Abstract
The study aims to explore the molecular mechanism involved in Parkinson’s disease (PD). Hsa_circ_0004381, microRNA-185-5p (miR-185-5p), and Rac family small GTPase 1 (RAC1) level were measured by real-time quantitative polymerase chain reaction (RT-qPCR). Furthermore, cell viability and apoptosis rate were assessed by Cell Counting Kit-8 (CCK-8) and flow cytometry assays, respectively. Protein levels of B cell lymphoma-2 (Bcl-2), Bcl-2-related X protein (Bax), cleaved-caspase 3 (c-caspase 3), and RAC1 were determined by western blot assay. The levels of tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and IL-6 were detected by enzyme-linked immunosorbent assay (ELISA). The ROS generation and LDH and SOD activity were detected by the corresponding kits. The binding relationship between miR-185-5p and hsa_circ_0004381 or RAC1 was predicted by Starbase and then verified by a dual-luciferase reporter and RNA Immunoprecipitation (RIP) assays. Hsa_circ_0004381 and RAC1 were increased, and miR-185-5p was decreased in MPP+-triggered SK-N-SH cells. Moreover, hsa_circ_0004381 silencing promoted cell viability, and repressed apoptosis, inflammatory response, and oxidative stress in MPP+-treated SK-N-SH cells. The mechanical analysis suggested that hsa_circ_0004381 served as a sponge of miR-185-5p to affect RAC1 expression. Hsa_circ_0004381 could contribute to MPP+-triggered neuron injury by targeting the miR-185-5p/RAC1 axis, which provided a novel insight into the pathogenesis and treatment of PD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the second most frequent neurodegenerative disorder after Alzheimer’s disease, Parkinson’s disease (PD) is caused by the loss of neurons in the substantia nigra pars compacta (Reich and Savitt 2019). The incidence of PD is steadily increased with age, affecting at least 2% of people over 65 years in China (Pringsheim et al. 2014; Ma et al. 2014). Clinically, the patients undergo motor injuries, such as resting tremors, postural instability, and bradykinesia, which decrease their quality of life (Sveinbjornsdottir 2016). An extensive body of recent research has described the biological pathology and etiology of PD, but the pathogenesis of PD is still not completely clear (Raza et al. 2019). Currently, N-methyl-4-phenyl pyridine (MPP+) was defined as the putative toxic metabolite of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), which was used to selectively destroy dopaminergic neurons in the substantia nigra and induce PD (Zhu et al. 2018). As is widely recognized, MPP+ is applied to establish experimental models of PD by exerting the neurotoxic effects (Zhu et al. 2018; Küçükdoğru et al. 2020; Feng et al. 2020). Hence, depicting the internal mechanism underlying MPP+-induced cell damage is highly desirable for developing alternative therapeutic strategies.
During the past several decades, non-coding RNAs (ncRNAs) constitute the majority of the human-transcribed genome that regulated cell physiology and shape cellular functions (Panni et al. 2020). As a new species of endogenous non-coding RNA, circular RNAs (circRNAs) are single-stranded RNA molecules formed by the back splicing events of exons or introns, with a covalent closed-loop structure (Patop et al. 2019; Ashwal-Fluss et al. 2014). This feature enables circRNA to be resistant to RNA exonuclease, so they can be more stably present in tissues and cells (Rybak-Wolf et al. 2015; Barrett and Salzman 2016). Widely expressed and generally conserved in mammalian cells, circRNAs have attracted great interest for their application to clinical treatment (Zhang et al. 2018). Furthermore, work in several laboratories has discovered that most circRNAs possess the capability of regulating gene expression at different levels, presenting a wide range of functional roles in various biological processes (Memczak et al. 2013; Salzman 2016). Lately, it has become evident that circRNAs are being found associated with brain development and neuron maintenance (Rybak-Wolf et al. 2015; Mehta et al. 2020). In terms of PD, Feng et al. found that circDLGAP4 could exert neuroprotective effects in PD by facilitating cell viability, suppressing apoptosis, and enhancing autophagy (Feng et al. 2020). Synchronously, Wang et al. reported that circSAMD4A could exacerbate MPP+-caused neuronal damage by regulating the miR-29c-3p-mediated AMPA/mTOR pathway in vitro (Wang et al. 2021). Of note, relevant studies displayed that hsa_circ_0004381 abundance was aberrantly upregulated in PD patients, which might act as a dynamic monitoring factor for the development of PD (Zhong et al. 2021). To our knowledge, the potential pathogenesis of hsa_circ_0004381 involving neuroinflammation in PD remains unclear.
Nowadays, a large number of scholars have proposed the competitive endogenous RNA (ceRNA) hypothesis that circRNAs could be mutually regulated by competition for binding to common microRNA (miRNA) response elements (MREs) (Thomson and Dinger 2016; Panda 2018). This hypothesis has received increasing attention as a unifying function for lncRNAs. Here, bioinformatics analysis exhibited that hsa_circ_0004381 has some binding sites with miR-185-5p for the first time. Apart from that, previous research suggested that miR-185-5p is a potential serum-based biomarker for the diagnosis of PD (Ding et al. 2016), which could mitigate neuronal injury by regulating the PI3K/AKT signaling pathway in the 6-hydroxydopamine-challenged PD cell model in vitro (Qin et al. 2021). Therefore, the purpose of this study was to preliminarily unearth whether the regulatory role of hsa_circ_0004381 on the neuronal injury was mediated by sponging miR-185-5p.
