Abstract
Circular RNAs (circRNAs) was suggested to play crucial regulatory roles in various human diseases, including Parkinson’s disease (PD). This research aimed to investigate the function and potential mechanism of circ_0070441 in PD. MPP+ (1-methyl-4-phenylpyridinium)-treated SH-SY5Y cells was used as an in vitro cellular PD model. The expressions of circ_0070441, microRNA (miR)-626 and insulin receptor substrate 2 (IRS2) were measured by quantitative real-time polymerase chain reaction (RT-qPCR) or western blot. Cell Counting Kit-8 (CCK-8) assay, Cytotoxicity Detection Kit (Lactate Dehydrogenase), flow cytometry and Caspase-3 Assay Kit were used to detect cell viability, LDH release, cell apoptosis and caspase-3 activity, respectively. The levels of inflammation-related factors were detected by enzyme-linked immunosorbent assay (ELISA). The correlation among circ_0070441, miR-626 and IRS2 were confirmed by dual-luciferase reporter assay, RNA immunoprecipitation (RIP) assay and RNA pull-down assay. The levels of circ_0070441 and IRS2 were increased while miR-626 expression was decreased in MPP+-treated SH-SY5Y cells in dose- and time-dependent manners. Depletion of circ_0070441 alleviated MPP+-triggered neuronal damage by regulating cell apoptosis and inflammation. Circ_0070441 acted as a sponge for miR-626, and IRS2 was a target of miR-626. Besides, the neuroprotective effects of circ_0070441 knockdown or miR-626 overexpression were partly overturned by the suppression of miR-626 or IRS2 overexpression. Moreover, circ_0070441 upregulated IRS2 expression by interacting with miR-626. In summary, circ_0070441 aggravated MPP+-triggered neurotoxic effect in SH-SY5Y cells by regulating miR-626/IRS2 axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a complex neurodegenerative disease, Parkinson’s disease (PD) is mainly resulted from the dopaminergic neurons degeneration (Kalia and Lang 2015). Rigidity, rest tremor and bradykinesia are typical clinical symptoms of PD, which are commonly accompanied by autonomic dysfunction and cognitive decline, and eventually affected the quality of life of PD patients (Armstrong and Okun 2020; Coelho and Ferreira 2012). The estimated incidence rates for PD are 5 to 346 per 100,000 person-years in Europe (Ascherio and Schwarzschild 2016). At present, aging, oxidative stress, inflammation and apoptosis are considered as main pathogenic factors (Yang et al. 2020; Hallett et al. 2019). Nevertheless, current therapeutics have no ability to cure or impede the progress of PD except for relieving the associated symptoms (Armstrong and Okun 2020). Therefore, it is imperative that deep understanding of the pathogenic mechanism of PD and identify potential targets to develop novel therapeutic strategies for PD.
1-methyl-4-phenylpyridine (MPP+) has been widely applied to establish simulate PD in vitro model, since it could induce PD-related neuropathological change, including complex I inhibition, as well as the stimulating of the reactive oxygen species (ROS) pathway (Rostamian Delavar et al. 2018; Ding et al. 2019; Peng et al. 2019). In current research, MPP+-triggered SH-SY5Y cells were exploited as a cellular model for PD.
Circular RNA (circRNAs) are a novel sort of non-coding RNAs with a covalently closed circular structure without poly-A tails, which is generated from the back-splicing process (Kristensen et al. 2019). Several studies have proposed the vital function of circRNAs in neurodegenerative diseases initiation and development, such as PD (D’Ambra et al. 2019). For example, RNA sequencing and Kyoto Encyclopedia of Genes and Genomes analysis in varying brain regions of PD mice model recognized several deregulated circRNAs in PD and revealed that mmu_circRNA_0003292 might participate in the management of PD progression (Jia et al. 2020). Besides, Ravanidis et al. uncovered that circRNAs are significantly altered in the peripheral blood mononuclear cells (PBMCs) of PD patients, and provide reasonable sensitivity and specificity for PD, which could be used as possible therapeutic or diagnostic targets for PD (Ravanidis et al. 2021). Hsa_circ_0070441 (Position: chr4:90743396-90743539; Spliced sequence length: 608 bp) was derived from synuclein alpha (SNCA), and was reported to be upregulated in PD cellular model (Sang et al. 2018). Besides, hsa_circ_0127305, which is also derived from SNCA mRNA, was reported to engage in the symptomatic management of Pramipexole in PD (Sang et al. 2018). Nevertheless, the mechanism for regulating circ_0070441 in PD have not been investigated.
Current research suggested that circRNAs perform specific function by acting as miRNA sponge to deregulating microRNAs (miRNAs) stability and activity, thereby regulating downstream target mRNA expression (Zhou et al. 2020). In neurodegenerative disorders, many miRNAs are dysregulated and participate in the occurrence and development of diseases (Juzwik et al. 2019). For instance, knockdown of miR-217/miR-138-5p alleviated MPP+-triggered toxicity in SH-SY5Y cells (Wang et al. 2019). Besides, miR-190 restrained inflammation and reduced neuronal injury in MPTP-treated PD mouse model by interacting with NLR family pyrin domain containing 3 (NLRP3) (Sun et al. 2019). Furthermore, miR-626 was lower expressed in the cerebrospinal fluid (Qin et al. 2021) and peripheral blood (Khoo et al. 2012) of PD patients, which might be a promising detection, diagnosis, and monitoring biomarker for PD. However, the mechanism for regulating miR-626 in PD remains largely unknown.
