Abstract
The present work describes the synthesis of 4-amino-6-(2-benzylidenehydrazinyl)-pyrimidine-5-carbonitrile derivatives, 4-amino-6-[(2-phenylethyl)amino]pyrimidine-5-carbonitrile, and 4-amino-6-(piperidin-1-yl)pyrimidine-5-carbonitrile. The compounds were characterized by FT-IR and 1H and 13C NMR spectroscopy and mass spectrometry. All the compounds were evaluated for in vitro antimicrobial activity against different bacterial and fungal strains. The minimum inhibitory concentrations (MICs) of all the compounds were validated. 4-Amino-6-[2-(3,4-dimethoxybenzylidene)hydrazinyl]pyrimidine-5-carbonitrile and 4-amino-6-(piperidin-1-yl)pyrimidine-5-carbonitrile, which have the lowest MIC values were selected for cell leakage analysis and bacterial growth curve study. It was found that both the compounds have potential to induce bacterial cell membrane rupture and disintegration. Field emission scanning electron microscopic analysis confirmed the effect of the selected compounds on the morphology of both Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria. The mechanism of interaction between the drug and the target protein of S. aureus and E. coli was studied by molecular docking.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
The scope for the development of new classes of antimicrobial agents is high, as many of the pathogenic microorganisms have acquired resistance against currently used drugs. Also, the emergence of new infectious diseases has significantly increased the demand for novel and potent molecules to combat microbial infections. Pyrimidines, a class of molecules comprising a heterocyclic ring and two nitrogen atoms in their structure, display a wide range of biological and pharmaceutical activities, including anti-inflammatory [1–3], analgesic [2], antioxidant [4], antiviral [5, 6], anti-amoebic [7], CDK2 inhibitory [8], antitubercular [9], anticancer [10–12], antidiabetic [13], antitumor [14], anti-hyperglycemia [15], and in vivo diuretic [16] activities.
A great number of pyrimidine derivatives have been studied for their antimicrobial activity over the past few years [17]. Different classes of mono-, di-, tri-, and tetra-substituted pyrimidines showed antimicrobial activities against various strains of bacteria and fungi [17]. A wide range of trisubstituted pyrimidines were earlier evaluated for their in vitro antimicrobial activity, and the activity of 2,4,6-trisubstituted pyrimidine derivatives against both bacteria and fungi compared with that of standard drugs [18]. Desai et al. [19] have synthesized and analyzed the in vitro antibacterial activity of a novel series of 2-amino-4-(4-chlorophenyl)-6-substituted-pyrimidines and observed potent antibacterial activity against both gram-negative (E. coli, Salmonella) and gram-positive (Bacillus pumilus, micrococcus) bacteria.
In search of novel and potent antimicrobial agents, in the present work we have synthesized novel pyrimidine carbonitrile derivatives and screened them for antimicrobial activity against various bacterial and fungal strains.
RESULTS AND DISCUSSION
The target 4-amino-6-(2-benzylidenehydrazinyl)pyrimidine-5-carbonitrile derivatives 4a–4d, 4-amino-6-[(2-phenylethyl)amino]pyrimidine-5-carbonitrile (5), and 4-amino-6-(piperidin-1-yl)pyrimidine-5-carbonitrile (6) were synthesized as shown in Schemes 1–3, respectively. Scheme 1 represents the synthesis of compounds 4a–4d by the reaction of 4-amino-6-chloro-pyrimidine-5-carbonitrile (1) with hydrazine hydrate in ethanol to obtain 4-amino-6-hydrazinylpyrimidine-5-carbonitrile (2) followed by the condensation with various substituted aldehydes. Compound 5 was obtained by the reaction of compound 1 with (2-phenylethyl)amine in DMF in the presence of Et3N (Scheme 2). Compound 6 was synthesized by the reaction of compound 1 with piperidine in ethanol (Scheme 3).