Materials and Methods
Clinical Samples and Cell Culture
Approval to conduct this research project was acquired from the Ethics Committee of The First Affiliated Hospital of Jinzhou Medical University. After obtaining the written informed consent, the fresh venous blood samples were provided by 60 healthy volunteers and 80 PD patients. Also, the sample characteristics of the PD patients and healthy volunteers are described in Table 1. Then, serum samples were gained by centrifugation.
Human neuroblastoma cell line SK-N-SH cell line (CL-0214; Procell, Wuhan, China) was grown in a special medium (CM-0214; Procell) under a humidified incubator with 5% CO2 at 37 °C. Meanwhile, to verify the stability of hsa_circ_0004381, SK-N-SH cells were treated with 3 U/μg of RNase R (Epicentre, Madison, WI, USA) for 30 min at 37 °C or exposed to 2 μg/mL of Actinomycin D (Act D, Sigma-Aldrich, St. Louis, MO, USA) for 0, 6, 12, 18, and 24 h. For MPP+ treatment, SK-N-SH cells were treated with 0, 0.25, 0.5, 1, or 2 mM MPP+ (Sigma-Aldrich) for 0–48 h to establish PD cell models. At length, 1-mM MPP+ treatment for 24 h was selected for subsequent experiments.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Using TRIzol reagent (Invitrogen, Paisley Scotland, UK), total RNAs from serum samples and cells were prepared. After generating cDNA using PrimeScript™ RT Master Mix Kit (Takara, Tokyo, Japan), cDNA amplification was carried out by SYBR Green PCR Kit (TaKaRa) and specific primers. Finally, the data were analyzed in light of the 2−ΔΔCt method, normalizing by GAPDH for hsa_circ_0004381 and Rac family small GTPase 1 (RAC1), and U6 for miR-185-5p. Specific primers were exhibited in Table 2.
Cell Transfection
In this assay, the oligonucleotides, including hsa_circ_0004381 small interference RNA (si-hsa_circ_0004381#1 and si-hsa_circ_0004381#2), miR-185-5p mimic (anti-miR-185-5p: 5′-TGGAGAGAAAGGCAGTTCCTGA-3′), miR-NC, miR-185-5p inhibitor (miR-185-5p: 5′-TCAGGAACTGCCTTTCTCTCCA-3′), and anti-miR-NC, were provided by RiboBio (Guangzhou, China). Besides, the hsa_circ_0004381 overexpression vector (hsa_circ_0004381) and its control (pCD-ciR) were obtained from Geneseed (Guangzhou, China). Also, the cDNA sequences of RAC1 were cloned and introduced into pcDNA3.1 vector (Invitrogen) to acquire the pcDNA3.1-RAC1 (RAC1). After that, plasmids (50 ng) and oligonucleotides (20 nM) were, respectively, transfected into cells by mean of Lipofectamine 3000 (Invitrogen).
Cell Viability Assay
The 2000 transfected cells were seeded in 96-well plates overnight, followed by treatment with 1 mM MPP+ for 24 h. After being mixed with 10 μL of cell counting kit (CCK-8, Dojindo, Osaka, Japan) for 4 h at 37 °C, the detection of apoptosis rate was performed using a microplate reader.
Cell Apoptosis Assay
After being treated with MPP+ (1 mM, 24 h), transfected cells were trypsinized and washed using PBS, and then mixed with binding buffer. Whereafter, the cells were dual-stained with 5 μL Annexin (V-fluorescein isothiocyanate) V-FITC and 10 μL Propidium Iodide (PI) (eBioscience, San Diego, CA, USA), and were subjected to flow cytometry analysis (FACSan, BD Bioscience, Heidelberg, Germany).
Western Blot Assay
Using RIPA buffer (Beyotime, Nantong, China), cell lysates were prepared on ice, followed by quantification using a BCA protein assay kit (Beyotime). After that, equal amounts of protein were subjected to standard 10% SDS-PAGE and blotted to the nitrocellulose membranes (Millipore, Molsheim, France). Subsequently, the primary antibodies, B cell lymphoma-2 (Bcl-2; 1:1000, ab32124), Bcl-2-related X protein (Bax; 1:1000, ab32503), cleaved-caspase-3 (c-caspase-3; 1:500, ab2302), RAC1 (1:1000, ab155938, Abcam), β-Actin (1:5000; ab8227; Abcam), and secondary antibody (1:2500, ab205718, Abcam), were used for western blotting. At last, the detection of band intensity was conducted using ECL detection kit (Amersham Biosciences, Pittsburg, PA, Sweden), and all antibodies were purchased from Abcam (Cambridge, MA, USA).
Enzyme-Linked Immunosorbent Assay (ELISA)
After treatment with 1 mM MPP+ for 24 h, the cell supernatants from various groups were collected by centrifugation. And then, the secretion of IL-1β, IL-6, and TNF-α was assessed using the ELISA kits (R&D Systems, Minneapolis, MN, USA).
Determination of Oxidative Stress
For reactive oxygen species (ROS) level, treated cells were reacted with 10 nM DCFH-DA (Beyotime) at 37 °C in the absence of light. After incubation for 45 min, the fluorescence intensity was analyzed by means of a microplate reader (Tecan Infinite M200, Tecan, Switzerland) with an excitation wavelength of 485 nm and an emission wavelength of 530 nm. For lactate dehydrogenase (LDH) and superoxide dismutase (SOD), treated cells were collected and dissolved in cell lysis buffer (Beyotime), followed by the determination of LDH and SOD activities using LDH kit (Beyotime) and SOD kit-WST (Dojindo, Kumamoto, Japan) and a microplate reader.