Insulin Receptor Substrate 2 (IRS2), a member of IRSs family, which exerts a pivotal function in somatic growth and metabolic regulation (White 2002). Important discoveries have emerged recently regarding the function of IRS2 in the development of neurodegenerative diseases (White 2014). In particular, Xie et al. indicated that IRS2 was regulated by SOX21-AS1/miR-7-5p axis to modulate neuronal damage in MMP+-triggered SH-SY5Y cells (Xie et al. 2021). Consequently, IRS2 might be a prospective diagnostic target for PD although the mechanism underlying IRS2 in PD remains to be explored.
In the present research, a well-studied PD cellular model (MPP+-triggered SH-SY5Y cells) was employed. Next, the function and regulatory mechanism underlying circ_0070441 in MPP+-stimulated SH-SY5Y cells. Our research uncovered that circ_0070441 functioned its neurotoxic effect in PD by miR-626/IRS2 pathway, indicating a new approach for PD therapy.
Materials and methods
Bioinformatics analysis
CircInteractome database (https://circinteractome.nia.nih.gov/) and TargetScanHuman 7.2 database (http://www.targetscan.org/vert_72/) were employed for the prediction of putative targets.
Cell culture and transfection
Human neuroblastoma cell line SH-SY5Y was procured from American Type Culture Collection (ATCC; Manassas, VA, USA), then incubated in Dulbecco’s modified Eagle medium (DMEM; Hyclone, Logan, UT, USA) and 10 % fetal bovine serum (FBS; Hyclone) in an incubator with 5 % CO2 at 37 °C.
SH-SY5Y cells were stimulated with increasing concentrations (0, 0.5, 1 and 2 mM) of MPP+ (Sigma, St Louis, MO, USA) for 24 h, or stimulated with 2 mM MPP+ for different times (0 h, 12, 24 and 48 h), to establish the in vitro PD model.
The small interfering RNA (siRNA) against circ_0070441 (si-circ_0070441#1 and si-circ_0070441#2), miR-626 mimics (miR-626), miR-626 inhibitor (anti-miR-626), IRS2 overexpression vector (pcDNA-ISR2) and their matching negative control (si-NC, miR-NC, anti-miR-NC, pcDNA) were procured from HanBio Technology (Shanghai, China). Prior to transfecting oligonucleotides into SH-SY5Y cells via Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), SH-SY5Y cells (2 × 105 cells/well) were inoculated into 6-well-plates for 24 h. The cells were harvested at 48 h for RT-qPCR analysis or cultured further with 24 h of 2 mM MPP+ treatment.
Quantitative real-time PCR (RT-qPCR)
Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, Thorold, Canada) was employed for the isolation of cytoplasmic and nuclear RNA fractions. Total RNA was lysed in Trizol reagent (Solarbio, Beijing, China). Subsequently, the cDNA was acquired using the AMV Reverse Transcriptase (Solarbio) or miRNAFirst Strand cDNA Synthesis (Sangon, Shanghai, China). Then, the relative RNA levels were quantified via SYBR Green PCR Master Mix (Takara, Dalian, China) and estimated via the 2−ΔΔCt method. Table 1 exhibited the primers used in current research. GAPDH and U6 was regarded as the internal controls.
Cell counting Kit-8 (CCK-8) assay
Transfected SH-SY5Y cells were inoculated into 96-well plates in triplicate and nurtured with 2 mM MPP+ for 24 h. Then the cells were nurtured with 10 µL of CCK-8 solution (Sangon) for 3 h at 37 °C. Cell viability was evaluated via a Microplate Reader (BioTek, Burlington, VT, USA).
Lactate Dehydrogenase (LDH) assay
Lactate Dehydrogenase level was analyzed by Cytotoxicity Detection Kit (LDH) (Roche, Indianapolis, IN, USA). After 24 h of 2 mM MPP+ stimulation, the supernatant of transfected cells was collected and mixed with the reaction fluid for 30 min in the dark. Microplate Reader (BioTek) was used to evaluate the absorbance.
Flow cytometry
AnnexinV-FITC/Propidium Iodide (PI) Apoptosis Detection kit (Beyotime, Shanghai, China) was employed to estimate cell apoptosis. 1 × 106 SH-SY5Y cells were resuspended in binding buffer and spotted with AnnexinV-FITC or PI solution for 15 min. The apoptosis rate was measured via CytoFLEX flow cytometer (Beckman Coulter, Miami, FL, USA).
Caspase-3 activity assay
After indicated treatment, caspase-3 activity was estimated by Caspase-3 Assay Kit (Abcam, Cambridge, CA, USA). Proteins were extracted from SH-SY5Y cells and quantified using the BCA Protein Assay Kit (Pierce, Appleton, WI, USA). Then the protein was reacted with 2× reaction buffer containing DEVD-p-NA substrate for 1 h. The optical density was monitored via Microplate Reader (BioTek).
