for the ligand ZINC95386847 with the protein complex.
MD simulations data of the ligand ZINC95386847 with protein cathepsin B enzyme for 100 ns: a RMSD plot of protein–ligand complex, b Protein (cα) RMSF plot, c Ligand RMSF plot, d 2D diagram of amino acids interactions with the ligand, e Bar diagram of protein–ligand contacts, where green, grey, and blue colors represent H-bonding, hydrophobic interactions, and water bridges, respectively, f radius of gyration (rGyr) of the ligand in Å, and g solvent accessible surface area (SASA) of ligand in Å2
Conclusion
Cathepsin B has an extensive role in developing and progressing multifactorial Alzheimer’s disease through its involvement in Aβ generation and neuroinflammation. Developing novel cathepsin B inhibitors against AD is an excellent therapeutic option. Computational tools serve as an aid to speed up the modern drug discovery process in a cost-effective manner for identifying new moieties, which can be used as a starting lead for discovering novel therapeutic drugs. This study employed combined structure and ligand-based drug design strategies to identify novel cathepsin B inhibitors. Based on the co-crystallized ligand in complex with PDB file, five pharmacophore models were constructed and validated for screening the ZINC-15 database. The resulting hits were checked for duplicates, PAINS moieties, and structural similarities. Molecular docking study was carried out for screening hits showing considerably good binding affinity. The top seven hits obtained were further filtered based on criteria involving BBB permeability, high gastrointestinal absorption, p-glycoprotein substrate, and toxicity considerations. Finally, the best hits, ZINC827855702, ZINC123282431, and ZINC95386847, were subjected to molecular dynamics simulation studies to assess the protein–ligand complex stability under a dynamic simulated biological environment. ZINC123282431 was obtained as the final virtual lead, displaying acceptable RMSD values, compelling protein–ligand complex stability throughout the simulation run, and satisfying all the parameters undertaken for the study. The ligand depicted an overall binding energy score of − 9.31 kcal/mol and comparable essential amino acids interactions (CYS29, HIS110, HIS111, PRO118, CYS119, GLY198, HIS199) required to inhibit the cathepsin B enzyme. Moreover, based on the evaluation of top hits, the study predicted imidazopyridine, pyrazole, chromane, quinoline, and phenanthrene moieties, which can be used as starting scaffold for building novel cathepsin B inhibitors for the treatment of Alzheimer’s disease.
Data availability
No datasets were generated or analysed during the current study.
References
Cummings J et al (2016) Drug development in Alzheimer’s disease: the path to 2025. Alzheimer’s Res Therap 8:1–12
Castellani RJ, Rolston RK, Smith MA (2010) Alzheimer disease. Disease-a-Month: DM 56(9):484
Ju Y, Tam KY (2022) Pathological mechanisms and therapeutic strategies for Alzheimer’s disease. Neural Regen Res 17(3):543
Zhang W et al (2020) Association of rare variants in neurodegenerative genes with familial Alzheimer’s disease. Ann Clin Transl Neurol 7(10):1985–1995
Roy K (2023) Computational modeling of drugs against Alzheimer’s disease, vol 203. Springer
Srivastava S, Ahmad R, Khare SK (2021) Alzheimer’s disease and its treatment by different approaches: a review. Eur J Med Chem 216:113320
Gasteiger E et al (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31(13):3784–3788
Patel S et al (2018) Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed Pharmacother 105:526–532
Pišlar A, Kos J (2014) Cysteine cathepsins in neurological disorders. Mol Neurobiol 49:1017–1030
Sharma A et al (2022) Cathepsin BA neuronal death mediator in Alzheimer’s disease leading to neurodegeneration. Mini Rev Med Chem 22(15):2012–2023
Kelley N et al (2019) The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci 20(13):3328
Hafner-Bratkovič I et al (2012) NLRP3 inflammasome activation in macrophage cell lines by prion protein fibrils as the source of IL-1β and neuronal toxicity. Cell Mol Life Sci 69:4215–4228
Campden RI, Zhang Y (2019) The role of lysosomal cysteine cathepsins in NLRP3 inflammasome activation. Arch Biochem Biophys 670:32–42
Schaschke N et al (1997) E-64 analogues as inhibitors of cathepsin B: on the role of the absolute configuration of the epoxysuccinyl group. Bioorg Med Chem 5(9):1789–1797
Chitranshi N et al (2021) Identification of novel cathepsin B inhibitors with implications in Alzheimer’s disease: computational refining and biochemical evaluation. Cells 10(8):1946
Zhou Z, Wang Y, Bryant SH (2010) QSAR models for predicting cathepsin B inhibition by small molecules—continuous and binary QSAR models to classify cathepsin B inhibition activities of small molecules. J Mol Graph Model 28(8):714–727
Greenspan PD et al (2001) Identification of dipeptidyl nitriles as potent and selective inhibitors of cathepsin B through structure-based drug design. J Med Chem 44(26):4524–4534
Mendieta L et al (2010) Novel peptidyl aryl vinyl sulfones as highly potent and selective inhibitors of cathepsins L and B. ChemMedChem 5(9):1556–1567
Albeck A (2000) Peptidyl epoxides as selective protease inactivators. Drug Dev Res 50(3–4):425–434
Rasnick D (1985) Synthesis of peptide fluoromethyl ketones and the inhibition of human cathepsin B. Anal Biochem 149(2):461–465
Sosič I et al (2013) Development of new cathepsin B inhibitors: combining bioisosteric replacements and structure-based design to explore the structure–activity relationships of nitroxoline derivatives. J Med Chem 56(2):521–533
Yoon MC et al (2022) Molecular features of CA-074 pH-dependent inhibition of cathepsin B. Biochemistry 61(4):228–238
Pan X et al (2005) Amentoflavone and its derivatives as novel natural inhibitors of human cathepsin B. Bioorg Med Chem 13(20):5819–5825