Dual-Luciferase Reporter Assay
Starbase (http://starbase.sysu.edu.cn/) software was used to predict the binding between miR-185-5p and hsa_circ_0004381 or RAC1, as verified using a dual-luciferase reporter assay in an embryonic kidney cell line (HEK293T, CL-005, Procell). To be brief, wild-type (WT) luciferase reporter constructs (hsa_circ_0004381 WT1, hsa_circ_0004381 WT2, and RAC1-3’UTR WT) were generated by directly amplifying and inserting the partial sequence of them containing the predicted miR-185-5p into pmirGLO reporter vector (Promega, Madison, WI, USA). Also, site-directed mutant (MUT) constructs (hsa_circ_0004381 MUT1, hsa_circ_0004381 MUT2, and RAC1-3′UTR MUT) were achieved using the QuikChange II site-directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). At length, cells were transfected with the construct (200 ng) and miR-185-5p or miR-NC (50 nM), followed by analysis using a dual-luciferase reporter assay kit (Promega).
RNA Immunoprecipitation (RIP)
After being cultured to reach 80% confluency, the harvested cells were mixed with the complete RIP lysis buffer (Millipore). Whereafter, cell lysates were incubated with anti-Ago2 (Millipore) or normal mouse IgG (Millipore) at 4 °C for 4 h before treating magnetic protein A/G beads for 2 h. Following digested using proteinase K (Invitrogen), total RNA was isolated for the measurement of miR-185-5p and hsa_circ_0004381 using RT-qPCR assay.
RNA Pull-Down Assay
Generally, the probe-coated magnetic beads were generated by mixing the biotinylated (Bio) hsa_circ_0004381 probe (5′-GCCAGACAGATCCTCTTTCCTGTG-3′) and Bio-NC probe (GenePharma, Shanghai, China) with M-280 Streptavidin magnetic beads (Invitrogen) at room temperature for 2 h. Subsequently, the harvested SK-N-SH cells (1 × 107 cells/well) were sonicated and incubated with probe-coated beads at 4 °C overnight. After total RNA extraction, the enrichment of hsa_circ_0004381 and miRNAs was assessed with RT-qPCR assay.
Statistical Analysis
All of the results represent mean ± standard deviation (SD) and were analyzed in light of GraphPad Prism7 (GraphPad software, San Diego, CA, USA). Pearson correlation analysis was applied to analyze the expression association. P < 0.05 was considered statistically significant by Student’s t-test for two groups or one-way analysis of variance (ANOVA) with Tukey’s tests for multiple groups.
Results
Hsa_circ_0004381 Was Expressed at a High Level in PD Patients and MPP. +-Induced PD Cell Model
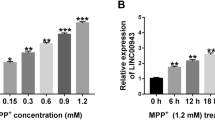
First of all, our analysis displayed that hsa_circ_0004381 is generated from the exon 6 and exon 7 of the ARID1B gene, and the end of exon 6 and exon 7 were back-spliced to form the circular structure (Fig. 1A). Then, to further validate the stability of hsa_circ_0004381, we treated SK-N-SH cells with RNase R and Act D (transcription inhibitor). As presented in Fig. 1B, RNase R treatment obviously reduced the expression of ARID1B mRNA, whereas it almost had no impact on the expression of hsa_circ_0004381 relative to the untreated group. Also, the Act D assay proved that the half-life of the hsa_circ_0004381 transcript exceeded 24 h, suggesting that this isoform is more stable than the linear ARID1B mRNA transcript in SK-N-SH cells (Fig. 1C). Whereafter, to investigate the role of hsa_circ_0004381 in PD, we collected the serum samples from 80 PD patients and 60 healthy volunteers. According to the data shown in Fig. 1D, the significant upregulation of hsa_circ_0004381 was noticed in serum samples from PD patients compared with the normal controls. Meanwhile, data exhibited that the PD and control groups did not differ significantly by age and sex (Table 1). Subsequently, we further explored the expression level of hsa_circ_0004381 in an in vitro model of PD, which was established in SK-N-SH cells treated with MPP+. Data displayed that the hsa_circ_0004381 level was gradually enhanced in SK-N-SH cells after treatment of 0.25–2 mM MPP+ for 24 h, especially in 1-mM MPP+-treated cells (Fig. 1E). Then, RT-qPCR assay further presented that hsa_circ_0004381 level was obviously increased in 1-mM MPP+-triggered SK-N-SH cells treated with more than or equal to 24 h (Fig. 1F). Hence, we selected 1-mM MPP+ treatment for 24 h to induce cell injury in SK-N-SH cells for subsequent experiments. These results proved the upregulation of hsa_circ_0004381 in MPP+-treated SK-N-SH cells in a dose-dependent and time-dependent manner, implying an underlying rale of hsa_circ_0004381 in neuronal cell damage in PD.
