Abstract
For this work, two series of new piperazine derivatives (3a–o) and triazolo-pyrazine derivatives (3p–t) were synthesized in a single-step reaction. All twenty adducts were obtained in good to high yields and fully characterized by 1H NMR, 13C NMR, IR, and mass spectrometry techniques. To further confirm the chemical identity of the adducts, a crystal of N-{[(4-chlorophenyl)-3-(trifluoromethyl)]-5,6-dihydro-[1,2,4]triazolo[4,3-a]}pyrazine-7(8H)-carboxamide (3t) was prepared and analyzed using X-ray crystallography. In vitro screening of the antimicrobial activity of all compounds (3a–t) was evaluated against five bacterial and two fungal strains. This study disclosed that N-{[(3-chlorophenyl)]-4-(dibenzo[b,f][1,4]thiazepin-11-yl)}piperazine-1-carboxamide (3o) was the superior antimicrobial with good growth inhibition against A. baumannii. Furthermore, the results from the performed molecular docking studies were promising, since the observed data could be used to develop more potent antimicrobials.
Graphic abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Piperazine and triazolo-pyrazine are nitrogen-containing heterocyclic compounds (Fig. 1) that play an important role in medicinal chemistry since both moieties can serve as frameworks for small molecule synthesis, drug design, and drug discovery. For instance, some of the piperazine and triazolo-pyrazine derivatives have shown diverse biological activities such as antidiabetic [1], anticancer [2], anticonvulsant [3], blood platelet aggregation inhibitors [4], human renin activity [5], DPP-IV inhibitor [6], anti-tubercular, and antioxidant potential [7].
It is well known that microbes develop antimicrobial resistance (AMR) to pharmaceutical drugs and the rate of this undesirable process is increasing rapidly over the years [8, 9]. In order to mitigate and/or overcome the AMR issue, newer drugs need to be designed and synthesized at the same rate at which the resistance is developing. Although many researchers across academic institutions, as well as pharmaceutical industries, are heavily involved in the development of new antimicrobial drugs to counteract AMR, there is still a great need for novel and more efficient drug candidates. A particular interest lies in small molecules that are easy to synthesize from simple starting materials under mild reaction conditions, such as the ones presented in this manuscript (Scheme 1).
In continuation of our effort to design, synthesize, and gain access to novel pharmacophores targeting multi-drug-resistant (MDR) strains, very recently we identified several promising N-substituted piperazines as novel antimicrobial agents [10]. Likewise, we have identified various other pharmacophores such as substituted chalcones, ureas, and N-heterocyclic carbenes and their corresponding metal complexes as feasible antimicrobial agents [11,12,13,14,15,16,17,18,19,20,21,22]. Owing to the importance of nitrogen-containing heterocycles, in various antimicrobial drug candidates, our research efforts have been directed towards the development of new antimicrobial agents from the aforementioned classes of nitrogen-containing heterocycles. In this work, we have synthesized fifteen different and previously unknown piperazine derivatives, as well as five novel piperazine-based triazolo[4,3-a]pyrazine adducts (Scheme 1). All prior compounds were screened against five bacteria and two fungi strains, and in general, the new adducts showed greater inhibition towards Acinetobacter baumannii versus the other screened microbial strains (Fig. 2). To our delight, the triazolo[4,3-a]pyrazine showed superior inhibition when compared to the piperazine derivatives, and therefore, these scaffolds will be further studied to understand the possible structure–activity relationship (SAR). At this point, the synthesis, characterization, and antimicrobial studies are reported below.
Results
Chemistry
The synthetic route utilized to prepare all piperazine and triazolo-pyrazine derivatives (3a–t) followed a mild and straightforward approach (Scheme 1). In order to prepare all the adducts, some starting materials were purchased from commercially available sources, for instance aryl isocyanates (Millipore Sigma) and 3-{(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]}pyrazine (TCI), whereas the mono-substituted piperazines (2a-e), namely 1-[bis(4-fluorophenyl)methyl]piperazine, 1-benzhydrylpiperazine, 1-[(4-fluorophenyl)(phenyl)methyl]-piperazine, 1-[(2-fluorophenyl)(4-fluorophenyl)methyl]piperazine and 11-(piperazin-1-yl-dibenzo[b,f][1, 4])thiazepine, were synthesized according to reported literature procedures [23, 24]. All piperazine and triazolo-pyrazine derivatives (3a–t) were synthesized in a single step from the reaction of isocyanates with 2a-f in toluene at 40–45 °C, for 1 h. This simple method affords high yields (75–85%), under catalysis-free and mild reaction conditions (Scheme 1).
Spectroscopic characterization
The chemical identities of all synthesized piperazine and triazolo-pyrazine derivatives (3a–t) were confirmed employing the following spectroscopic techniques: 1H NMR, 13C NMR, IR, and mass spectra. The FT-IR spectra for the piperazine and triazolo-pyrazine derivatives were recorded in the region from 400 to 4000 cm−1. The IR spectral data of compounds (3a–t) displayed peaks in the region of 3347–3233 cm−1 due to N–H stretching and peaks at 2885–2775 cm−1 due to aromatic C–H stretching. The peaks at 1678–1631 cm−1 of piperazine and triazolo-pyrazine derivatives may be assigned to the ν(C = O) of the urea groups. Peaks corresponding to C = C stretching were also observed at 1601–1494 cm−1. Chemical structures of all the piperazine and triazolo-pyrazine derivatives were further confirmed through 1H-NMR and 13C-NMR spectra. 1H-NMR and 13C-NMR spectra were appropriate to the reported chemical structures of the synthesized compounds. Mass spectra of the piperazine and triazolo-pyrazine derivatives showed the characteristic molecular ion peaks (M + H)+ in accordance with their molecular formulas.
X-ray crystallography
The single-crystal X-ray structure of compound 3t was acquired. Crystals suitable for X-ray analysis were grown by slow evaporation of dichloromethane at room temperature. A perspective view of compound 3t showing the atomic numbering is depicted in Fig. 3. The crystal data and refinement details of the compound 3t are summarized in Table 1, whereas selected bond lengths and bond angles are shown in Table S1 (supporting information). The compound crystallized in the monoclinic crystal system with space group P121/n1, Z = 4, V = 1442.416 Å3, and unit cell parameters a = 8.3200(5) A°, b = 15.3468(7) A°, c = 11.7575(7) A °, α = 90°, β = 106.095(1) and γ = 90°. The X-ray structure of compound 3t discloses that the molecule is coplanar in the solid state. The C–C bond lengths in aromatic rings are in the normal range of 1.352(5)-1.435(5) Å, which is characteristic of delocalized aromatic rings. The C–C–C bond angles in aromatic rings are around 120° with the variation being less than 3°, which is characteristic of sp2-hybridized carbons. In compound 3t, there is an absence of any lattice-held water molecules or organic solvent molecules in the unit cell of the determined structure. The O(1)–C(2) is 1.241(5) Å, which is typical for the C = O bonds. The C–Cl bond was 1.752(2) Å. The molecular packing diagram displays four layers of molecules. In each layer, the molecules are parallel. NH moiety in each molecule is not involved in hydrogen bonding and nevertheless appears to interact with the π-electrons of a benzene ring. The molecular packing diagram also displays the presence of one intermolecular hydrogen bond. One of the hydrogens of the aromatic ring of one molecule is involved in intermolecular hydrogen bonding with the oxygen of the C = O entity of another molecule. This hydrogen bonding stabilizes the crystal packing.