The structures of compounds 2, 4a–4d, 5, and 6 were confirmed by IR and NMR spectroscopy and mass spectrometry. The IR spectra of the all the synthesized compounds displayed bands at 3409 to 3076 cm–1, assigned due to NH stretching vibrations and 2359 to 2201 cm–1, assigned to C≡N stretching vibrations. The bands at 1666 to 1651 cm–1 in the spectra of compounds 4a–4d are characteristic of the C=N group.
The 1H NMR spectrum of compound 2 displayed signals at 7.63 and 7.96 ppm, assignable to the hydrazine moiety –NHNH2 and the NH2 group attached to the pyrimidine ring. The 1H NMR spectra of compounds 4a–4d contained a singlet NH signal at 11.14 to 11.69 ppm. The amino group of pyrimidine derivatives 4a–4d gives two-hydrogen singlets in the range 7.00 to 11.27 ppm, and the singlet signal at 7.45 to 9.37 ppm was assigned to the imine CH proton. The 1H NMR spectrum of compound 5 shows peaks at 7.38 ppm, assignable to two –NH2 protons and at 5.39 ppm, assignable to the proton of the NH group bonded to the alkyl moiety. The 1H NMR of compound 6 displays a singlet at 8.01 ppm from one pyrimidine CH proton, a broad singlet at 7.21 ppm from two NH2 protons, a triplet at 3.76 ppm from four protons of the piperidine ring, and a multiplet at 1.73–1.48 ppm from the remaining six piperidine protons.
In the13C NMR analysis for compounds 2, 4a–4d, 5, and 6, there were signals at 117 attributed to the C≡N group attached to the aromatic ring. There were signals between 157.9 to 168.3 ppm, which were assigned to the pyrimidine C=N carbons. The mass spectral data were consistent with the proposed structure of the synthesized compounds. The crystal structure of compound 6 was reported in [28].
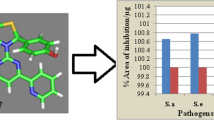
The synthesized compounds 4a–4d, 5, and 6 were screened for their antimicrobial activity against four bacteria and two fungal strains as per CLSI guidelines. The test bacteria comprised two Gram-positive bacteria S. aureus (ATCC 29213) and B. subtilis (NCTC 8236) and two Gram-negative bacteria E. coli (ATCC 10536) and S. typhi. The two fungal strains comprised A. flavus (NCIM539) and A. niger (NCIM1196). Ampicillin and fluconazole were used as standard drugs for antibacterial and antifungal activity screening, respectively. The antibacterial and antifungal activity was measured in terms the minimum inhibitory concentration (MIC) (Tables 1 and 2).
All the synthesized compounds showed good to moderate levels of antibacterial and antifungal activity. Compounds 4c and 6 were more potent than the other compounds against bacterial strains, and compound 6 proved to be the most potent against all microbial pathogens. The MIC values of 4c and 6 fell in the range 25–50 μg/mL for both bacteria and fungi, which compares with the respective values for standard drugs. Compound 4b exhibited a lower antibacterial activity compared to 4c and 6, while compounds 4a and 4d did not give a significant zone of inhibition against the test fungal strains. Compound 4d was the least active against the test bacterial strains. Compounds 4a and 5 showed moderate antimicrobial activity compared to standard used. The resulting data showed that compounds 4c and 6 hold promise as potential antibacterial and antifungal agents, and just these compounds were chosen for further research.
The ability of compounds 4c and 6 to induce bacterial cell lysis was evaluated by the cell leakage assay. Nucleotides leaked from bacterial cells were measured by plotting the optical density at 320 nm at different exposure times (up to 24 h with 4-h intervals). The results confirmed that both the test compounds induce a time-dependent increase in the rate of leakage of cell nucleotides in both E. coli and S. aureus. Results are depicted in Fig. 1.
The effect of compounds 4c and 6 on the bacterial growth curve was evaluated on E. coli and S. aureus bacteria. The control bacterial culture showed a typical growth pattern with a lag phase of 4 h and a log phase of 8–10 h. Treatment with the IC50 concentrations of compounds 4c and 6 induced significant changes in the normal bacterial growth pattern. The lag phase of bacteria significantly decreased to 5–6 h (Fig. 2). This result provided clear evidence for the antibacterial potential of both test compounds against the pathogens studied.