Sabe VT et al (2021) Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur J Med Chem 224:113705
Reddy AS et al (2007) Virtual screening in drug discovery—a computational perspective. Curr Protein Pept Sci 8(4):329–351
Yang S-Y (2010) Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discovery Today 15(11–12):444–450
Yamamoto A et al (2000) Substrate specificity of bovine cathepsin B and its inhibition by CA074, based on crystal structure refinement of the complex. J Biochem 127(4):635–643
Sunseri J, Koes DR (2016) Pharmit: interactive exploration of chemical space. Nucleic Acids Res 44(W1):W442–W448
Kchouk S, Hegazy L (2022) Pharmacophore modeling for biological targets with high flexibility: LXRβ case study. Med Drug Discov 15:100135
Bhagwati S, Siddiqi MI (2020) Identification of potential soluble epoxide hydrolase (sEH) inhibitors by ligand-based pharmacophore model and biological evaluation. J Biomol Struct Dyn 38(16):4956–4966
Mysinger MM et al (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55(14):6582–6594
Heller SR et al (2015) InChI, the IUPAC international chemical identifier. J Cheminf 7(1):1–34
O’Boyle NM et al (2011) Open Babel: an open chemical toolbox. J Cheminf 3(1):1–14
Pouliot M, Jeanmart S (2016) Pan assay interference compounds (PAINS) and other promiscuous compounds in antifungal research. J Med Chem 59(2):497–503
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):42717
Berthold MR et al (2009) KNIME-the Konstanz information miner: version 2.0 and beyond. AcM SIGKDD Explor Newslett 11(1):26–31
Tian S et al (2015) The application of in silico drug-likeness predictions in pharmaceutical research. Adv Drug Deliv Rev 86:2–10
Wager TT et al (2010) Moving beyond rules: the development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem Neurosci 1(6):435–449
Ashton M et al (2002) Identification of diverse database subsets using property-based and fragment-based molecular descriptions. Quant Struct-Act Relat 21(6):598–604
Yamashima T et al (1998) Inhibition of ischaemic hippocampal neuronal death in primates with cathepsin B inhibitor CA-074: a novel strategy for neuroprotection based on ‘calpain–cathepsin hypothesis.’ Eur J Neurosci 10(5):1723–1733
Hook V et al (2020) Cathepsin B in neurodegeneration of Alzheimer’s disease, traumatic brain injury, and related brain disorders. Biochim Biophys Acta 1868(8):140428
Pettersen EF et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Šali A et al (1995) Evaluation of comparative protein modeling by MODELLER. Proteins: Struct Funct Bioinf 23(3):318–326
Mackay D, Cross A, Hagler A (1989) The role of energy minimization in simulation strategies of biomolecular systems. Prediction of protein structure and the principles of protein conformation. Springer, pp 317–358
Hevener KE et al (2009) Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model 49(2):444–460
Pinto VDS et al (2019) In silico study to identify new antituberculosis molecules from natural sources by hierarchical virtual screening and molecular dynamics simulations. Pharmaceuticals 12(1):36
Xiong G et al (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49(W1):W5–W14
Salo-Ahen OM et al (2020) Molecular dynamics simulations in drug discovery and pharmaceutical development. Processes 9(1):71
Aier I, Varadwaj PK, Raj U (2016) Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci Rep 6(1):34984
Cob-Calan NN et al (2019) Molecular docking and dynamics simulation of protein β-tubulin and antifungal cyclic lipopeptides. Molecules 24(18):3387
Tripathi N et al (2022) Virtual screening and molecular simulation study of natural products database for lead identification of novel coronavirus main protease inhibitors. J Biomol Struct Dyn 40(8):3655–3667
Ghahremanian S et al (2022) Molecular dynamics simulation approach for discovering potential inhibitors against SARS-CoV-2: a structural review. J Mol Liq 354:118901
Parida PK, Paul D, Chakravorty D (2020) The natural way forward: molecular dynamics simulation analysis of phytochemicals from Indian medicinal plants as potential inhibitors of SARS-CoV-2 targets. Phytother Res 34(12):3420–3433
Jiang Z et al (2019) Effects of an electric field on the conformational transition of the protein: a molecular dynamics simulation study. Polymers 11(2):282
Ausaf Ali S et al (2014) A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr Protein Pept Sci 15(5):456–476
Acknowledgements
JJ and RS would like to express their gratitude for the financial research assistance provided to them through teaching assistantships by the Ministry of Education (MoE), New Delhi, India. NGB is genuinely grateful to the MoE for giving him the PMRF fellowship. We gladly acclimate the resources and assistance offered by the “PARAM Shivay Facility” at the Indian Institute of Technology (BHU), Varanasi, as part of the National Supercomputing Mission, Government of India. We also wish to thank the Centre for Computing and Information Services (CCIS), Indian Institute of Technology (BHU), Varanasi, for the computational assistance.
Author information
Authors and Affiliations
Contributions
JJ: design, conceptualization, data curation, molecular modeling and original draft preparation. NGB: computational modeling, guidance, editing and reviewing of the manuscript. RS: molecular docking. AK: supervision. SKS: supervision, reviewing, administering, and editing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jangra, J., Bajad, N.G., Singh, R. et al. Identification of novel potential cathepsin-B inhibitors through pharmacophore-based virtual screening, molecular docking, and dynamics simulation studies for the treatment of Alzheimer’s disease. Mol Divers (2024). https://doi.org/10.1007/s11030-024-10821-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-024-10821-z