Software: APEX2 and SAINT (Bruker 2014) [18], SHELXS97 and SHELXL2013 (Sheldrick 2008) [19], and JANA2006 [Petricek et at. 2014].
Antimicrobial activity
The newly synthesized title compounds (3a–t) were evaluated for their antimicrobial activity against five bacterial [one gram-positive (Staphylococcus aureus) and four gram-negative (Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter baumannii)] and two fungal (Candida albicans and Cryptococcus neoformans) strains [Table 2]. The antimicrobial screening showed that some of the screened compounds exhibited good inhibition of antimicrobial growth against various tested microbial strains. The results indicated that among all twenty compounds, 3o displayed the best activity against Acinetobacter baumannii. Compound 3o has a chloro group and thiazepine moieties attached to the piperazine core, which is accounted for the enhanced antimicrobial activity. Among the various substituted compounds, compounds 3n-q against Staphylococcus. aureus, compound 3 g against Pseudomonas aeruginosa, compounds 3o, 3r, and 3t against Klebsiella pneumoniae, compound 3e, 3f, and 3o-s against Acinetobacter baumannii and compound 3n against Candida albicans showed good inhibition of antimicrobial growth (Table 2). Unfortunately, all compounds 3a–t did not show any useful inhibition of antimicrobial growth towards both Escherichia coli and Cryptococcus neoformans microbial strains. Overall, the reported class of compounds displayed the best inhibition towards A. baumannii, albeit the relationships between the structure of the heterocyclic scaffolds and the detected antimicrobial properties are still not clear and merits further studies. However, it is worth noting that the new triazolo-pyrazine derivatives did in fact showed higher inhibition when compared with piperazine derivatives (Table 2).
Molecular docking
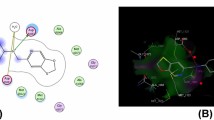
To investigate the mechanism of antimicrobial activity and detailed intermolecular interactions between the synthesized compounds, molecular docking studies were performed on the crystal structure of Acinetobacter baumannii PBP1a in complex with penicillin G (PDB ID 3UDI, 2.6 Å X-ray resolution) using the surflex-dock program of sybyl-X 2.0 software. All the twenty inhibitors along with the ligand were docked into the active site of the enzyme, as shown in Fig. 4a and b. The predicted binding energies of the compounds are listed in Table 3. The docking study revealed that all the compounds have shown good docking scores against the enzyme.
In order to make our study more meaningful, we selected one adduct from the piperazine derivatives (3 k) and one from the triazolo-pyrazine series (3t) to perform docking studies; see below. In Fig. 5a–c, compound (3 k) makes three hydrogen bonding interactions at the active site of the enzyme (PDB ID: 3UDI), among them three bonding interactions raised from oxygen atom carbonyl group with hydrogen atoms of LYS669, THR670, and SER487 (C = O––H-LYS669, 2.15 Å; C = O––H-THR670, 1.91 Å; C = O––H-SR487, 2.71 Å), and the remaining interaction came from the fluorine atom present on the 4th position of the phenyl ring makes interaction with the hydrogen atom of ARG488 (F–––H-ARG488, 2.63 Å). As depicted in Fig. 6a–c, compound (3t) makes three hydrogen bonding interactions at the active site of the enzyme (PDB ID: 3UDI), among them a bonding interaction raised from 2nd nitrogen atom of triazole ring with a hydrogen of ALA676 (-N–– H-ALA676, 2.30 Å), an oxygen atom of carbonyl group makes interaction with the hydrogen atom of TYR707 (C = O–––H-TYR707, 2.07 Å), and remaining one more interaction came from the hydrogen atom of CONH group with the oxygen atom of ARG704 (-CONH–– O-ARG704, 2.20 Å). The binding interaction of 3UDI_ligand with enzyme active sites shows six bonding interactions, and the docked view of the same is depicted in Fig. 7a-c. Figure 8a, b represents the hydrophobic and hydrophilic amino acids surrounded by the studied compound (3 k) and (3t).
All the compounds showed a consensus score in the range 6.77–1.44, indicating the summary of all forces of interaction between ligands, and the enzyme. Also, it was observed that the studied compounds have shown the same type of interactions with amino acid residues SER487, TYR707, and THR670 as that of reference 3UDI_ligand. This indicates that molecules preferentially bind to the enzyme in comparison with the reference 3UDI_ligand (Table 3).
Conclusions
In summary, we have synthesized fifteen piperazine derivatives (3a–o) and five triazolo-pyrazine derivatives (3p–t), from aryl isocyanates in a single-step reaction. The obtained yields were high, and all compounds were characterized by 1H NMR, 13C NMR, IR, and mass spectrometry techniques. Furthermore, the X-ray structure of the compound (3t) was elucidated and reported herein. The freshly prepared adducts were evaluated for antimicrobial activity. The outcome of this screening suggested that some of the compounds displayed moderate inhibition of antimicrobial growth. Among all the tested samples, compound (3o) exhibited the best antibacterial growth inhibition against A. baumannii. Hence, compound (3o) could be considered as promising antimicrobial lead and might form the structural backbone for further design and development of various piperazine derivatives to be developed and screened. Finally, molecular docking studies, of the adducts, were performed on the crystal structure of A. baumannii PBP1a in complex with penicillin G (PDB ID 3UDI, 2.6 Å X-ray resolution), and the results of such study are reported in this manuscript.
Experimental
General considerations
All readily available chemicals, including isocyanates and pyrazine starting material, were bought from either Millipore Sigma or TCI and were used without further purification. All solvents used in this work were of analytical grade and were used as received. All the reactions were carried out under aerobic conditions in oven-dried glassware with magnetic stirring. Heating was accomplished by either a heating mantle or silicone oil bath. Reactions were monitored by thin-layer chromatography (TLC) performed on 0.25-mm Merck TLC silica gel plates, using UV light as a visualizing agent. Purification of reaction products was carried out by flash column chromatography using silica gel 60 (230–400 mesh). Yields refer to isolated pure material. High vacuum or concentration in vacuo refers to the removal of volatile solvent using a rotary evaporator attached to a dry diaphragm pump (10–15 mm Hg) followed by pumping to a constant weight with an oil pump (< 300 mTorr). 1H spectra were recorded on JEOL Eclipse Plus 500 (500 MHz) and are reported relative to DMSO-d6 (δ 2.50). 1H NMR coupling constants (J) are reported in Hertz (Hz), and multiplicities are indicated as follows: s (singlet), d (doublet), t (triplet), quint (quintet), m (multiplet). Proton-decoupled 13C NMR spectra were recorded on JEOL Eclipse Plus 500 (125 MHz) and reported relative to DMSO-d6 (δ 39.52). IR spectra were recorded on an Alpha-P BrukerFT/IR spectrometer. Liquid chromatography-mass spectra (LC–MS) were recorded on an Agilent technologies quadrupole LC–MS system. X-ray diffraction data for compound (3t) were collected using Mo-Kα radiation and a Bruker SMART APEXII diffractometer [25]. The structure was solved by the direct method using SHELXS-97 and refined by full-matrix least squares on F2 for all data using SHELXL-97at 100 ºK [25]. An analytical absorption correction based on the shape of the crystal was performed. All hydrogen atoms were added at calculated positions and refined using a riding model. Anisotropic thermal displacement parameters were used for all non-hydrogen atoms. Further details about the data collection and reliability factors are listed in Table 1. CCDC–1992766 (for 3t) contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from the Cambridge Crystallographic Data Centre via http://www.ccdc.cam.ac.uk/data_request/cif.