Hsa_circ_0004381 expression was elevated in PD patients and MPP+-induced PD cell model. A The schematic diagram revealed the formation of hsa_circ_0004381 from the ARID1B gene. Back-splicing occurs at the end of exon 6 and exon 7 to allow these two exons to form the closed-loop structure. B RT-qPCR assay was performed to detect the expression of hsa_circ_0004381 and ARID1B mRNA in SK-N-SH cells treated with or without RNase R. C RT-qPCR analysis of hsa_circ_0004381 and ARID1B mRNA after treatment with Actinomycin D at the indicated time points in SK-N-SH cells. D RT-qPCR analysis of hsa_circ_0004381 expression in the serum samples from 80 PD patients and 60 healthy volunteers. E hsa_circ_0004381 level was detected in SK-N-SH cells treated with various concentrations of MPP+ (0, 0.25, 0.5, 1, and 2 mM) for 24 h. F hsa_circ_0004381 level was assessed in SK-N-SH cells stimulated with 1 mM MPP.+ for 0, 6, 12, 24, and 48 h. *P < 0.05
Hsa_circ_0004381 Could Repress Cell Viability, and Boost Apoptosis, Inflammatory Response, and Oxidative Stress in MPP. +-Treated SK-N-SH Cells
Then, to identify the function of hsa_circ_0004381 in MPP+-induced PD model, we knocked down or overexpressed hsa_circ_0004381 in SK-N-SH cells. As shown in Fig. 2A, the introduction of si-hsa_circ_0004381#1 or si-hsa_circ_0004381#2 could significantly reduce the expression level of hsa_circ_0004381 in comparison with the control group. Especially in si-hsa_circ_0004381#1-transfected cells, so it was chosen for the following assays. Also, the overexpression efficiency of pCD-hsa_circ_0004381 (hsa_circ_0004381) was detected and presented in Fig. 2B. However, the introduction of si-hsa_circ_0004381 or pCD-hsa_circ_0004381 had no significant effect on the expression of ARID1B mRNA in SK-N-SH cells. After MPP+ treatment, we found that the silencing of hsa_circ_0004381 could weaken MPP+-triggered increase in hsa_circ_0004381 level in SK-N-SH cells; inversely, the upregulation of hsa_circ_0004381 could intensify MPP+-induced enhancement in hsa_circ_0004381 level (Fig. 2C). After that, CCK-8 assay displayed that declined cell viability on account of MPP+ treatment was obviously reinforced by hsa_circ_0004381 deficiency or apparently attenuated by upregulation in SK-N-SH cells (Fig. 2D). Apart from that, the results from flow cytometry assay suggested that the treatment of MPP+ could markedly improve the apoptosis rate of SK-N-SH cells, which was mitigated by the transfection of si-hsa_circ_0004381 or aggravated by hsa_circ_0004381 overexpression (Fig. 2E). Synchronously, in order to further verify the effect of hsa_circ_0004381 on apoptosis rate in MPP+-treated SK-N-SH cells, Bcl-2 (an anti-apoptosis factor) and Bax and c-caspase 3 (pro-anti-apoptosis factors) were determined by western blot assay. As displayed in Fig. 2F, MPP+-induced enhancement in Bax and c-caspase 3 protein levels were blocked via hsa_circ_0004381 knockdown or was aggravated after pcDNA3.1-hsa_circ_0004381 in SK-N-SH cells. Also, anti-apoptosis protein Bcl-2 presented an opposite trend in SK-N-SH cells. In terms of the inflammatory response, secretions of pro-inflammatory cytokines (TNF-α, IL-1β, and IL-6) were highly induced due to the treatment of MPP+, while the downregulation of hsa_circ_0004381 abated these effects and the forced expression of hsa_circ_0004381 intensified these effects in SK-N-SH cells (Fig. 2G–I). Besides, our data also exhibited that MPP+-induced oxidative stress was weakened by hsa_circ_0004381 silencing, as evidenced by decreased LDH level and ROS generation, and increased SOD activity (Fig. 2J–L). Contrarily, MPP+-triggered oxidative stress was aggravated through hsa_circ_0004381 overexpression, as described by higher LDH level and ROS generation and lower SOD activity (Fig. 2J–L). Together, these results suggested that hsa_circ_0004381 could exacerbate MPP+-caused neuronal cell injury in vitro.