Enzyme-linked immunosorbent assay (ELISA)
The treated SH-SY5Y cells were inoculated into 24-well plates, and the levels of interleukin 1 beta (IL-1β), tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) in the supernatant were tested by corresponding ELISA kits (R&D Systems, Minneapolis, MN, USA).
Western blot assay
Total protein was isolated using the RIPA Buffer (Thermo Fisher Scientific, Waltham, MA, USA). Quantification of protein samples was evaluated by BCA Protein Assay Kit (Pierce). Equal amount of protein was separated by 12 % sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and electro-transferred onto the polyvinylidene fluoride (PVDF) membranes (Corning, Inc., Corning, NY, USA). After incubating with 5 % skimmed milk, the membranes were hatched with primary antibodies against Bax (1:5000, ab32503, Abcam), Bcl-2 (1:2000, ab182858, Abcam), β-actin (1:2000, ab8227, Abcam) or IRS2 (1:1000, MAB6347, R&D System), then probed with the secondary antibody (1:25,000, ab205718, Abcam). The blots were visualized using an ECL system (Beyotime).
Dual luciferase reporter assay
Circ_0070441 and IRS2 3’UTR sequences containing wild-type (WT) or mutant type (MUT) miR-626 binding sites were cloned into pGL3 vector (Promega, Madison, WI, USA) separately to construct the circ_0070441-WT, circ_0070441-MUT, IRS2 3’UTR-WT or IRS2 3’UTR-MUT reporter plasmids. After co-transfecting the recombinant reporter plasmids and miR-626 or miR-NC into SH-SY5Y cells via Lipofectamine 3000 (Invitrogen) reagent, the luciferase activity was measured using Dual-Lucy Assay Kit (Solarbio).
RNA immunoprecipitation (RIP) assay
RIP assay was executed through the EZ-Magna RNA Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). SH-SY5Y cells were lysed in RIP lysis buffer, then subject to the RIP buffer contained magnetic beads with Ago2 antibody or IgG antibody conjugation. Followed by extracting the immunoprecipitated RNA by incubating the mixture with protease K. And the enrichments of circ_0070441, miR-626 or IRS2 were assessed using RT-qPCR analysis.
RNA pull-down assay
A Pierce Magnetic RNA-Protein Pull down Kit (Thermo Fisher Scientific) was employed to implement the RNA pull-down assay. In brief, biotinylated miR-626 and its negative control Bio-NC were synthesized (HanBio Technology, Shanghai, China) to incubate the lysate samples of SH-SY5Y cells. Then the RNA-RNA complexes were pulled down using streptavidin magnetic beads. After purification, the enrichment of circ_0070441 was assessed by RT-qPCR.
Statistical analysis
Analysis of data was carried out by Graphpad Prism 7.0 software (GraphPad, San Diego, CA, USA). All data from at least three independent repetitions were presented as mean ± standard deviation. P values were analyzed by Student’s t-test (two groups) or one-way ANOVA (multiple groups). When P <0.05, it was supposed to be statistically significant.
Results
Circ_0070441 expression is elevated in MPP+-triggered SH-SY5Y cells
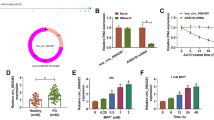
To explicate the function of circ_0070441 in PD, we first determined the expression of circ_0070441 in SH-SY5Y cells with MPP+ induced. As illustrated in Fig. 1A and B, MPP+ treatment provoked a dose- and time-dependent decline in cell viability. Meanwhile, circ_0070441 level was significantly decreased with the increasing of time and MPP+ concentration in MPP+-triggered SH-SY5Y cells (Fig. 1C and D). Then, MPP+ (2 mM)-triggered SH-SY5Y cells were employed as the cellular model for PD.