Synthesis
General experimental procedure for the synthesis of all twenty derivatives.
To a solution of isocyanate (0.25 g, 1.823 mmol) in toluene (2.5 mL) was added a solution of the respective piperazine or triazolo-pyrazine derivative (1.823 mmol) in toluene (1.0 mL). The reaction mixture was heated at 40–45 °C for 30–60 min. Then, the reaction mixture was cooled down to room temperature (around 25 °C) and the resulting solids were filtered and washed with toluene (2.0 mL). The wet solids were then taken into toluene (2.0 mL), stirred at room temperature for about 30 min, filtered, and washed with toluene (1.0 mL) to obtain the crude adducts. Lastly, all the crude derivatives were purified by column chromatography using hexane/ethyl acetate (9:1) to afford pure piperazine and triazolo-pyrazine derivatives. All compounds are stable in air and light over a period of several months.
Synthesis of 4-{[bis(4-fluorophenyl)methyl]-N-(2-fluorophenyl)}piperazine-1-carboxamide (3a). Compound (3a) was synthesized from 2-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-[bis (4-fluorophenyl)methyl]piperazine (0.525 g, 1.823 mmol) according to the general procedure. White solid. Yield: 85% (0.66 g). 1H NMR (CDCl3, 500 MHz): δ 8.10–8.06 (m, 1H), 7.38–7.35 (m, 4H), 7.08 (t, J = 8.02 Hz, 1H), 7.05–6.92 (m, 6H), 6.59 (s, 1H), 4.26 (s, 1H), 3.51 (t, J = 5.15 Hz, 4H), 2.42 (t, J = 5.15 Hz, 4H). 13C NMR (CDCl3, 125 MHz): δ 162.03 (d, 1JC,F = 245.92 Hz), 154.33, 152.61 (d, 1JC,F = 239.92 Hz), 137.78 (d, 4JC,F = 2.40 Hz), 129.31 (d, 3JC,F = 8.40 Hz), 127.53 (d, 3JC,F = 9.60 Hz), 124.61 (d, 4JC,F = 2.40 Hz), 122.93 (d, 3JC,F = 8.40 Hz), 121.47, 115.71 (d, 2JC,F = 20.39 Hz), 114.64 (d, 2JC,F = 19.19 Hz), 74.36, 51.45, 44.25. IR (KBr): ῡ = 3347, 2816, 1640, 1503, 1218, 824. LC–MS for C24H22F3N3O: m/z = 426 [M + H]+.
Synthesis of 4-[benzhydryl-N-(2-fluorophenyl)]piperazine-1-carboxamide (3b). Compound (3b) was synthesized from 2-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-benzhydrylpiperazine (0.46 g, 1.823 mmol) according to the general procedure. White solid. Yield: 80% (0.57 g). 1H NMR (CDCl3, 500 MHz): δ 8.11–8.08 (m, 1H),7.44 (d, J = 6.87 Hz, 4H), 7.31 (t, J = 7.45 Hz, 4H), 7.23–7.20 (m, 2H), 7.09 (t, J = 8.02 Hz,1H), 7.06–7.02 (m, 1H), 6.99–6.92 (m, 1H), 6.60 (d, J = 4.01 Hz, 1H), 4.27 (s, 1H), 3.52 (t, J = 5.15 Hz, 4H), 2.46 (t, J = 5.15 Hz, 4H). 13C NMR (CDCl3, 125 MHz): δ 154.35, 152.60 (d, 1JC,F = 239.92 Hz), 142.26, 128.73, 127.94, 127.59 (d, 3JC,F = 9.60 Hz), 127.28, 124.58 (d, 4JC,F = 3.60 Hz), 122.84 (d, 3JC,F = 7.20 Hz), 121.47, 114.61 (d, 2JC,F = 19.19 Hz), 76.03, 51.57, 44.26. IR (KBr): ῡ = 3233, 2816, 1645, 1501, 1265, 858. LC–MS for C24H24FN3O: m/z = 390 [M + H]+.
Synthesis of N-{(2-fluorophenyl)-4-[(4-fluorophenyl)(phenyl)methyl]}piperazine-1-carboxamide (3c). Compound (3c) was synthesized from 2-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-[(4-fluorophenyl)(phenyl)methyl]piperazine (0.49 g, 1.823 mmol) according to the general procedure. White solid. Yield: 83% (0.62 g). 1H NMR (CDCl3, 500 MHz): δ 8.09 (t, J = 8.02 Hz, 1H), 7.40–7.39 (m, 4H), 7.31 (t, J = 6.87 Hz, 2H), 7.24–7.21 (m, 1H), 7.09 (t, J = 7.45 Hz, 1H), 7.05–6.92 (m, 4H), 6.59 (d, J = 2.86 Hz, 1H), 4.26 (s, 1H), 3.51 (t, J = 5.15 Hz, 4H), 2.44 (t, J = 5.15 Hz, 4H). 13C NMR (CDCl3, 125 MHz): δ 161.98 (d, 1JC,F = 245.92 Hz), 154.33, 152.59 (d, 1JC,F = 241.12 Hz), 142.02, 138.03, 129.38 (d, 3JC,F = 8.40 Hz), 128.83, 127.85, 127.58 (d, 3JC,F = 9.60 Hz), 127.44, 124.61 (d, 2JC,F = 2.40 Hz), 122.87 (d, 3JC,F = 7.20 Hz), 121.44, 115.61 (d, 2JC,F = 21.59 Hz), 114.62 (d, 2JC,F = 19.19 Hz), 75.20, 51.52, 44.27. IR (KBr): ῡ = 3331, 2853, 1631, 1519, 1262, 757. LC–MS for C24H23F2N3O: m/z = 408 [M + H]+.
Synthesis of N-{(2-fluorophenyl)-4-[(2-fluorophenyl)(4-fluorophenyl)methyl]}piperazine-1-carboxamide (3d). Compound (3d) was synthesized from 2-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-[(2-fluorophenyl)(4-fluorophenyl)methyl]piperazine (0.52 g, 1.823 mmol) according to the general procedure. White solid. Yield: 82% (0.64 g). 1H NMR (CDCl3, 500 MHz): δ 8.07 (t, J = 8.02 Hz, 1H), 7.59 (t, J = 6.87 Hz, 4H), 7.43–7.40 (m, 2H), 7.22–7.18 (m, 1H), 7.15–7.12 (m, 1H), 7.08 (t, J = 8.02 Hz,1H), 7.05–6.92 (m, 5H), 6.61 (s, 1H), 4.72 (s, 1H), 3.52 (t, J = 4.58 Hz, 4H), 2.50–2.43 (m, 4H). 13C NMR (CDCl3, 125 MHz): δ 162.04 (d, 1JC,F = 245.92 Hz), 160.69 (d, 1JC,F = 245.92 Hz), 154.35, 152.65 (d, 1JC,F = 241.12 Hz), 136.96, 129.62 (d, 3JC,F = 8.40 Hz), 128.89, 128.74 (d, 3JC,F = 8.40 Hz), 128.55 (d, 4JC,F = 2.40 Hz), 127.53 (d, 3JC,F = 9.60 Hz), 124.59, 122.94 (d, 3JC,F = 7.20 Hz), 121.53, 115.82 (d, 2JC,F = 25.19 Hz), 115.71 (d, 2JC,F = 21.59 Hz), 114.65 (d, 2JC,F = 19.19 Hz), 65.99, 51.36, 44.23. IR (KBr): ῡ = 3337, 2885, 1641, 1504, 1263, 758. LC–MS for C24H22F3N3O: m/z = 426 [M + H]+.