The effects of hsa_circ_0004381 knockdown on cell viability, apoptosis, inflammatory response, and oxidative stress in MPP+-treated SK-N-SH cells. A RT-qPCR assay was used to determine the expression level of hsa_circ_0004381 and ARID1B mRNA in SK-N-SH cells transfected with si-NC, si-hsa_circ_0004381#1, or si-hsa_circ_0004381#2. B hsa_circ_0004381 level and ARID1B mRNA level were detected in pcDNA3.1- or hsa_circ_0004381-transfected SK-N-SH cells by RT-qPCR assay. C–L SK-N-SH cells were treated with control, MPP+, MPP+ + si-NC, MPP+ + si-hsa_circ_0004381#1, MPP+ + pcDNA3.1, and MPP.+ + hsa_circ_0004381. C hsa_circ_0004381 level was determined in treated SK-N-SH cells by RT-qPCR assay. D CCK-8 analysis of cell viability in treated SK-N-SH cells. E Flow cytometry analysis of apoptosis rate in treated SK-N-SH cells. F Protein levels of Bcl-2, Bax, and c-caspase 3 in treated SK-N-SH cells. G–I TNF-α, IL-1β, and IL-6 were detected in treated SK-N-SH cells by enzyme-linked immunosorbent assay (ELISA). J–L The special assay kits were adopted to detect the products of ROS, LDH, and SOD in treated SK-N-SH cells. *P < 0.05
Hsa_circ_0004381 Directly Bound miR-185-5p
To further understand the potential mechanism by which hsa_circ_0004381 functioned, we investigated the putative hsa_circ_0004381-interacting miRNAs using Starbase, circbank, and circAtlas database. As shown in Figure S1, eight potential target miRNAs (miR-134-5p, miR-3612, miR-185-5p, miR-4640-5p, miR-296-5p, miR-149-5p, miR-3150-3p, miR-5691) of hsa_circ_0004381 were predicted. Then, the Bio-hsa_circ_0004381 probe or Bio-NC probe were designed and used to conduct RNA pull-down assay. Data exhibited that miR-185-5p could be abundantly pulled down by the hsa_circ_0004381 probe in SK-N-SH cells among the eight candidate miRNAs (Figure S1). Data suggested that miR-185-5p possessed some binding sites with hsa_circ_0004381 (Fig. 3A). To verify whether the interaction of miR-185-5p with hsa_circ_0004381 was mediated by the putative binding sites, we conducted a dual-luciferase reporter assay in HEK293T cells. In the first, miR-185-5p mimic (miR-185-5p) was synthesized and introduced into HEK293T cells, followed by the measurement of transfection efficiency (Fig. 3B). Then, a dual-luciferase reporter assay displayed that miR-185-5p upregulation could efficiently decline the luciferase activity of hsa_circ_0004381 WT1 and hsa_circ_0004381 WT2 reporter vector but not that of hsa_circ_0004381 MUT1 and hsa_circ_0004381 MUT2 (Fig. 3C). Whereafter, to further validate the mutual effect of hsa_circ_0004381 and miR-185-5p at the endogenous level, RIP assay was performed using Ago2 which is the core component of the RNA-induced silencing complex (RISC). As exhibited in Fig. 3D, hsa_circ_0004381 and miR-185-5p were both significantly enriched in Ago2 pellets of SK-N-SH cell extracts when compared with the IgG control group. Furthermore, RT-qPCR assay displayed that elevated hsa_circ_0004381 could distinctly hinder the expression level of miR-185-5p in SK-N-SH cells (Fig. 3E); on the contrary, the knockdown of hsa_circ_0004381 could greatly enhance miR-185-5p expression (Fig. 3F). Interestingly, we found that the expression level of miR-185-5p was significantly decreased in a dose-dependent and time-dependent manner (Fig. 3G and H), suggesting the involvement of miR-185-5p in neuronal cell damage in PD. In addition, negatively correlated with hsa_circ_0004381 expression, miR-185-5p was downregulated in the serum samples from PD patients (Fig. 3I and J). Overall, these results indicated that hsa_circ_0004381 regulated the abundance of miR-185-5p via binding to miR-185-5p.
hsa_circ_0004381 directly bound miR-185-5p. A The binding sequence between hsa_circ_0004381 and miR-185-5p and the sequence of hsa_circ_0004381 MUT. B RT-qPCR analysis for miR-185-5p expression in miR-NC- or miR-185-5p-transfected HEK293T cells. C A dual-luciferase reporter assay was used to verify the binding between hsa_circ_0004381 and miR-185-5p in HEK293T cells. D RIP assay was carried out in SK-N-SH cell extracts to examine miR-185-5p endogenously associated with hsa_circ_0004381. E RT-qPCR analysis for miR-185-5p level in pCD-ciR or hsa_circ_0004381-transfected SK-N-SH cells. F miR-185-5p level was detected in SK-N-SH cells transfected with si-NC, si-hsa_circ_0004381#1, or si-hsa_circ_0004381#2 using RT-qPCR assay. G RT-qPCR analysis for miR-185-5p expression in SK-N-SH cells treated with 0, 0.25, 0.5, 1, and 2 mM of MPP+ for 24 h. H miR-185-5p level was determined in SK-N-SH cells treated with 1 mM MPP.+ for 0, 6, 12, 24, and 48 h by RT-qPCR assay. I miR-185-5p expression in the serum samples from 80 PD patients and 60 healthy volunteers were assessed using RT-qPCR assay. J The expression association between miR-185-5p and hsa_circ_0004381 in PD patients’ serum samples was analyzed by Pearson correlation analysis. *P < 0.05
Hsa_circ_0004381 Knockdown Could Attenuate MPP.+-Induced Neuronal Cell Damage by Targeting miR-185-5p
Given the regulatory role of hsa_circ_0004381 in miR-185-5p expression in SK-N-SH cells, we further explored that the influence of hsa_circ_0004381 on MPP+-triggered cell injury was correlative with miR-185-5p. First of all, the knockdown efficiency of miR-185-5p inhibitor was measured and displayed in Fig. 4A. Then, RT-qPCR assay showed that the re-introduction of anti-miR-185-5p could partly counteract hsa_circ_0004381 deficiency-caused improvement in miR-185-5p level in MPP+-treated SK-N-SH cells (Fig. 4B). Functional analysis discovered that the promotion of cell viability caused by hsa_circ_0004381 silencing was significantly overturned by miR-185-5p downregulation in MPP+-treated SK-N-SH cells (Fig. 4C). Simultaneously, flow cytometry assay presented that miR-185-5p inhibitor could remarkably abrogate the suppressive effect of si-hsa_circ_0004381 on apoptosis rate in MPP+-treated SK-N-SH cells (Fig. 4D and E), as depicted by lower Bcl-2 and higher Bax, and c-caspase 3 (Fig. 4F–I). Moreover, hsa_circ_0004381 knockdown-mediated decline in the inflammatory response in MPP+-treated SK-N-SH cells was alleviated by miR-185-5p downregulation, as described by augmented secretions of pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 (Fig. 4J–L). In addition, the re-introduction of anti-miR-185-5p also could reverse the negative action of hsa_circ_0004381 silencing on oxidative stress, accompanied by ROS and LDH increase, and SOD decrease (Fig. 4M–O). Collectively, these results suggested that hsa_circ_0004381 could regulate MPP + -triggered neuronal cell injury by interacting with miR-185-5p in vitro.