Circ_0070441 is up-regulated in MPP+-treated SH-SY5Y cells. (A) Cell viability of SH-SY5Y cells incubated with different doses (0, 0.5, 1 or 2 mM) of MPP+ for 24 h was examined by CCK-8 assay. (B) SH-SY5Y cells incubated with 2 mM MPP+ for 0 h, 12 h, 24 or 48 h, and cell viability was analyzed by CCK-8 assay. (C and D) Circ_0070441 expression in SH-SY5Y cells treated with MPP+ for different doses (0, 0.5, 1 or 2 mM) or different times (0 h, 12 h, 24 or 48 h) was analyzed by RT-qPCR. *P < 0.05
Circ_0070441 knockdown mitigates MPP+-triggered inflammation and neuronal apoptosis in SH-SY5Y cells
To explore the function of circ_0070441 in in vitro PD model, loss-of-function assays were executed by introducing si-circ_0070441 (si-circ_0070441#1 or si-circ_0070441#2) or si-NC into SH-SY5Y cells. RT-qPCR assay indicated that circ_0070441 expression was suppressed more than half in cells with si-circ_0070441#1 or si-circ_0070441#2 transfection (Fig. 2A). And si-circ_0070441#1 exhibited a higher transfection efficiency, so it was chosen for further investigated. MPP+ treatment significantly limited the viability of SH-SY5Y cells in contrast with the control group, whereas circ_0070441 knockdown alleviated the declined viability of SH-SY5Y cells (Fig. 2B). Besides, circ_0070441 knockdown significantly mitigated the increased LDH level in SH-SY5Y cells that stimulated with MPP+ (Fig. 2C). Furthermore, flow cytometry, western blot and Caspase-3 activity detection assays were employed to determine apoptosis. MPP+ treatment facilitated the apoptosis of SH-SY5Y cells, as identified by the elevated apoptosis rate (Fig. 2D), the increased level of pro-apoptotic protein Bax and decreased level of anti-apoptotic protein Bcl-2 (Fig. 2E), as well as the increased caspase-3 activity (Fig. 2F). Transfection of si-circ_0070441# significantly attenuated the effect of MPP+ on apoptosis (Fig. 2D-F). Meanwhile, we evaluated the inflammation response in MPP+-treated and circ_0070441 knockdown SH-SY5Y cells by ELISA. MPP+ treatment strikingly promoted the secretion of pro-inflammatory cytokines (TNF-α, IL-1β and IL-6), even though circ_0070441 knockdown reversed it (Fig. 2G and I). Thus, knockdown of circ_0070441 partly mitigated the MPP+-triggered inflammation and neuronal apoptosis in SH-SY5Y cells.
Circ_0070441 knockdown inhibits the inflammatory response and neuronal apoptosis in MPP+-treated SH-SY5Y cells. (A) SH-SY5Y cells were transfected with si-NC, si-circ_0070441#1 or si-circ_0070441#2, and circ_0070441 expression was evaluated by RT-qPCR. (B-I) SH-SY5Y cells transfected with si-NC or si-circ_0070441#1 were treated with 2 mM MPP+ for 24 h. (B) The viability of SH-SY5Y cells was determined by CCK-8 analysis. (C) The release of LDH was measured by cytotoxicity detection kit. (D) Cell apoptosis was evaluated using flow cytometry. (E) The protein levels of Bax and Bcl-2 in transfected cells were determined by western blot. (F) The activity of caspase-3 was examined by Caspase-3 assay kit. (G-I) The levels of TNF-α, IL-1β and IL-6 in transfected cells were detected via ELISA. *P < 0.05
Circ_0070441 directly interacts with miR-626
Current research indicated that circRNA exerted its regulatory function in multiple diseases by acting as a miRNA sponge, which is mainly occurs in the cytoplasm (Hansen et al. 2013). RT-qPCR assay indicated that circ_0070441 was largely enriched in the cytoplasm of SH-SY5Y cells, but not the nucleus (Fig. 3A). Subsequently, the probable miRNA of circ_0070441 by CircInteractome software, and uncovered that circ_0070441 sequences harbored the complementary binding sites of miR-626 (Fig. 3B). Thereafter, dual-luciferase reporter assay, RIP and RNA pull down assays were utilized to validate the correlation between miR-626 and circ_0070441. As displayed in Fig. 3C, miR-626 expression was enhanced 20 fold by miR-626 mimic. Besides, miR-626 mimic conspicuously diminished the luciferase activity of circ_0070441-WT group, rather than the circ_0070441-MUT group (Fig. 3D), indicating the presence of miR-626 binding elements in circ_0070441 sequence. Besides, Ago2 RIP assay disclosed the prominent enrichments of circ_0070441 and miR-626 in Ago2-immunoprecipitate group in relative to IgG-immunoprecipitate group (Fig. 3E). Furthermore, miR-626 level in the Bio-circ_0070441 group was remarkably higher than that in Bio-NC group, implying that miR-626 was pulled down by Bio-circ_0070441 (Fig. 3F). Additionally, MPP+ stimulation significantly decreased miR-626 level at a dose-dependent and time-dependent manner (Fig. 3G H). Moreover, knockdown of circ_0070441 markedly elevated miR-626 level (Fig. 3H).