Synthesis of 4-[(dibenzo[b,f][1,4]thiazepin-11-yl)-N-(2-fluorophenyl)]piperazine-1-carboxamide (3e). Compound (3e) was synthesized from 2-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 11-(piperazin-1-yl-dibenzo [b,f] [1, 4])thiazepine (0.53 g, 1.823 mmol) according to the general procedure. White solid. Yield: 81% (0.64 g). 1H NMR (CDCl3, 500 MHz): δ 8.10–8.06 (m, 1H), 7.54 (d, J = 8.02 Hz, 1H), 7.42–7.40 (m, 1H), 7.38–7.31 (m, 4H), 7.22–7.19 (m, 1H), 7.12–7.04 (m, 3H), 7.00–6.95 (m, 1H), 6.92 (dd, J = 7.45, 1.15 Hz, 1H), 6.63 (d, J = 4.01 Hz, 1H), 3.67–3.52 (m, 8H). 13C NMR (CDCl3, 125 MHz): δ 160.72, 154.46, 152.71 (d, 1JC,F = 241.12 Hz), 148.63, 140.17, 133.96, 132.44, 132.39, 131.24, 129.21, 128.98, 128.61, 128.06, 127.41 (d, 3JC,F = 9.60 Hz), 125.38, 124.67 (d, 4JC,F = 3.60 Hz), 123.39, 123.17 (d, 3JC,F = 8.40 Hz), 121.63, 114.74 (d, 2JC,F = 20.39 Hz), 43.74. IR (KBr): ῡ = 3270, 2842, 1646, 1593, 1237, 750. LC–MS for C24H21FN4OS: m/z = 433 [M + H]+.
Synthesis of 4-{[bis(4-fluorophenyl)methyl]-N-(3-fluorophenyl)}piperazine-1-carboxamide (3f). Compound (3f) was synthesized from 3-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-[bis(4-fluorophenyl)methyl]piperazine (0.52 g, 1.823 mmol) according to the general procedure. White solid. Yield: 82% (0.64 g). 1H NMR (CDCl3, 500 MHz): δ 7.36–7.33 (m, 4H), 7.27–7.24 (m, 1H), 7.19–7.14 (m, 1H), 7.01–6.96 (m, 5H), 6.72–6.68 (m, 1H), 6.58 (s, 1H), 4.23 (s, 1H), 3.47 (t, J = 5.15 Hz, 4H), 2.38–2.36 (m, 4H). 13C NMR (CDCl3, 125 MHz): δ 163.15 (d, 1JC,F = 244.72 Hz), 162.03 (d, 1JC,F = 245.92 Hz), 154.67, 140.78 (d, 3JC,F = 10.80 Hz), 137.74, 129.92 (d, 3JC,F = 9.60 Hz), 129.32 (d, 3JC,F = 8.40 Hz), 129.16, 128.34, 125.41, 115.71 (d, 2JC,F = 21.59 Hz), 115.08 (d, 4JC,F = 2.40 Hz), 109.75 (d, 2JC,F = 21.59 Hz), 107.29 (d, 2JC,F = 26.39 Hz), 74.36, 51.46, 44.28. IR (KBr): ῡ = 3336, 2802, 1638, 1503, 1235, 777. LC–MS for C24H22F3N3O: m/z = 426 [M + H]+.
Synthesis of 4-[benzhydryl-N-(3-fluorophenyl)]piperazine-1-carboxamide (3 g). Compound (3 g) was synthesized from 3-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-benzhydrylpiperazine (0.46 g, 1.823 mmol) according to the general procedure. White solid. Yield: 83% (0.59 g). 1H NMR (CDCl3, 500 MHz): δ 7.43 (d, J = 7.45 Hz, 4H), 7.31–7.26 (m, 5H), 7.22–7.15 (m, 3H), 6.98–6.96 (m, 1H), 6.70 (td, J = 8.02, 2.29, 1H), 6.49 (s, 1H), 4.26 (s, 1H), 3.48 (t, J = 5.15 Hz, 4H), 2.42 (t, J = 5.15 Hz, 4H). 13C NMR (CDCl3, 125 MHz): δ 163.19 (d, 1JC,F = 243.52 Hz), 154.65, 142.26, 140.80 (d, 3JC,F = 10.80 Hz), 129.92 (d, 3JC,F = 8.40 Hz), 128.77, 127.98, 127.32, 114.98, 109.71 (d, 2JC,F = 21.59 Hz), 107.23 (d, 2JC,F = 26.39 Hz), 76.06, 51.61, 44.34. IR (KBr): ῡ = 3285, 2805, 1636, 1541, 1242, 746. LC–MS for C24H24FN3O: m/z = 390 [M + H]+.
Synthesis of N-{(3-fluorophenyl)-4-[(4-fluorophenyl)(phenyl)methyl]}piperazine-1-carboxamide (3 h). Compound (3 h) was synthesized from3-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-[(4-fluorophenyl)(phenyl)methyl]piperazine (0.49 g, 1.823 mmol) according to the general procedure. White solid. Yield: 81% (0.60 g). 1H NMR ((CD3)2CO, 500 MHz): δ 8.12 (s, 1H), 7.54–7.51 (m, 3H), 7.47 (d, J = 7.45, 2H), 7.31 (t, J = 7.45, 2H), 7.21–7.19 (m, 3H), 7.07 (t, J = 9.16, 2H), 6.69–6.66 (m, 1H), 4.37 (s, 1H), 3.56 (t, J = 5.15 Hz, 4H), 2.39 (t, J = 4.58 Hz, 4H). 13C NMR ((CD3)2CO, 125 MHz): δ 163.73 (d, 1JC,F = 239.92 Hz), 162.58 (d, 1JC,F = 243.52 Hz), 155.44, 143.52 (d, 3JC,F = 10.80 Hz), 143.41, 139.76 (d, 4JC,F = 2.40 Hz), 130.47 (d, 3JC,F = 9.60 Hz), 130.44 (d, 3JC,F = 8.40 Hz), 129.44, 128.64, 127.94, 115.99 (d, 2JC,F = 20.39 Hz), 115.47 (d, 4JC,F = 2.40 Hz), 108.80 (d, 2JC,F = 21.59 Hz), 106.87 (d, 2JC,F = 26.39 Hz), 75.60, 52.46, 44.91. IR (KBr): ῡ = 3327, 2808, 1635, 1541, 1241, 779. LC–MS for C24H23F2N3O: m/z = 408 [M + H]+.
Synthesis of N-{(3-fluorophenyl)-4-[(2-fluorophenyl)(4-fluorophenyl)methyl]}piperazine-1-carboxamide (3i). Compound (3i) was synthesized from 3-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 1-[(2-fluorophenyl)(4-fluorophenyl)methyl]piperazine (0.52 g, 1.823 mmol) according to the general procedure. White solid. Yield: 84% (0.65 g). 1H NMR (CDCl3, 500 MHz): δ 7.57 (t, J = 6.87, 1H), 7.41–7.39 (m, 2H), 7.28–7.12 (m, 4H), 7.00–76.98 (m, 4H), 6.73 (s, 1H), 6.68 (td, J = 8.59, 2.29, 1H), 4.69 (s, 1H), 3.47 (t, J = 4.58 Hz, 4H), 2.44–2.36 (m, 4H). 13C NMR (CDCl3, 125 MHz): δ 163.10 (d, 1JC,F = 243.52 Hz), 162.02 (d, 1JC,F = 245.92 Hz), 160.68 (d, 1JC,F = 247.12 Hz), 154.78, 140.83 (d, 3JC,F = 10.80 Hz), 136.93, 129.87 (d, 3JC,F = 9.60 Hz), 129.61 (d, 3JC,F = 8.40 Hz), 129.15, 128.85, 128.74 (d, 3JC,F = 8.40 Hz), 128.57 (d, 4JC,F = 3.60 Hz), 128.34, 125.41, 124.57, 115.80 (d, 2JC,F = 25.19 Hz), 115.61 (d, 2JC,F = 21.59 Hz), 115.22, 109.72 (d, 2JC,F = 21.59 Hz), 107.36 (d, 2JC,F = 26.39 Hz), 66.00, 51.36, 44.25. IR (KBr): ῡ = 3316, 2814, 1634, 1602, 1244, 757. LC–MS for C24H22F3N3O: m/z = 426 [M + H]+.