Hsa_circ_0004381-mediated cell viability, apoptosis, inflammatory response, and oxidative stress were modulated by miR-185-5p. A RT-qPCR analysis for miR-185-5p expression in anti-miR-NC- or anti-miR-185-5p-transfected SK-N-SH cells. B–O SK-N-SH cells were treated with MPP+ + si-NC, MPP+ + si-hsa_circ_0004381#1, MPP+ + si-hsa_circ_0004381#1 + anti-miR-NC, and MPP.+ + si-hsa_circ_0004381#1 + anti-miR-185-5p. B miR-185-5p level was determined in treated SK-N-SH cells by RT-qPCR assay. C–E CCK-8 assay and flow cytometry assay were applied to assess cell viability and apoptosis rate in treated SK-N-SH cells. F–I Western blot assay was used to measure the protein levels of Bcl-2, Bax, and c-caspase 3 in treated SK-N-SH cells. J–L ELISA was performed to detect the levels of TNF-α, IL-1β, and IL-6 in treated SK-N-SH cells. M–O The products of ROS, LDH, and SOD in treated SK-N-SH cells were analyzed in treated SK-N-SH cells. *P < 0.05
RAC1 Was a Direct and Functional Target of miR-185-5p
Then, to further identify the mechanism of action of miR-185-5p, we search the potential target genes of miR-185-5p using the online software Starbase. As a result, we found hundreds of potential target mRNAs of miR-185-5p from Starbase. Then, we selected 8 mRNAs associated with the process of Parkinson’s disease, which was subjected to RT-qPCR analysis responding to miR-185-5p upregulation. As presented in Figure S1C, RAC1 showed the highest fold change. Hence, we selected RAC1 for further research. Also, the bioinformatics analysis suggested RAC1 as one candidate with the predicated binding sites on miR-185-5p (Fig. 5A). Subsequently, a dual-luciferase reporter assay exhibited that the luciferase activity in HEK293T cells transfected with RAC1-3′UTR WT and miR-185-5p was evidently reduced versus that in cells with RAC1-3′UTR WT and miR-NC, whereas there was no impact in the cells transfected with RAC1-3′UTR MUT (Fig. 5B). In order to determine the authentic effect of miR-185-5p on RAC1 expression, the overexpression efficiency of miR-185-5p mimic was assessed and presented in Fig. 5C. Also, western blot assay displayed that the upregulation of miR-185-5p could repress the protein level of RAC1 in SK-N-SH cells, while the downregulation of miR-185-5p could elevate RAC1 expression (Fig. 5D and E). Furthermore, we observed that RAC1 level was increased in MPP+-stimulated SK-N-SH cells in a certain concentration- and time-dependent manner, implying a latent role of RAC1 in neuronal cell damage in PD (Fig. 5F and G). Meanwhile, RT-qPCR analysis suggested that RAC1 was expressed at a high level in the serum samples of PD patients (Fig. 5H), and was negatively correlated to miR-185-5p expression (Fig. 5I). However, a consistent expression pattern of RAC1 and hsa_circ_0004381 was analyzed in the serum samples from PD patients (Fig. 5J). Additionally, western blot analysis discovered that the silencing of hsa_circ_0004381 could dampen the protein level of RAC1 in MPP+-treated SK-N-SH cells, which was partly counteracted by miR-185-5p downregulation (Fig. 5K), indicating that hsa_circ_0004381 could act as a sponge of miR-185-5p to affect RAC1 expression in MPP+-treated SK-N-SH cells. All in all, these observations indicate that RAC1 served as a direct target gene of miR-185-5p.
miR-185-5p could suppress RAC1 expression. A Schematic of a putative target sequence for miR-185-5p in RAC1 and mutated miR-185-5p-binding sites. B The binding was proved by a dual-luciferase reporter assay in HEK293T cells. C miR-185-5p level was detected in miR-NC- or miR-185-5p-transfected SK-N-SH cells by RT-qPCR assay. D and E The effects of miR-185-5p upregulation or downregulation on RAC1 protein level in SK-N-SH cells by western blot assay. F and G Western blot measured the expression level of RAC1 after 0–2-mM MPP+ treatment for 24 h, and 1-mM MPP+ treatment for 0–48 h. H RAC1 level was detected in the serum samples from 40 PD patients and 40 healthy volunteers by RT-qPCR assay. I and J Pearson correlation analysis was used to evaluate the expression correlation of RAC1 with miR-142 and hsa_circ_0004381 in serum samples from PD patients. K RAC1 protein level was detected in SK-N-SH cells treated with MPP+ + si-NC, MPP+ + si-hsa_circ_0004381#1, MPP+ + si-hsa_circ_0004381#1 + anti-miR-NC, and MPP.+ + si-hsa_circ_0004381#1 + anti-miR-185-5p. *P < 0.05
miR-185-5p Could Abolish MPP.+-Caused Neuronal Cell Injury by Modulating RAC1
To provide further mechanistic insight into the link between miR-185-5p and RAC1 on MPP+-induced cell injury, we conducted the rescue assays in SK-N-SH cells. We first assessed the transfection efficiency of pcDNA3.1-RAC1 (RAC1) in SK-N-SH cells (Fig. 6A). Whereafter, western blot assay suggested that miR-185-5p elicited an obvious decrease in RAC1 level in MPP+-treated SK-N-SH cells, while the co-transfection of RAC1 could partially counteract these effects (Fig. 6B). Functionally, increased cell viability due to the upregulation of miR-185-5p was obviously overturned by the overexpression of RAC1 in MPP+-treated SK-N-SH cells (Fig. 6C). Synchronously, the repression of apoptosis rate caused by miR-185-5p was partly abrogated by RAC1 upregulation in MPP+-treated SK-N-SH cells (Fig. 6D and E), as described by reduced Bcl-2, and improved Bax and c-caspase 3 (Fig. 6F). Apart from that, transfection of miR-185-5p led to a substantial decline in TNF-α, IL-1β, and IL-6 levels in MPP+-treated SK-N-SH cells, which was apparently abolished by RAC1 overexpression, indicating that the inhibitory role of miR-185-5p on inflammatory response was reversed by RAC1 introduction (Fig. 6G–I). Besides, oxidative stress was hindered by miR-185-5p in MPP+-treated SK-N-SH cells and then was partly relieved by RAC1 overexpression, accompanied by higher ROS and LDH, and lower SOD (Fig. 6J–L). All of these data suggested that miR-185-5p could mitigate MPP+-triggered neuronal cell injury by targeting RAC1.