Circ_0070441 directly interacts with miR-626. (A) Nuclear and cytoplasmic separation assay was employed to determine the level of circ_0070441 in nucleus and cytoplasm. (B) The predicted binding sites between circ_0070441 and miR-626 was displayed. (C) The level of miR-626 was measured in SH-SY5Y cells transfected with miR-NC or miR-626. (D) The luciferase activity of SH-SY5Y cells co-transfected with miR-NC or miR-626 and circ_0070441-WT or circ_0070441-MUT was determined by dual-luciferase reporter assay. (E) The enrichments of miR-626 and circ_0070441 in SH-SY5Y cells incubated with Anti-IgG or Anti-Ago2 were determined by RNA pull-down assay. (F) RNA pull down assay for the enrichment of circ_0070441 in SH-SY5Y cells incubated with Bio-NC or Bio-miR-626. (G) SH-SY5Y cells were stimulated with 0, 0.5, 1 or 2 mM of MPP+ for 24 h, and miR-626 level was determined by RT-qPCR. (H) RT-qPCR assay for the level of miR-626 in SH-SY5Y cells incubated with 2 mM MPP+ for 0 h, 12 h, 24 or 48 h. (I) The expression level of miR-626 in SH-SY5Y cells transfected with si-NC or si-circ_0070441 was examined. *P < 0.05
Circ_0070441 knockdown alleviates MPP+-induced inflammation and neuronal apoptosis by sponging miR-626
To elucidate whether circ_0070441 mediated MPP+-induced inflammation and neuronal apoptosis by regulating miR-626, we performed the loss-of-function experiments. The expression of miR-626 was suppressed by half in cells with anti-miR-626 transfection (Fig. 4A). Circ_0070441 depletion elevated the cell viability of MPP+-treated SH-SY5Y cells, while co-introduction of si-circ_0070441#1 and anti-miR-626 decreased the cell viability (Fig. 4B). Besides, miR-626 inhibitor partly overturned the effect of circ_0070441 downregulation on LDH release (Fig. 4C). Furthermore, circ_0070441 depletion decreased cell apoptosis rate, inhibited Bax protein level and caspase-3 activity, but elevated the protein level of Bcl-2, whereas these impacts were partly abrogated by downregulating of miR-626 (Fig. 4D F). Moreover, inhibition of circ_0070441 induced a considerable decrease in the release of inflammatory factors in MPP+-disposed SH-SY5Y cells, whereas these effects were partly transposed by anti-miR-626 (Fig. 4G and I). Hence, downregulation of circ_0070441 weakened MPP+-triggered inflammation and neuronal apoptosis by sponging miR-626.
Circ_0070441 knockdown alleviates MPP+-induced inflammatory response and neuronal apoptosis by regulating miR-626. (A) SH-SY5Y cells were transfected with anti-miR-NC or anti-miR-626, and the level of miR-626 was determined. (B-I) SH-SY5Y cells were introduced with si-NC, si-circ_0070441#1, si-circ_0070441#1 + anti-miR-NC or si-circ_0070441#1 + anti-miR-626, and stimulated with 2 mM MPP+ for 24 h. Cell viability (B), LDH release (C), apoptosis rate (D), Bax and Bcl-2 protein levels (E), caspase-3 activity (F), the production of inflammatory cytokines (TNF-α, IL-1β and IL-6) (G-I) in transfected cells were examined by CCK-8, cytotoxicity detection kit, flow cytometry, western blot, Caspase-3 assay kit, or ELISA assays, respectively. *P < 0.05
IRS2 is targeted by miR-626
We next predicted the downstream target of miR-626 using the TargetScanHuman 7.2, and found that IRS2 3’UTR sequence contained the putative miR-626 binding sites (Fig. 5A). Besides, miR-626 mimic significantly decreased the luciferase activity of IRS2 3’UTR-WT group, implying the presence of miR-626 binding sites in IRS2 3’UTR sequence (Fig. 5B). Besides, IRS2 and miR-626 were significantly enriched in Ago2-immunoprecipitate group instead of IgG-immunoprecipitate group (Fig. 5C), which further confirmed the existence of IRS2 and miR-626 in RNA-induced silencing complex. In addition, knockdown of miR-626 elevated IRS2 mRNA and protein levels, while up-regulation of miR-626 blocked IRS2 mRNA and protein levels (Fig. 5D and E). Moreover, MPP+ treatment provoked a notable elevation in IRS2 mRNA and protein levels with the increase of concentration (Fig. 5F and G) and time (Fig. 5H and I).
IRS2 is a target of miR-626. (A) The putative binding sites between miR-626 and IRS2 3’UTR were shown. (B) SH-SY5Y cells were co-transfected with miR-NC or miR-626 and IRS2 3’UTR-WT or IRS2 3’UTR-MUT, and the luciferase activity of cells were determined. (C) The enrichments of IRS2 and miR-626 in Anti-IgG or Anti-Ago2 group were detected using RIP assay. (D and E) SH-SY5Y cells were introduced with anti-miR-NC, anti-miR-626, miR-NC or miR-626, and IRS2 mRNA and protein levels were evaluated by RT-qPCR and western blot. (F and G) The mRNA and protein levels of IRS2 were examined in SH-SY5Y cells stimulated with 0, 0.5, 1 or 2 mM of MPP+ for 24 h. (H and I) RT-qPCR and western blot assays for the mRNA and protein level of miR-626 in SH-SY5Y cells incubated with 2 mM MPP+ for 0 h, 12 h, 24 or 48 h. *P < 0.05
MiR-193a-3p attenuates MPP+-triggered inflammation and neuronal apoptosis by targeting IRS2
To verify whether miR-626 affected MPP+-triggered neuronal damage by modulating IRS2, SH-SY5Y cells were introduced with miR-NC, miR-626, miR-626 + pcDNA or miR-626 + pcDNA-IRS2, and then stimulated with 2 mM MPP+. IRS2 mRNA and protein levels were prominently elevated in the pcDNA-IRS2 group (Fig. 6A and B). Besides, the elevated IRS2 weakened the promotion influence of miR-626 overexpression on cell viability (Fig. 6C). Additionally, miR-626 mimic suppressed the release of LDH, while co-introduction of miR-626 and pcDNA-IRS2 promoted LDH release (Fig. 6D). Furthermore, IRS2 overexpression partly overturned the suppression influences of miR-626 on cell apoptosis (Fig. 6E and G), as well as the suppression effects on the secretion of inflammatory cytokines (Fig. 6H-J). These results uncovered that miR-626 regulated inflammation and neuronal apoptosis by modulating IRS2 in MPP+-treated SH-SY5Y cells.