Synthesis of 4-[(dibenzo[b,f][1,4]thiazepin-11-yl)-N-(3-fluorophenyl)]piperazine-1-carboxamide (3j). Compound (3j) was synthesized from 3-fluoro phenyl isocyanate (0.25 g, 1.823 mmol) and 11-(piperazin-1-yl-dibenzo [b,f] [1, 4])thiazepine (0.538 g, 1.823 mmol) according to the general procedure. White solid. Yield: 83% (0.65 g). 1H NMR (CDCl3, 500 MHz): δ 7.54 (d, J = 7.45 Hz, 1H), 7.41 (dd, J = 7.45, 1.15 Hz, 1H), 7.38–7.35 (m, 1H), 7.33–7.32 (m, 2H), 7.30–7.26 (m, 1H), 7.22–7.17 (m, 2H), 7.10 (dd, J = 8.02, 1.15 Hz, 1H), 7.03–7.01 (m, 1H), 6.94–6.91 (m, 1H), 6.74–6.70 (m, 2H), 3.61–3.47 (m, 8H). 13C NMR (CDCl3, 125 MHz): δ 163.15 (d, 1JC,F = 244.72 Hz), 160.74, 154.88, 148.60, 140.65 (d, 3JC,F = 10.80 Hz), 140.09, 133.91, 1332.41 (d, 4JC,F = 3.60 Hz), 131.26, 129.99 (d, 3JC,F = 9.60 Hz), 129.33, 128.97, 128.64, 128.09, 125.35, 123.40, 115.27, 109.95 (d, 2JC,F = 21.59 Hz), 107.46 (d, 2JC,F = 25.19 Hz), 43.74. IR (KBr): ῡ = 3274, 2840, 1640, 1600, 1237, 738. LC–MS for C24H21FN4OS: m/z = 433 [M + H]+.
Synthesis of 4-{[bis(4-fluorophenyl)methyl]-N-(3-chlorophenyl)}piperazine-1-carboxamide (3 k). Compound (3 k) was synthesized from 3-chloro phenyl isocyanate (0.25 g, 1.628 mmol) and 1-[bis(4-fluorophenyl)methyl]piperazine (0.469 g, 1.628 mmol) according to the general procedure. White solid. Yield: 75% (0.54 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.67–8.65 (m, 1H), 7.62–7.60 (m, 1H), 7.46–7.43 (m, 4H), 7.35–7.34 (m, 1H), 7.26–7.21 (m, 1H), 7.18–7.13 (m, 4H), 6.97–6.95 (m, 1H), 4.43 (s, 1H), 3.45 (br, 4H), 2.29 (br, 4H). 13C NMR (DMSO-d6, 125 MHz): δ 161.11 (d, 1JC,F = 2443.52 Hz), 154.52, 142.12, 138.39, 132.70, 129.94, 129.0.47 (d, 3JC,F = 7.20 Hz), 121.22, 118.71, 117.63, 115.39 (d, 2JC,F = 20.39 Hz), 72.65, 51.16, 43.77. IR (KBr): ῡ = 3323, 2806, 1637, 1503, 1218, 824. LC–MS for C24H22ClF2N3O: m/z = 442 [M + H]+.
Synthesis of 4-[benzhydryl-N-(3-chlorophenyl)]piperazine-1-carboxamide (3 l). Compound (3 l) was synthesized from 3-chloro phenyl isocyanate (0.25 g, 1.628 mmol) and 1-benzhydrylpiperazine (0.41 g, 1.628 mmol) according to the general procedure. White solid. Yield: 80% (0.53 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.67 (s, 1H), 7.64–7.63 (m, 1H), 7.44 (d, J = 7.45 Hz, 4H), 7.38–7.36 (m, 1H), 7.30 (t, J = 7.45 Hz, 4H), 7.25–7.18 (m, 3H), 6.96 (dd, J = 8.02, 1.15 Hz, 1H), 4.0.34 (s,1H), 3.47 (t, J = 4.58 Hz, 4H), 2.32 (t, J = 4.58 Hz, 4H). 13C NMR (DMSO-d6, 125 MHz): δ 154.55, 142.52, 142.16, 132.72, 129.93, 128.60, 127.65, 126.97, 121.21, 118.73, 117.64, 74.76, 51.38, 43.80. IR (KBr): ῡ = 3295, 2805, 1640, 1535, 1250, 744. LC–MS for C24H24ClN3O: m/z = 406 [M + H]+.
Synthesis of N-{(3-chlorophenyl)-4-[(4-fluorophenyl)(phenyl)methyl]}piperazine-1-carboxamide (3 m). Compound (3 m) was synthesized from3-chloro phenyl isocyanate (0.25 g, 1.628 mmol) and 1-[(4-fluorophenyl)(phenyl)methyl]piperazine (0.44 g, 1.628 mmol) according to the general procedure. White solid. Yield: 80% (0.55 g). 1H NMR (CDCl3, 500 MHz): δ 7.42–7.37 (m, 5H), 7.32–7.29 (m, 2H), 7.23–7.21 (m, 1H), 7.17–7.12 (m, 2H), 1.00–6.97 (m, 3H), 6.54 (s, 1H), 4.24 (s, 1H), 3.46 (t, J = 4.58 Hz, 4H), 2.39–2.36 (m, 4H). 13C NMR (CDCl3, 125 MHz): δ 161.98 (d, 1JC,F = 245.92 Hz), 154.70, 141.99, 140.35, 138.03. 134.50, 129.87, 129.40 (d, 3JC,F = 8.40 Hz), 128.85, 127.87, 127.45, 123.13, 120.06, 117.98, 115.61 (d, 2JC,F = 21.59 Hz), 75.20, 51.52, 44.31. IR (KBr): ῡ = 3303, 2802, 1638, 1507, 1236, 782. LC–MS for C24H23ClFN3O: m/z = 424 [M + H]+.