miR-185-5p could boost cell viability, and block apoptosis, inflammatory response, and oxidative stress by regulating RAC1 in MPP+-treated SK-N-SH cells. A Western blot analysis of RAC1 expression in pcDNA3.1- or RAC1-transfected SK-N-SH cells. B–J SK-N-SH cells were treated with control, MPP+, MPP+ + miR-NC, MPP+ + miR-185-5p, MPP+ + miR-185-5p + pcDNA3.1, and MPP.+ + miR-185-5p + RAC1. B RAC1 protein level was detected in treated SK-N-SH cells by western blot assay. C–E Cell viability and apoptosis rate were evaluated by CCK-8 assay and flow cytometry assay in treated SK-N-SH cells. F Protein levels of Bcl-2, Bax, and c-caspase 3 of treated SK-N-SH cells were tested by western blot assay. G–I TNF-α, IL-1β, and IL-6 levels of treated SK-N-SH cells were measured by ELISA. J–L The analysis of ROS, LDH, and SOD products was carried out using the corresponding kits in treated SK-N-SH cells. *P < 0.05
Discussion
Accumulative evidence has indicated that circRNAs play important regulators in PD-related pathological processes, offering potential attractive biomarkers for PD diagnosis and prognosis (Jia et al. 2020; Ravanidis and Bougea 2021). Nevertheless, the exercise mechanism behind circRNAs implicating PD progression is still lacking in-depth research. In recent years, the exposure of dopamine neurons to MPP+ has been described to selectively kill dopaminergic neurons in vivo and in vitro through diverse toxic mechanisms (Choi et al. 2015; González-Polo et al. 2001). It has been widely reported that MPP+ was generally used to induce a cellular model of PD in vitro (Prommahom and Dharmasaroja 2021; Xie et al. 2016). The present work displayed that hsa_circ_0004381, a typical circRNA origination from exons 6 and 7 of the host gene ARID1B, has been documented to be abnormally highly expressed in PD patients using circRNA microarray, coherently with the former study (Zhong et al. 2021). Furthermore, our data are the first to verify the upregulation of hsa_circ_0004381 in the MPP+-challenged PD cell model, suggesting that the dysregulation of hsa_circ_0004381 might take part in neuronal cell injury in PD.
Recent studies have manifested that neuron dysfunction was recognized as an important contributor to the pathogenesis of PD (Lev et al. 2003; Michel et al. 2016). The current study suggested that the deficiency of hsa_circ_0004381 could overturn MPP+-triggered decrease in cell viability and increase in cell apoptosis in SK-N-SH cells. Simultaneously, the defining hallmarks of PD are inflammatory damage and excessive generation of oxidative stress (Trist et al. 2019; Puspita et al. 2017; Gelders and Baekelandt 2018). In this paper, MPP+-induced inflammatory response and oxidative stress could be relieved by hsa_circ_0004381 downregulation in SK-N-SH cells. Therefore, these findings suggested that hsa_circ_0004381 is an ideal candidate or a promising breakthrough for insight into the molecular mechanisms of PD.