MiR-626 attenuates MPP+-triggered inflammatory response and neuronal apoptosis by targeting IRS2. (A and B) IRS2 mRNA and level were detected in SH-SY5Y cells transfected with pcDNA or pcDNA-IRS2. (C-J) SH-SY5Y cells were transfected with miR-NC, miR-626, miR-626 + pcDNA or miR-626 + pcDNA-IRS2, followed by incubating with 2 mM MPP+ for 24 h. Cell viability (C), LDH release (D), apoptosis rate (E), Bax and Bcl-2 protein levels (F), caspase-3 activity (G), the production of inflammatory cytokines (TNF-α, IL-1β and IL-6) (H-J) in transfected cells were examined by CCK-8, cytotoxicity detection kit, flow cytometry, western blot, Caspase-3 assay kit, or ELISA assays, respectively. *P < 0.05
Circ_0070441 upregulates IRS2 expression by sponging miR-626
To clarify the correlation among circ_0070441, miR-626 and IRS2, SH-SY5Y cells were introduced with si-circ_0070441 alone or combine with anti-miR-626. And we found that circ_0070441 knockdown strikingly reduced IRS2 mRNA and protein levels in contrast with the si-NC group, whereas knockdown of miR-626 partly abolished the suppression effect of si-circ_0070441#1 on IRS2 expression (Fig. 7A and B). Taken together, circ_0070441 downregulation protected SH-SY5Y cells from MPP+-induced inflammation and neuronal apoptosis through regulating miR-626/IRS2 axis.
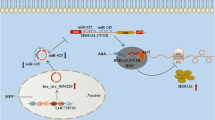
Circ_0070441 upregulates IRS2 expression by sponging miR-626. (A and B) The mRNA and protein levels of IRS2 were measured by RT-qPCR and western blot in SH-SY5Y cells transfected with si-NC, si-circ_0070441#1, si-circ_0070441#1 + anti-miR-NC or si-circ_0070441#1 + anti-miR-626. (C) Schematic of how circ_0070441/miR-626/IRS2 axis mediates inflammatory response and neuronal apoptosis in PD. *P < 0.05
Discussion
MPP+ triggered oxidative stress and mitochondrial dysfunction to produce dopaminergic neuron degeneration, which mimics the primary features of PD pathogenesis (Schildknecht et al. 2017). Therefore, a PD in vitro cellular model was employed to explore the molecular mechanism of PD development. In current research, MPP+ treatment provoked a dose- and time-dependent diminution in cell proliferation, as well as provoked cell apoptosis and inflammation in SH-SY5Y cells. In past studies, MPTP was thought to affect RNA profiles in mice brain tissue and mediated neuroinflammatory processes (Soreq et al. 2012). Therefore, exploring gene expression under MPP+ can effectively simulate in vivo processes.
As a newly identified RNA type, circRNAs were proposed to play a vital function in multiple diseases, whereas the research on their function in PD was limited (D’Ambra et al. 2019; Gámez-Valero et al. 2020). As previously described, circRNA circ_0127305, which is also derived from SNCA gene, could mediate the suppressive effect of pramipexol on PD progression (Sang et al. 2018). Besides, downregulation of circ_0127305 or SNCA inhibited apoptosis but promoted autophagy in MPP+-treated SH-SY5Y cells (Sang et al. 2018). In current research, we uncovered that circ_0070441 was elevated in SH-SY5Y cells with the treatment of MPP+ in a concentration-dependent gradient or a time-dependent gradient. Downregulation of circ_0070441 remitted the neurotoxic influence of MPP+ on SH-SY5Y cells through promoting cell viability, apoptosis and inflammation. Hence, we speculate that circ_0070441 may play a neurotoxic function in MPP+-treated SH-SY5Y cells to promote PD pathogenesis. However, it is worth noting that in a recent study, Hanan et al. used RNA sequencing to screen for differentially expressed circRNA in the brain tissues of PD patients and control donors (Hanan et al. 2020), but did not screen circ_0070441. Although the specific reasons are still unclear, the selection of circ_0070441 focused in this study came from a published article (Sang et al. 2018) that revealed that circ_0070441 was significantly upregulated in MPP+-induced PD cell models, which has also been further confirmed in our study. After functional test verification, we determined that circ_0070441 could exacerbate the neurotoxicity of SH-SY5Y cells triggered by MPP+. Therefore, we confirmed that circ_0070441 was an important regulator for the progression of PD.