The effect of compound 4c and 6 on the morphology of S. aureus and E. coli cells was evaluated by field emission scanning electron microscopy (FESEM) (Figs. 3 and 4). The FE-SEM images of control untreated cells showed smooth cell surfaces with normal morphological characteristics. Treatment with both test compounds led to a significant deterioration in the cell wall, which in turn led to disintegration of cell membrane. These findings gave further evidence showing that compounds 4c and 6 cause lysis of S. aureus and E. coli cells.
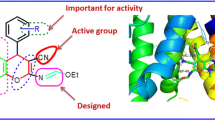
Penicillin-binding proteins (PBPs) are the targets for the antibiotics, as they play a major role in the bacterial cell wall synthesis. Beta-lactam antibiotics bind to the active site of the PBPs and thus inhibit the cross-linking of peptidoglycans, leading to bacterial death [20]. In the present study, based on the MIC assay and docking analysis, we selected to study the mode of binding of the synthesized molecules with PBPs. The ligands were allowed to interact with a PBP of B. subtilis. The standard antibiotic drug ampicillin was used as control.
In silico docking analysis was performed between ligands 4c and 6, ampicillin, and the PBP4a of B. subtilis. The crystal structure of the PBP4a of B. subtilis was retrieved from the RCSB-PDB database (PDB ID-1W5D) in the .pdb format. The structural refinement of the protein was performed using Galaxy Refine [21] and further subjected to Ramachandran plot analysis using PROCHECK software [22]. The refined protein was loaded to AutoDock vina [23] of the PyRx software for docking analysis. The structures of ligands 4c, 6, and ampicillin were drawn in Marvin sketch and saved in the .sdf format. Energy minimization was performed using the Open Babel [24] in PyRx0.8. The grid box was set to the XYZ coordinates of 45.67, 37.89, and 93.54, respectively, and the box dimensions were 73.67, 85.31, and 47.62 along the XYZ axis, respectively, to cover the entire protein. The protein–ligand interaction of the conformation complex with the lowest AutoDock vina score was visualized using PyMOL 1.3, and the interaction was analyzed using LIGPLOT+ software [25].
All PDBs contain serine in the active site. The PBP4a of B. subtilis (1W5D) has serine at position 52 [26]. The results of docking of compounds 4c, 6, and ampicillin to 1W5D, the interactions with Ser52 are highly conserved. Ligand 4c forms two hydrogen bonds (3.14 and 3.17 Å) with Ser52 and one with Thr412 (3.17 Å) (Fig. 5, A/A1). The higher the number of hydrogen bonds, the more specific is binding of a ligand to a target protein [27]. Ligand 6 forms one hydrogen bond with Ser52 (3.13 Å) (Fig. 5, B/B1). However, the reference ligand ampicillin forms one hydrogen bond with Ser52 (3.32 Å) and another with Ser299 (3.32 Å) (Fig. 5, C/C1). Along with hydrogen bonding, the ligands enter hydrophobic interactions, where Tyr150, Asn301, and Ser414 are highly conserved in all the three interactions depicted in Fig. 5.
As judged from the MIC values and docking results (Tables 1 and 3), ligand 4c has a higher binding affinity to PBPs compared to ligand 6 and the standard drug ampicillin.
In summary, we have synthesized 4-amino-6-(2-benzylidenehydrazinyl)-pyrimidine-5-carbonitrile derivatives 4a–4d, 4-amino-6-[(2-phenylethyl)amino]pyrimidine-5-carbonitrile (5), and 4-amino-6-(piperidin-1-yl)pyrimidine-5-carbonitrile (6). All the synthesized compounds were screened for their antimicrobial activity against S. aureus and B. subtilis (Gram-positive) and E. coli and S. typhi (Gram-negative) bacterial strains and A. flavus and A. niger fungal strains. All the compounds showed good to moderate levels of antibacterial and antifungal activity, and compounds 4c and 6 proved the most potent antimicrobial agents with lowest MIC values. Compounds 4c and 6 have a potential to induce bacterial cell membrane rupture and disintegration. The mode of action was explored by FESEM imaging, which clearly indicated the membrane damaging effects of compounds 4c and 6. In silico molecular docking gave evidence showing that compound 4c has a higher binding affinity to PBPs compared to compound 6 and the standard drug ampicillin. Compounds 4c and 6 hold promise as potent antimicrobial agents.