Synthesis of N-{(3-chlorophenyl)-4-[(2-fluorophenyl)(4-fluorophenyl]}methyl)piperazine-1-carboxamide (3n). Compound (3n) was synthesized from 3-chloro phenyl isocyanate (0.25 g, 1.628 mmol) and 1-[(2-fluorophenyl)(4-fluorophenyl)methyl]piperazine (0.46 g, 1.628 mmol) according to the general procedure. White solid. Yield: 75% (0.54 g). 1H NMR (CDCl3, 500 MHz): δ 7.57 (td, J = 7.45, 1.15 Hz, 1H), 7.41–7.38 (m, 3H), 7.24–7.18 (m, 1H), 7.16–7.09 (m, 3H), 7.01–6.91 (m, 4H), 6.74 (s, 1H), 4.69 (s, 1H), 3.46 (t, J = 5.15 Hz, 4H), 2.44–2.35 (m, 4H). 13C NMR (CDCl3, 125 MHz): δ 162.03 (d, 1JC,F = 245.92 Hz), 160.68 (d, 1JC,F = 245.92 Hz), 154.89, 140.30, 136.92, 134.45, 129.84, 129.61 (d, 3JC,F = 7.20 Hz), 128.84, 128.75 (d, 3JC,F = 8.40 Hz), 128.58 (d, 4JC,F = 3.60 Hz), 124.59 (d, 4JC,F = 3.60 Hz), 123.21, 120.34, 118.28, 115.91, 115.62 (d, 2JC,F = 20.39 Hz), 65.99, 51.35, 44.26. IR (KBr): ῡ = 3306, 2809, 1638, 1508, 1224, 754. LC–MS for C24H22ClF2N3O: m/z = 442 [M + H]+.
Synthesis of N-[(3-chlorophenyl)-4-(dibenzo[b,f][1,4]thiazepin-11-yl)]piperazine-1-carboxamide (3o). Compound (3o) was synthesized from 3-chloro phenyl isocyanate (0.25 g, 1.628 mmol) and 11-(piperazin-1-yl-dibenzo [b,f] [1, 4])thiazepine (0.48 g, 1.628 mmol) according to the general procedure. White solid. Yield: 82% (0.60 g). 1H NMR (CDCl3, 500 MHz): δ 7.53 (d, J = 7.45 Hz, 1H), 7.43–7.40 (m, 2H), 7.38–7.34 (m, 1H), 7.32–7.31 (m, 2H), 7.21–7.14 (m, 3H), 7.10 (dd, J = 8.02, 1.15 Hz, 1H), 6.99 (dt, J = 7.16, 1.72 Hz, 1H), 6.92 (td, J = 7.45, 1.15 Hz, 1H), 6.77 (s, 1H), 3.59–3.45 (m, 8H). 13C NMR (CDCl3, 125 MHz): δ 160.71, 154.96, 148.60, 140.22, 140.07, 134.48, 133.89, 132.41, 132.38, 131.24, 129.89, 129.32, 128.96, 128.63, 128.07, 125.34, 123.36, 123.33, 120.34, 118.29, 43.73. IR (KBr): ῡ = 3267, 2837, 1641, 1601, 1238, 759. LC–MS for C24H21ClN4OS: m/z = 449 [M + H]+.
Synthesis of N-{[(3-chlorophenyl)-3-(trifluoromethyl)]-5,6-dihydro-[1,2,4]triazolo[4,3-a]}pyrazine-7(8H)-carboxamide (3p). Compound (3p) was synthesized from 3-(chlorophenyl)isocyanate (0.25 g, 1.628 mmol) and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine (0.31 g, 1.628 mmol) according to the general procedure. White solid. Yield: 85% (0.48 g). 1H NMR ((CD3)2CO, 500 MHz): δ 8.50 (s, 1H), 7.73–7.72 (m, 1H), 7.43 (d, J = 8.02, 1H), 7.25 (t, J = 8.02, 2H), 7.01 (dd, J = 8.02, 1.15 Hz, 1H), 5.02 (s, 2H), 4.38 (t, J = 5.73 Hz, 2H), 4.16 (t, J = 5.73 Hz, 2H). 13C NMR ((CD3)2CO, 125 MHz): δ 155.16, 151.72, 143.85 (q, 2JC,F = 39.59 Hz), 142.40, 142.32, 134.41, 130.69, 123.00, 120.16, 120.07, 119.69 (q, 1JC,F = 272.31 Hz), 118.60, 118.51, 44.37, 42.09, 41.22. IR (KBr): ῡ = 3312, 2874, 1664, 1501, 1279, 775. LC–MS for C13H11ClF3N5O: m/z = 446 [M + H]+. Matching previously reported data [25].
Synthesis of N-{[(2-fluorophenyl)-3-(trifluoromethyl)]-5,6-dihydro-[1,2,4]triazolo[4,3-a]}pyrazine-7(8H)-carboxamide (3q). Compound (3q) was synthesized from 2-(fluorophenyl)isocyanate (0.25 g, 1.823 mmol) and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine (0.35 g, 1.823 mmol) according to the general procedure. White solid. Yield: 78% (0.47 g). 1H NMR (CDCl3, 500 MHz): δ 7.74(t, J = 7.45 Hz, 1H), 7.32–7.31 (m, 1H), 7.08–7.00 (m, 3H), 4.95 (, 2H), 4.17 (t, J = 5.15 Hz, 2H), 4.00 (t, J = 5.15 Hz, 2H). 13C NMR (CDCl3, 125 MHz): δ 154.40, 153.83 (d, 1JC,F = 243.52 Hz), 150.12, 143.77 (q, 2JC,F = 39.59 Hz), 126.32 (d, 3JC,F = 10.80 Hz), 124.88 (d, 3JC,F = 7.20 Hz), 124.54 (d, 4JC,F = 3.60 Hz), 123.45, 118.28 (q, 1JC,F = 271.11 Hz), 115.22 (d, 2JC,F = 19.19 Hz), 43.57, 41.99, 40.49. IR (KBr): ῡ = 3248, 2775, 1637, 1498, 1279, 756. LC–MS for C13H11F4N5O: m/z = 330 [M + H]+.
Synthesis of N-{[(3-fluorophenyl)-3-(trifluoromethyl)]-5,6-dihydro-[1,2,4]triazolo[4,3-a]}pyrazine-7(8H)-carboxamide (3r). Compound (3r) was synthesized from 3-(fluorophenyl)isocyanate (0.25 g, 1.823 mmol) and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine (0.35 g, 1.823 mmol) according to the general procedure. White solid. Yield: 83% (0.5 g). 1H NMR ((CD3)2CO, 500 MHz): δ 8.55 (s, 1H), 7.54–7.51 (m, 1H), 7.27–7.25 (m, 2H), 6.76–6.72 (m, 1H), 5.02 (s, 2H), 4.37 (t, J = 5.15 Hz, 2H), 4.15 (t, J = 5.15 Hz, 2H). 13C NMR ((CD3)2CO, 125 MHz): δ 163.69 (d, 1JC,F = 241.12 Hz), 155.16, 151.75, 143.88 (q, 2JC,F = 39.59 Hz), 142.81 (d, 3JC,F = 12.0 Hz), 130.66 (d, 3JC,F = 9.60 Hz), 119.73 (q, 1JC,F = 268.71 Hz), 115.87 (d, 3JC,F = 2.40 Hz), 109.56 (d, 2JC,F = 21.59 Hz), 107.28(d, 2JC,F = 27.59 Hz), 44.39, 42.09, 41.22. IR (KBr): ῡ = 3358, 2872, 1678, 1538, 1280, 757. LC–MS for C13H11F4N5O: m/z = 330 [M + H]+.