Mechanically, circRNAs could sponge specific miRNAs, which share the same miRNA response elements (MRE), thereby inducing the degradation of target miRNAs (Salmena et al. 2011). Herein, we predicted that miR-626 is a target of circ_0070441, and this prediction was further confirmed by the decreased luciferase activity of circ_0070441-WT and miR-626 co-transfected group, as well as the enrichment of circ_0070441 in Bio-miR-626 group. Besides, our data showed that circ_0070441 acted as a miRNA sponge for miR-626, thereby repressing the level of miR-626 in cells. Accumulating studies have illuminated the pivotal role of miR-626 in human cancers, including bladder cancer (Dong et al. 2019), oral squamous cell carcinoma (Shi et al. 2019), lung cancer (Liu et al. 2021), and hepatocellular carcinoma (Zhu et al. 2020). Moreover, recent research disclosed that miR-626 was lower expressed in the cerebrospinal fluid of PD patients, which could be used as a biomarker for PD (Qin et al. 2019). In consistent with previous research, MPP+ treatment induced a remarkable downregulation in the expression of miR-626, and knockdown of miR-626 partly diminished the neuroprotective effect of circ_0070441 knockdown on MPP+-treated SH-SY5Y cells.
Additionally, we predicted that IRS2 contained the complementary binding sites of miR-626. And miR-626 could inhibit ISR2 expression at transcription and translation levels. IRS2, a cytoplasmic signaling molecule that mediates the effects of insulin, was informed to be significantly conducive to the progress of neurodegenerative diseases (White 2014). Wakabayashi et al. suggested that long-term high-fat diet feeding promoted brain amyloid-β (Aβ) deposition in IRS-2-deficient mice (Wakabayashi et al. 2019). Besides, IRS2 downregulation could remit the neurotoxic influence of MPP+ on SH-SY5Y cells (Xie et al. 2021). Hence, IRS2 might be a novel biomarker for PD therapy. Herein, in consistent with previous research, IRS2 exerted a neurotoxic role in PD, since IRS2 overexpression partly overturned the neuroprotective influence of miR-626 on MPP+-triggered SH-SY5Y cells. Furthermore, our results uncovered that circ_0070441 exerts as a miRNA sponge for miR-626, thereby upregulating the level of IRS2.
However, there are some limitations existing in this research. For instance, current research is performed in vitro, and in vivo assays are necessary to perform to prove this mechanism. Another limitation of the present research is the lack of correlation analysis of clinicopathologic features and circ_0070441/miR-626/IRS2 axis.
In conclusion, circ_0070441 upregulated IRS2 expression by sponging miR-626, thereby promoting apoptosis, inflammation and neuron damage in MPP+-triggered SH-SY5Y cells (Fig. 7C). These findings suggested a novel ceRNA-mediated regulation mechanism in PD, which may provide a new therapeutic target for PD.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Armstrong MJ, Okun MS (2020) Diagnosis and treatment of Parkinson disease: a review. JAMA 323(6):548–560
Ascherio A, Schwarzschild MA (2016) The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol 15(12):1257–1272
Coelho M, Ferreira JJ (2012) Late-stage Parkinson disease. Nat Rev Neurol 8(8):435–442
D’Ambra E, Capauto D, Morlando M (2019) Exploring the regulatory role of circular RNAs in neurodegenerative disorders. Int J Mol Sci 20(21):5477
Ding XM, Zhao LJ, Qiao HY, Wu SL, Wang XH (2019) Long non-coding RNA-p21 regulates MPP(+)-induced neuronal injury by targeting miR-625 and derepressing TRPM2 in SH-SY5Y cells. Chem Biol Interact 307:73–81
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q et al (2019) Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer 18(1):95
Gámez-Valero A, Guisado-Corcoll A, Herrero-Lorenzo M, Solaguren-Beascoa M, Martí E (2020) Non-coding RNAs as sensors of oxidative stress in neurodegenerative diseases. Antioxidants (Basel) 9(11):1095
Hallett PJ, Engelender S, Isacson O (2019) Lipid and immune abnormalities causing age-dependent neurodegeneration and Parkinson’s disease. J Neuroinflammation 16(1):153
Hanan M, Simchovitz A, Yayon N, Vaknine S, Cohen-Fultheim R, Karmon M et al (2020) A Parkinson’s disease CircRNAs Resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol Med 12(9):e11942
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Jia E, Zhou Y, Liu Z, Wang L, Ouyang T, Pan M et al (2020) Transcriptomic profiling of circular RNA in different brain regions of Parkinson’s disease in a mouse model. Int J Mol Sci 21(8):3006
Juzwik CA, S SD, Zhang Y, Paradis-Isler N, Sylvester A, Amar-Zifkin A et al (2019) microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog Neurobiol 182:101664
Kalia LV, Lang AE (2015) Parkinson’s disease. Lancet 386(9996):896–912