EXPERIMENTAL
Commercial reagents and solvents were used without any further purification. The melting points were measured in open capillaries using a Guna melting point apparatus and are uncorrected. The IR spectra were run on a Nicolet iz10 FTIR spectrophotometer. The 1H and 13C NMR spectra were recorded on a Bruker 400 spectrometer at 400 MHz in DMSO-d6 (Sigma-Aldrich), internal standard TMS. The reaction progress was monitored on TLC silica gel plates. The mass spectra were obtained on an Agilent Technologies 6110 Quadrupole LC/MS instrument. Field emission scanning electron microscopy (FESEM) was performed on a GEMINI SEM 300 Nano VP scanning electron microscope.
4-Amino-6-hydrazinylpyrimidine-5-carbonitrile (2). A mixture of 2.0 g (0.01298 mmol) of 4-amino-6-chloropyrimidine-5-carbonitrile (1) and 3.3 g (0.10384 mmol) of hydrazine hydrate was refluxed in 40 mL of dry ethanol (0.2 mmol) for 12 h. On completion of the reaction (by TLC), the reaction mixture was cooled to room temperature. The precipitate that formed was filtered off, washed with ethanol, dried, and recrystallized from ethanol. Yield 84%, off-white solid, mp 270–273°C. IR spectrum, ν, cm–1: 3409 (NH), 3083 (Ar–CH), 2208 (C≡N), 1666 (C=N), 1321 (C–N). 1H NMR spectrum, δ, ppm: 3.73 d (1H, NH, J 16.0 Hz), 7.08 s (1H, Pm-H), 7.62 d (2H, NH2, J 8.0 Hz), 7.96 d (2H, NH2, J 16.0 Hz). 13C NMR spectrum, δ, ppm: 117.0, 157.9, 166.4, 174.9. Mass spectrum (LCMS), m/z: 151.1 [M]+. Found, %: C 40.90; H 4.17; N 55.92. C5H6N6. Calculated, %: C 40.00; H 4.03; N 55.97.
4-Amino-6-(2-benzylidenehydrazinyl)pyrimidine-5-carbonitriles 4a–4d (general procedure). Substituted aldehyde 3a–3d (0.0079 mmol) was added to a solution of 0.2 g (0.0066 mmol) of 4-amino-6-hydrazinylpyrimidine-5-carbonitrile (2) in 10 mL of ethanol, and the reaction mixture was refluxed for 12 h. On completion of the reaction (by TLC), the reaction mixture was cooled to room temperature. The precipitate that formed was filtered off, washed with ethanol, and dried to obtain target product 4a–4d.
4-Amino-6-[2-(3H-indol-3-yl-methylene)hydrazinyl]pyrimidine-5-carbonitrile (4a). Yield 80%, light yellow solid, mp 247–248°C. IR spectrum, ν, cm–1: 3361 (NH), 3142 (Ar–H), 2201 (C≡N), 1651 (C=N), 1335 (C–N). 1H NMR spectrum, δ, ppm: 8.04 s (1H, imine-CH), 7.07–8.54 m (6H, Ar-H), 8.91 s (1H, Pm-H), 11.27 s (2H, NH2), 11.60 s (1H, NH). 13C NMR spectrum, δ, ppm: 68.3, 112.8, 117.8, 121.2, 123.9, 125.6, 126.9, 131.3, 132.5, 138.6, 142.7, 155.6, 160.8, 166.8. Mass spectrum (LCMS), m/z: 279.1 [M]+. Found, %: C 60.16; H 4.72; N 35.18. C14H13N7. Calculated, %: C 60.20; H 4.69; N 35.10.