Synthesis of N-{[(4-fluorophenyl)-3-(trifluoromethyl)]-5,6-dihydro-[1,2,4]triazolo[4,3-a]}pyrazine-7(8H)-carboxamide (3 s). Compound (3 s) was synthesized from (4-fluorophenyl)isocyanate (0.25 g, 1.823 mmol) and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine (0.35 g, 1.823 mmol) according to the general procedure. White solid. Yield: 80% (0.48 g). 1H NMR ((CD3)2CO, 500 MHz): δ 8.40 (s, 1H), 7.54–7.52 (m, 2H), 7.05–7.01 (m, 2H), 5.01 (s, 2H), 4.36 (t, J = 5.15 Hz, 2H), 4.14 (t, J = 5.73 Hz, 2H). 13C NMR ((CD3)2CO, 125 MHz): δ 159.31 (d, 1JC,F = 238.72 Hz), 155.52, 151.83, 143.87 (q, 2JC,F = 39.39 Hz), 137.08, 129.74, 129.01, 122.51 (d, 3JC,F = 7.20 Hz), 119.76 (q, 1JC,F = 269.91 Hz),115.69 (d, 2JC,F = 21.59 Hz), 44.42, 42.07, 41.21. IR (KBr): ῡ = 3356, 2846, 1672, 1507, 1251, 839. LC–MS for C13H11F4N5O: m/z = 330 [M + H]+. Matching previously reported data [25].
Synthesis of N-{[(4-chlorophenyl)-3-(trifluoromethyl)]-5,6-dihydro-[1,2,4]triazolo[4,3-a]}pyrazine-7(8H)-carboxamide (3t). Compound (3t) was synthesized from 4-(chlorophenyl)isocyanate (0.25 g, 1.628 mmol) and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine (0.35 g, 1.628 mmol) according to the general procedure. White solid. Yield: 78% (0.44 g). 1H NMR ((CD3)2CO, 500 MHz): δ 7.58–7.55 (m, 2H), 7.28–7.26 (m, 2H), 5.01 (s, 2H), 4.37 (t, J = 5.73 Hz, 2H), 4.15 (t, J = 5.73 Hz, 2H). 13C NMR ((CD3)2CO, 125 MHz): δ 155.24, 151.75, 143.85 (q, 2JC,F = 39.39 Hz), 139.84, 129.19, 127.69, 121.99, 121.89, 119.74 (q, 1JC,F = 268.71 Hz), 44.40, 42.07, 41.20. IR (KBr): ῡ = 3366, 2883, 1670, 1494, 1236, 836. LC–MS for C13H11ClF3N5O: m/z = 346 [M + H]+.
Molecular docking simulations
Method details. Molecular docking was used to clarify the binding mode of the compounds to provide straightforward information for further structural optimization. The crystal structure of Acinetobacter baumannii PBP1a in complex with penicillin G (PDB ID 3UDI, 2.6 Å X-ray resolution) was extracted from the Brookhaven Protein Database (PDB http://www.rcsb.org/pdb). The proteins were prepared for docking by adding polar hydrogen atom with Gasteiger–Huckel charges and water molecules were removed. The 3D structure of the ligands was generated by the SKETCH module implemented in the SYBYL program (Tripos Inc., St. Louis, USA) and its energy-minimized confirmation was obtained with the help of the Tripos force field using Gasteiger–Huckel [26] charges and molecular docking was performed with Surflex-Dock program that is interfaced with Sybyl-X 2.0. [27] and other miscellaneous parameters were assigned with the default values given by the software.
Antimicrobial studies
Samples were prepared in DMSO and water to a final testing concentration of 32 μg/mL or 20 μM (unless otherwise indicated in the datasheet), in a 384-well, non-binding surface plate (NBS) for each bacterial/fungal strain, and in duplicate (n = 2), and keeping the final DMSO concentration to a maximum of 1% DMSO [24, 25, 25,26,27]. All the sample preparation for antimicrobial studies were done using liquid handling robots.
Antimicrobial assay
Primary antimicrobial screening study by whole-cell growth inhibition assays, using the compounds (3a–t) at a single concentration, in duplicate (n = 2). The inhibition of growth is measured against five bacteria: Escherichia coli (E. coli) ATCC 25,922, Klebsiella pneumoniae (K. pneumoniae) ATCC 700,603, Acinetobacter baumannii (A. baumannii) ATCC 19,606, Pseudomonas aeruginosa (P. Aeruginosa) ATCC 27,853) and Staphylococcus aureus (S. aureus) ATCC 43,300, and two fungi: Candida albicans (C. albicans) ATCC 90,028 and Cryptococcus neoformans (C. Neoformans) ATCC 208,821 [28].
Procedure
All bacteria were cultured in Cation-adjusted Mueller Hinton broth (CAMHB) at 37 °C overnight. A sample of each culture was then diluted 40-fold in fresh broth and incubated at 37 °C for 1.5–3 h. The resultant mid-log phase cultures were diluted (CFU/mL measured by OD600) and then added to each well of the compound containing plates, giving a cell density of 5 × 105 CFU/mL and a total volume of 50 μL. All the plates were covered and incubated at 37 °C for 18 h without shaking.
Analysis
Inhibition of bacterial growth was determined by measuring absorbance at 600 nm (OD600), using a Tecan M1000 Pro monochromator plate reader. The percentage of growth inhibition was calculated for each well, using negative control (media only) and positive control (bacteria without inhibitors) on the same plate as references. The significance of the inhibition values was determined by modified Z-scores, calculated using the median and median absolute deviation (MAD) of the samples (no controls) on the same plate. Samples with inhibition value above 80% and Z-score above 2.5 for either replicate (n = 2 on different plates) were classed as active. Samples with inhibition values between 15 and 80% and Z-score above 2.5 for either replicate (n = 2 on different plates) were classed as partial active. Samples with inhibition values below 15% and Z-score above 2.5 for either replicate (n = 2 on different plates) were classed as inactive.
Antifungal assay
Procedure
Fungi strains were cultured for three days on Yeast Extract-Peptone Dextrose (YPD) agar at 30 °C. A yeast suspension of 1 × 106 to 5 × 106 CFU/mL (as determined by OD530) was prepared from five colonies. The suspension was subsequently diluted and added to each well of the compound-containing plates giving a final cell density of fungi suspension of 2.5 × 103 CFU/mL and a total volume of 50 μL. All plates were covered and incubated at 35 °C for 24 h without shaking.
Analysis
Growth inhibition of C. albicans was determined by measuring absorbance at 530 nm (OD530), while the growth inhibition of C. neoformans was determined by measuring the difference in absorbance between 600 and 570 nm (OD600-570), after the addition of resazurin (0.001% final concentration) and incubation at 35 °C for additional 2 h. The absorbance was measured using a Biotek Synergy HTX plate reader. The percentage of growth inhibition was calculated for each well, using negative control (media only) and positive control (fungi without inhibitors) on the same plate. The significance of the inhibition values was determined by modified Z-scores, calculated using the median and MAD of the samples (no controls) on the same plate. Samples with inhibition value above 80% and Z-score above 2.5 for either replicate (n = 2 on different plates) were classed as active. Samples with inhibition values between 15 and 80% and Z-score above 2.5 for either replicate (n = 2 on different plates) were classed as partial active.