Khoo SK, Petillo D, Kang UJ, Resau JH, Berryhill B, Linder J et al (2012) Plasma-based circulating MicroRNA biomarkers for Parkinson’s disease. J Parkinsons Dis 2(4):321–331
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20(11):675–691
Liu Q, Cao G, Wan Y, Xu C, He Y, Li G (2021) Hsa_circ_0001073 targets miR-626/LIFR axis to inhibit lung cancer progression. Environ Toxicol 36(6):1052–1060
Peng T, Liu X, Wang J, Liu Y, Fu Z, Ma X et al (2019) Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson’s disease via regulating miR-100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif Cells Nanomed Biotechnol 47(1):2764–2774
Qin LX, Tan JQ, Zhang HN, Tang JG, Jiang B, Shen XM et al (2019) Preliminary study of hsa-miR-626 change in the cerebrospinal fluid of Parkinson’s disease patients. J Clin Neurosci 70:198–201
Qin LX, Tan JQ, Zhang HN, Tang JG, Jiang B, Shen XM et al (2021) Preliminary study of hsa-mir-626 change in the cerebrospinal fluid in Parkinson’s disease. Neurol India 69(1):115–118
Ravanidis S, Bougea A, Karampatsi D, Papagiannakis N, Maniati M, Stefanis L et al (2021) Differentially expressed circular RNAs in peripheral blood mononuclear cells of patients with Parkinson’s disease. Mov Disord 36(5):1170–1179
Rostamian Delavar M, Baghi M, Safaeinejad Z, Kiani-Esfahani A, Ghaedi K, Nasr-Esfahani MH (2018) Differential expression of miR-34a, miR-141, and miR-9 in MPP+-treated differentiated PC12 cells as a model of Parkinson’s disease. Gene 662:54–65
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358
Sang Q, Liu X, Wang L, Qi L, Sun W, Wang W et al (2018) CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging 10(6):1281–1293
Schildknecht S, Di Monte DA, Pape R, Tieu K, Leist M (2017) Tipping points and endogenous determinants of nigrostriatal degeneration by MPTP. Trends Pharmacol Sci 38(6):541–555
Shi J, Bao X, Liu Z, Zhang Z, Chen W, Xu Q (2019) Serum miR-626 and miR-5100 are promising prognosis predictors for oral squamous cell carcinoma. Theranostics 9(4):920–931
Soreq L, Ben-Shaul Y, Israel Z, Bergman H, Soreq H (2012) Meta-analysis of genetic and environmental Parkinson’s disease models reveals a common role of mitochondrial protection pathways. Neurobiol Dis 45(3):1018–1030
Sun Q, Wang S, Chen J, Cai H, Huang W, Zhang Y et al (2019) MicroRNA-190 alleviates neuronal damage and inhibits neuroinflammation via Nlrp3 in MPTP-induced Parkinson’s disease mouse model. J Cell Physiol 234(12):23379–23387
Wakabayashi T, Yamaguchi K, Matsui K, Sano T, Kubota T, Hashimoto T et al (2019) Differential effects of diet- and genetically-induced brain insulin resistance on amyloid pathology in a mouse model of Alzheimer’s disease. Mol Neurodegener 14(1):15
Wang M, Sun H, Yao Y, Tang X, Wu B (2019) MicroRNA-217/138-5p downregulation inhibits inflammatory response, oxidative stress and the induction of neuronal apoptosis in MPP(+)-induced SH-SY5Y cells. Am J Transl Res 11(10):6619–6631
White MF (2002) IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab 283(3):E413–E422
White MF (2014) IRS2 integrates insulin/IGF1 signalling with metabolism, neurodegeneration and longevity. Diabetes Obes Metab 16(Suppl 1):4–15
Xie Y, Zhang S, Lv Z, Long T, Luo Y, Li Z (2021) SOX21-AS1 modulates neuronal injury of MMP(+)-treated SH-SY5Y cells via targeting miR-7-5p and inhibiting IRS2. Neurosci Lett 746:135602
Yang L, Mao K, Yu H, Chen J (2020) Neuroinflammatory responses and Parkinson’ disease: pathogenic mechanisms and therapeutic targets. J Neuroimmune Pharmacol 15(4):830–837
Zhou DN, Ye CS, Deng YF (2020) CircRNAs: potency of protein translation and feasibility of novel biomarkers and therapeutic targets for head and neck cancers. Am J Transl Res 12(5):1535–1552
Zhu P, Liang H, Huang X, Zeng Q, Liu Y, Lv J et al (2020) Circular RNA Hsa_circ_0004018 inhibits Wnt/β-catenin signaling pathway by targeting microRNA-626/DKK3 in hepatocellular carcinoma. Onco Targets Ther 13:9351–9364
Funding
The project was funded by Ningxia Hui Autonomous Region Key Research and Development Project (No. 2017BY037) and Ningxia Natural Science Foundation Project (No. NZ17189).
Author information
Authors and Affiliations
Contributions
XC and JG designed, supervised the study, conducted the experiments and drafted the manuscript. DX, TZ and HH collected and analyzed the data. HM and TM contributed the methodology and analyzed the data. MQ and JH operated the software and edited the manuscript. XC was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuqing Cao and Jiangtao Guo are co-authors.
Rights and permissions
About this article
Cite this article
Cao, X., Guo, J., Mochizuki, H. et al. Circular RNA circ_0070441 regulates MPP+-triggered neurotoxic effect in SH-SY5Y cells via miR-626/IRS2 axis. Metab Brain Dis 37, 513–524 (2022). https://doi.org/10.1007/s11011-021-00869-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-021-00869-3