4-Amino-6-[2-(3-ethoxy-4-hydroxybenzylidene)hydrazinyl]pyrimidine-5-carbonitrile (4b). Yield 75%, white solid, mp 220–224°C. IR spectrum, ν, cm–1: 3326, 3143 (NH), 3520 (OH), 2204 (C≡N), 1656 (C=N), 1237 (C–N), 1177 (C–O). 1H NMR spectrum, δ, ppm: 1.30 t (3H, CH3, J 8.0 Hz), 4.07–4.19 m (2H, OCH2), 6.80 s (1H, OH), 6.98 s (1H, Ar-H), 7.00 br.s (2H, NH2), 7.61 s (1H, Ar-H), 7.95 s (1H, Ar-H), 8.05 s (1H, Pm-H), 9.37 s (1H, =CH), 11.57 s (1H, NH). 13C NMR spectrum, δ, ppm: 64.3, 79.2, 100.2, 114.9, 116.5, 118.1, 123.2, 128.2, 143.8, 148.3, 151.3, 161.1, 162.4, 168.3. Mass spectrum (LCMS), m/z: 300.2 [M]+. Found, %: C 55.73; H 5.86; N 27.85; O 10.68. C14H16N6O2. Calculated, %: C 55.99; H 5.37; N 27.98; O 10.66.
4-Amino-6-[2-(3,4-dimethoxybenzylidene)hydrazinyl]pyrimidine-5-carbonitrile (4c). Yield 78%, light yellow solid, mp 215–218°C. IR spectrum, ν, cm–1: 3076 (NH), 2359 (C≡N), 1662 (C=N), 1239 (C–N), 1259 (C–O). 1H NMR spectrum, δ, ppm: 3.97 d [6H, 2(OCH3), J 8.0 Hz], 6.98 d (1H, Ar-H, J 8.0 Hz), 7.49 d (1H, Ar-H, J 8.0 Hz), 7.67 s (1H, Ar-H), 8.02 s (1H, =CH), 8.07 s (1H, Pm-H), 8.19 br.s (2H, NH2), 11.69 br.s (1H, NH). 13C NMR spectrum, δ, ppm: 56.6, 68.8, 109.0, 112.1, 113.1, 122.3, 125.1, 128.6, 143.8, 149.8, 151.2, 161.1, 162.0, 167.8. Mass spectrum (LCMS), m/z: 300.22 [M]+. Found, %: C 55.63; H 5.52; N 27.93; O 10.62. C14H16N6O2. Calculated, %: C 55.99; H 5.37; N 27.98; O 10.66.
4-Amino-6-(2-butylidenehydrazinyl)pyrimidine-5-carbonitrile (4d). Yield 78%, white solid, mp 240–242°C. IR spectrum, ν, cm–1: 3331, 3174 (NH), 2207 (C≡N), 1654 (C=N), 1267 (C–N), 2870, 2932 (CH). 1H NMR spectrum, δ, ppm: 0.92 t (3H, CH3, J 8.0 Hz), 1.55–2.24 m (4H, CH2), 7.15 s (2H, NH2), 7.45 s (1H, imine-CH), 8.01 s (1H, Pm-H), 11.14 s (1H, NH). 13C NMR spectrum, δ, ppm: 13.9, 19.8, 34.1, 68.2, 117.2, 158.3, 161.4, 162.6, 167.5. Mass spectrum (LCMS), m/z: 206.1 [M]+. Found, %: C 49.93; H 6.25; N 43.78. C8H12N6. Calculated, %: C 49.99; H 6.29; N 43.72.