References
Wei Y, Xia S, He C, Xiong W, Xu H (2016) Highly enantioselective production of a chiral intermediate of sitagliptin by a novel isolate of Pseudomonas pseudoalcaligenes. Biotechnol Lett 38:841–846
Hu G, Wang C, Xin X, Li S, Li Z, Zhao Y, Gong P (2019) Design, synthesis and biological evaluation of novel 2,4-diaminopyrimidine derivatives as potent antitumor agents. New J Chem 43:10190
Kelley JL, Linn JA, Bankston DD, Burchall CJ, Soroko FE, Cooper BR (1995) 8-Amino-3-benzyl-1,2,4-triazolo[4,3-a]pyrazines. synthesis and anticonvulsant activity. J Med Chem 38:3676
Sado T, Inoue A (1990) Jpn Kokai Tokkyo Koho JP. Chem Abstr 113:78422k
Bradbury RH, Major JS, Oldham AA, Rivett JE, Roberts DA, Slater AM, Timms D, Waterson D (1990) 1,2,4-Triazolo[4,3-a]pyrazine derivatives with human renin inhibitory activity. 2. synthesis, biological properties and molecular modeling of hydroxyethylene isostere derivatives. J Med Chem 33:2335
Kim D, Wang L, Beconi M, Eiermann GJ, Fisher MH, He H, Hickey GJ, Kowalchick JE, Leiting B, Lyons K (2005) (2R)-4-Oxo-4-[3-(Trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin- 7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: a potent, orally active dipeptidyl peptidase iv inhibitor for the treatment of type 2 diabetes. J Med Chem 48:141
S. Bhati, V. Kumar, S. Singh, J. Singh, (2019) J Mol Struct, 1191: 197e205.
WHO in Antimicrobial Resistance 2014 Bull World Health Organ 61 383
Smith R (1999) In Antimicrobial resistance: the importance of developing long term policy. Bull. World Health Organ. 77:862
Patil M, Poyil AN, Joshi SD, Patil SA, Patil SA, Bugarin A (2019) Design, synthesis, and molecular docking study of new piperazine derivative as potential antimicrobial agents. Bioorgan chem 92:103217
Patil V, Barragan E, Patil SA, Patil SA, Bugarin A (2016) Direct synthesis and antimicrobial evaluation of structurally complex chalcones. ChemistrySelect 1:3647
Patil M, Poyil AN, Joshi SD, Patil SA, Patil SA, Bugarin A (2019) Synthesis, molecular docking studies, and antimicrobial evaluation of new structurally diverse ureas. Bioorgan chem 87:302–311
Patil M, Noonikara Poyil A, Joshi SD, Patil SA, Patil SA, Bugarin A (2019) New urea derivatives as potential antimicrobial agents: synthesis, biological evaluation, and molecular docking studies. Antibiotics 8:178
Shahini CR, Achar G, Budagumpi S, Müller-Bunz H, Tacke M, Patil SA (2018) Benzoxazole and dioxolane substituted benzimidazole–based N–heterocyclic carbene–silver(I) complexes: synthesis, structural characterization and in vitro antimicrobial activity. J Organomet Chem 868:1
Shahini CR, Achar G, Budagumpi S, Tacke M, Patil SA (2017) Non-symmetrically p-nitrobenzyl-substituted N-heterocyclic carbene-silver(I) complexes as metallopharmaceutical agents. Appl Organomet Chem 31:e3819
Shahini CR, Achar G, Budagumpi S, Tacke M, Patil SA (2017) Synthesis, structural investigation and antibacterial studies of non–symmetrically p–nitrobenzyl substituted benzimidazole N–heterocyclic carbene–silver(I) complexes. Inorg Chim Acta 466:432
Subramanya Prasad TV, Shahini CR, Patil SA, Huang X, Bugarin A, Patil SA (2017) Non-symmetrically p-nitrobenzyl-and p-cyanobenzyl-substituted N-heterocyclic carbene-silver (I) complexes: synthesis, characterization and antibacterial studies. J Coord Chem 70(4):600–614
Patil S, Dietrich K, Deally A, Gleeson B, Müller-Bunz H, Paradisi F, Tacke M (2010) Synthesis, cytotoxicity and antibacterial studies of novel symmetrically and nonsymmetrically 4-(Methoxycarbonyl)benzyl-substituted n-heterocyclic carbene-silver acetate complexes. Helv Chim Acta 93:2347
Patil S, Dietrich K, Deally A, Gleeson B, Müller-Bunz H, Paradisi F, Tacke M (2010) Synthesis, cytotoxicity and antibacterial studies of symmetrically and non-symmetrically benzyl- or p-cyanobenzyl-substituted N-Heterocyclic carbene-silver complexes. Appl Organomet Chem 24:781
Patil S, Claffey J, Deally A, Gleeson B, Hogan M, Menéndez-Méndez LM, Müller-Bunz H, Paradisi F, Tacke M (2010) Synthesis, cytotoxicity and antibacterial studies of p-methoxybenzyl-substituted and benzyl-substituted N-Heterocyclic carbene-silver complexes. Eur J Inorg Chem 2010:1020
Patil S, Deally A, Gleeson B, Hackenberg F, Müller-Bunz H, Paradisi F, Tacke M, Allg Z (2011) Synthesis, cytotoxicity and antibacterial studies of novel symmetrically and non-symmetrically p-Nitrobenzyl-substituted N-Heterocyclic carbene-silver(i) acetate complexes. Anorg Chem 637:386
Patil S, Deally A, Gleeson B, Müller-Bunz H, Paradisi F, Tacke M (2011) Novel benzyl-substituted N-heterocyclic carbene–silver acetate complexes: synthesis, cytotoxicity and antibacterial studies. Metallomics 3:74
Shivprakash S, Reddy GC (2014) Stereoselective synthesis of (Z)-1-benzhydryl-4-cinnamylpiperazines via the Wittig reaction. Syn Commun 44(5):600–609
Gudisela MR, Srinivasu N, Mulakayala C, Bommu P, Rao MB, Mulakayala N (2017) Design, synthesis and anticancer activity of N-(1-(4-(dibenzo [b, f][1, 4] thiazepin-11-yl) piperazin-1-yl)-1-oxo-3-phenylpropan-2-yl derivatives. Bioorgan med chem lett 27(17):4140–4145
Sheldrick GM (2008) A short history of SHELX. Acta Cryst A 64:122
Mannam MR, Devineni SR, Pavuluri CM, Chamarthi NR, Kottapalli RSP (2019) Phosphorus. Sulfur Silicon Relat Elem 194(9):922–932
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219
Tripos International, Sybyl-X 2.0, Tripos International, St. Louis, MO, USA. 2012
Edwards IA, Elliott AG, Kavanagh AM, Zuegg J, Blaskovich MAT, Cooper MA, Infect ACS (2016) Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. Dis 2:442
Funding
This research was partially funded by Florida Gulf Coast University. The ACS Petroleum Research Fund supported this research (Grant # 58269-ND1) as well as the Seidler Grant. Author SAP thanks the DST-Nanomission, India (SR/NM/NS-20/2014), DST-SERB, India [SERB/F/1423/2017–18 (File No. YSS/2015/000010)], and Jain University, India, for financial support received. Antimicrobial screening was performed by CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Welcome Trust (UK) and The University of Queensland (Australia). Dr. Shrinivas Joshi thanks the Vision Group on Science and Technology, Bangalore, India [VGST letter Ref No. VGST/GRD-567/2016–17/2017–18/183 dated 31–05-2018] for financial support.
Author information
Authors and Affiliations
Contributions
MP and ANP contributed to conceptualization ; MP and ANP contributed to methodology ; SDJ contributed to software; AB, SAP and SAP contributed to validation; AB and SAP contributed to formal analysis; MP and ANP contributed to investigation; AML contributed to resources; AB contributed to data curation; SAP contributed to writing—original draft preparation; AB and AML contributed to writing—review and editing; SAP contributed to visualization; AB and SAP contributed to supervision, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patil, M., Noonikara-Poyil, A., Joshi, S.D. et al. Synthesis, molecular docking studies, and in vitro antimicrobial evaluation of piperazine and triazolo-pyrazine derivatives. Mol Divers 26, 827–841 (2022). https://doi.org/10.1007/s11030-021-10190-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10190-x