4-Amino-6-[(2-phenylethyl)amino]pyrimidine-5-carbonitrile (5). A mixture of 1.0 g (0.00649 mmol) of 4-amino-6-chloropyrimidine-5-carbonitrile (1), 10 mL (10 V) of DMF, 0.94 g (0.00779 mmol) of ethyl(phenyl)amine, and 1.0 g (0.00974 mmol) of triethylamine were refluxed for 6 h. Upon cooling, the mixture was poured into 20 mL of ice-cold water. The precipitate that formed was filtered off, washed with water, dried, and recrystallized from ethanol to obtain the target product. Yield 74%, white solid, mp 134–135°C. IR spectrum, ν, cm–1: 3396, 3322 (NH), 2201 (C≡N), 1663 (C=N), 1289 (C–N), 1450 (CH). 1H NMR spectrum, δ, ppm: 1.49 d (4H, 2CH2, J 8.0 Hz), 5.39 t (1H, NH, J 8.0 Hz), 7.19–7.31 m (5H, Ar-H), 7.37 d (2H, NH2, J 4.0 Hz), 7.97 s (Pm-H). 13C NMR spectrum, δ, ppm: 24.3, 52.1, 68.5, 116.3, 126.3, 126.4, 128.2, 145.5, 161.2, 163.2, 165.4. Mass spectrum (LCMS), m/z: 240.1 [M]+. Found, %: C 61.63; H 5.59; N 33.13. C13H14N6. Calculated, %: C 61.40; H 5.55; N 33.05.
4-Amino-6-(piperidin-1-yl)pyrimidine-5-carbonitrile (6). A mixture of 1.0 g (0.0065 mol) of 4-amino-6-chloropyrimidine-5-carbonitrile (1) and 2.75 g (0.0325 mol) of piperidine was refluxed in 20 mL dry ethanol for 6 h. The reaction mixture was then cooled and stirred for 2 h at room temperature. The precipitate that formed was filtered off, washed with ethanol, dried, and recrystallized from acetone to obtain the target product. Yield 74%, colorless crystals, mp 140–143°C. IR spectrum, ν, cm–1: 3426, 3308 (NH), 2190 (C=N), 1646 (C=N), 1223 (CN). 1H NMR spectrum, δ, ppm: 1.48–1.73 m (6H, 3CH2), 3.76 t (4H, 2CH2, J 8.0 Hz), 7.21 br.s (2H, NH2), 8.01 s (1H, Pm-H). 13C NMR spectrum, δ, ppm: 24.9, 26.8, 58.9, 118.1, 159.9, 164.3, 168.5. Mass spectrum (LCMS), m/z: 205.1 [M]+. Found, %: C 59.28; H 6.49; N 34.43. C10H13N5. Calculated, %: C 59.10; H 6.45; N 34.46.
REFERENCES
Dasari, S.R., Tondepu, S., Vadali, L.R., and Seelam, N., Synth. Commun., 2020, vol. 50, p. 2950. https://doi.org/10.1080/00397911.2020.1787449
Tolba, M.S., Ahmed, M., El-Dean, A.M.K., Hassanien, R., and Farouk, M., J. Heterocycl. Chem., 2017, vol. 66. https://doi.org/10.1002/jhet.3056
Abu-Hashem, A.A. and Youssef, M.M., Molecules, 2011, vol. 16, p. 1956. https://doi.org/10.3390/molecules16031956
Salem, M.S. and Errayes, A.O., J. Chem. Res., 2016, vol. 40, p. 299. https://doi.org/10.3184/174751916X14605482579576
Ramiz, M.M.M., El-Sayed, W.A., Hagag, E., and Abdel-Rahman, A.A.H., J. Heterocycl. Chem., 2011, vol. 48, p. 1028. https://doi.org/10.1002/jhet.686
Bai, S., Liu, S., Zhu, Y., and Wu, Q., Tetrahedron Lett., 2018, vol. 59, p. 3179. https://doi.org/10.1016/j.tetlet.2018.07.020
Parveen, H., Hayat, F., Mukhtar, S., Salahuddin, A., Khan, A., Islam, F., and Azam, A., Eur. J. Med. Chem., 2011, vol. 46, p. 4669. https://doi.org/10.1016/j.ejmech.2011.05.055
Ibrahim, D.A. and El-Metwally, A.M., Eur. J. Med. Chem., 2010, vol. 45, p. 1158. https://doi.org/10.1016/j.ejmech.2009.12.026
Desai, N.C., Trivedi, A.R., Vaghani, H.V., Somani, H.C., and Bhatt, K.A., Med. Chem. Res., 2016, vol. 25, p. 329. https://doi.org/10.1007/s00044-015-1485-7
Kassab, A.E. and Gedawy, E.M., Eur. J. Med. Chem. 2013, vol. 63, p. 224. https://doi.org/10.1016/j.ejmech.2013.02.011
Al-Issa, S.A., Saudi. Pharm. J., 2013, vol. 21, p. 305. https://doi.org/10.1016/j.jsps.2012.09.002
Ghorab, M.M. and Alsaid, M.S., Biomed. Sci., 2016, vol. 27, p. 110.
Barakat, A., Soliman, S.M., Al-Majid, A.M., Lotfy, G., Ghabbour, H.A., Fun, H.-K., Yousuf, S., Choudhary, M.I., and Wadood, A., J. Mol. Struct., 2015, vol. 1098, p. 365. https://doi.org/10.1016/j.molstru.2015.06.037
Hassan, A.S., Mady, M.F., Awad, H.M., and Hafez, T.S., Chin. Chem. Lett., 2017, vol. 28, p. 388. https://doi.org/10.1016/j.cclet.2016.10.022
Fatahala, S.S., Mahgub, S., Taha, H., and Hameed, R.H.A-E., J. Enzyme Inhib. Med. Chem., 2018, vol. 33, p. 809. https://doi.org/10.1080/14756366.2018.1461854
Majeed, J. and Shaharyar, M., J. Enzyme Inhib. Med. Chem., 2011, vol. 26, p. 819. https://doi.org/10.3109/14756366.2011.557022
Sharma, V., Chitranshi, N., and Agarwal, A.K., Int. J. Med. Chem., 2014, vol. 2014, Article ID 202784. https://doi.org/10.1155/2014/202784
Ashok, D., Kumar, R.S., Mohan Gandhi, D., and Jayashree, A., Russ. J. Gen. Chem., 2016, vol. 86, p. 1396. https://doi.org/10.1134/S1070363216060268
Desai, V., Desai, C.M., and Patel, D., J. Institut. Chemists, 2005, vol. 11, p. 104.
Oztürk, H., OzkirimLi, E., and Özgür, A., PLoS One, 2015. https://doi.org/10.1371/journal.pone.0117874
Heo, L., Park, H., and Seok, C., Nucleic Acids Res., 2013, vol. 41, p. W384. https://doi.org/10.1093/nar/gkt458
Laskowski, R.A., Macarthur, M.W., Moss, D.S., and Thornton, J.M., J. Appl. Crystallogr., 1993, vol. 26, p. 283. https://doi.org/10.1107/S0021889892009944
Trott, O. and Olson, A.J. J., Comput. Chem., 2010, vol. 31, p. 1.
O’Boyle, N.M., Banck, M., James, C.A., Morley, C., Vandermeersch, T., and Hutchison, G.R., J. Chem. Inform., 2011, vol. 3, p. 1. https://doi.org/10.1186/1758-2946-3-33
Laskowski, R.A. and Swindells, M.B., J. Chem. Inf. Model., 2011, vol. 51, p. 2778. https://doi.org/10.1021/ci200227u
Sauvage, E., Duez, C., Herman, R., Kerff, F., Petrella, S., Anderson, J.W., Adediran, S.A., Pratt, R.F., Frиre, J.M., and Charlier, P., J. Mol. Biol., 2007, vol. 371, p. 528. https://doi.org/10.1016/j.jmb.2007.05.071
Klebe, G., Drug Design, 2013, p. 61. https://doi.org/10.1007/978-3-642-17907-5-4
Radhika, B., Shraddha, K.N., and Begum, N.S., IUCrData, 2020, vol. 5, x200385. https://doi.org/10.1107/S2414314620003855
ACKNOWLEDGMENTS
The authors are grateful to the Zeiss Microscopy Customer Center, Carl Zeiss India (Bangalore), for providing the FESEM facility.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest
Supplementary information
Rights and permissions
About this article
Cite this article
Bhat, R., Begum, N.S. Synthesis, Characterization, Antimicrobial Activity Screening, and Molecular Docking Study of Pyrimidine Carbonitrile Derivatives. Russ J Org Chem 57, 1352–1360 (2021). https://doi.org/10.1134/S1070428021080169
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1070428021080169