Previous literature has discovered that circRNAs mainly function in human disease by targeting corresponding miRNAs to increase mRNA expression (Han et al. 2018; Hansen et al. 2013). In this research, miR-185-5p is of particular interest in the paper, considered a neuroprotective regulator in the 6-hydroxydopamine-induced PD cell model in vitro (Qin et al. 2021). Apart from that, the upregulation of miR-185-5p was confirmed to suppress autophagy and apoptosis of dopaminergic cells in PD (Wen et al. 2018). Our findings highlighted that hsa_circ_0004381 directly interacted with miR-185-5p to suppress its expression for the first time. Moreover, silencing of hsa_circ_0004381 attenuated MPP+-caused SK-N-SH cell damage through targeting miR-185-5p. Canonically, miRNAs could exert the functional role via regulating their target genes. Analogously, we are the first to identify that RAC1 was a downstream target of miR-185-5p in this study. RAC1 belongs to the small G protein Rho subfamily that plays a central role in skin homeostasis, containing barrier function and inflammatory responses (Winge and Marinkovich 2019). Meanwhile, the dysregulation of RAC1 was correlated with apoptosis pathways through ROS production (Bhat et al. 2016). Also, it has been proven that the downregulation of RAC1 could prevent apoptosis in cell models of MPP+-challenged PD (Lu et al. 2020). Hence, RAC1 was chosen for an in-depth investigation in this chapter. The current work indicated that miR-185-5p repressed MPP+-triggered cell injury by regulating RAC1. In our present study, we demonstrated the hsa_circ_0004381/miR-185-5p/RAC1 axis in the MPP+-induced PD cell model. Frankly speaking, there are still some shortcomings in this study. For example, the circRNA/miRNA/mRNA regulatory networks are intricate, and a circRNA might serve as an inhibitor of multiple miRNAs. Thus, there might be alternative miRNA/mRNA axes that remain to be discovered in the regulation of hsa_circ_0004381 in PD. In addition, we did not perform animal experiments, and more clinical assays need to be conducted in the future.
Conclusion
In summary, our data discovered that hsa_circ_0004381 could act as a sponge of miR-185-5p to increase RAC1 expression, thereby promoting the MPP+-induced neuronal injury in the PD cell model (Fig. 7). These findings may provide an available preclinical basis for PD treatment.
Availability of Data and Materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
References
Ashwal-Fluss R, Meyer M, Pamudurti NR et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Barrett SP, Salzman J (2016) Circular RNAs: analysis, expression and potential functions. Development 143(11):1838–1847. https://doi.org/10.1242/dev.128074
Bhat SS, Parray AA, Mushtaq U et al (2016) Actin depolymerization mediated loss of SNTA1 phosphorylation and Rac1 activity has implications on ROS production, cell migration and apoptosis. Apoptosis 21(6):737–748. https://doi.org/10.1007/s10495-016-1241-6
Choi SJ, Panhelainen A, Schmitz Y et al (2015) Changes in neuronal dopamine homeostasis following 1-methyl-4-phenylpyridinium (MPP+) exposure. J Biol Chem 290(11):6799–6809. https://doi.org/10.1074/jbc.M114.631556
Ding H, Huang Z, Chen M et al (2016) Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Parkinsonism Relat Disord 22(68–73). https://doi.org/10.1016/j.parkreldis.2015.11.014
Feng Z, Zhang L, Wang S et al (2020) Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun 522(2):388–394. https://doi.org/10.1016/j.bbrc.2019.11.102
Gelders G, Baekelandt V, Van der Perren A (2018) Linking neuroinflammation and neurodegeneration in Parkinson’s disease. 4784268. https://doi.org/10.1155/2018/4784268
González-Polo RA, Mora A, Clemente N et al (2001) Mechanisms of MPP(+) incorporation into cerebellar granule cells. Brain Res Bull 56(2):119–123. https://doi.org/10.1016/s0361-9230(01)00610-4
Han B, Chao J, Yao H (2018) Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 187(31–44). https://doi.org/10.1016/j.pharmthera.2018.01.010
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388. https://doi.org/10.1038/nature11993
Jia E, Zhou Y, Liu Z et al (2020) Transcriptomic profiling of circular RNA in different brain regions of Parkinson’s disease in a mouse model. 21(8). https://doi.org/10.3390/ijms21083006
Küçükdoğru R, Türkez H, Arslan ME et al (2020) Neuroprotective effects of boron nitride nanoparticles in the experimental Parkinson’s disease model against MPP+ induced apoptosis. Metab Brain Dis 35(6):947–957. https://doi.org/10.1007/s11011-020-00559-6
Lev N, Melamed E, Offen D (2003) Apoptosis and Parkinson’s disease. Prog Neuropsychopharmacol Biol Psychiatry 27(2):245–250. https://doi.org/10.1016/s0278-5846(03)00019-8
Lu W, Lin J, Zheng D et al (2020) Overexpression of microRNA-133a inhibits apoptosis and autophagy in a cell model of Parkinson’s disease by downregulating Ras-related C3 botulinum toxin substrate 1 (RAC1). Med Sci Monit 26(e922032). https://doi.org/10.12659/msm.922032
Ma CL, Su L, Xie JJ et al (2014) The prevalence and incidence of Parkinson’s disease in China: a systematic review and meta-analysis. J Neural Transm (vienna) 121(2):123–134. https://doi.org/10.1007/s00702-013-1092-z
Mehta SL, Dempsey RJ, Vemuganti R (2020) Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol 186(101746). https://doi.org/10.1016/j.pneurobio.2020.101746
Memczak S, Jens M, Elefsinioti A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338. https://doi.org/10.1038/nature11928
Michel PP, Hirsch EC, Hunot S (2016) Understanding dopaminergic cell death pathways in Parkinson disease. Neuron 90(4):675–691. https://doi.org/10.1016/j.neuron.2016.03.038
Panda AC (2018) Circular RNAs act as miRNA sponges. Adv Exp Med Biol 1087(67–79). https://doi.org/10.1007/978-981-13-1426-1_6
Panni S, Lovering RC, Porras P et al (2020) Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech 1863(6):194417. https://doi.org/10.1016/j.bbagrm.2019.194417
Patop IL, Wüst S, Kadener S (2019) Past, present, and future of circRNAs. 38(16):e100836. https://doi.org/10.15252/embj.2018100836
Pringsheim T, Jette N, Frolkis A et al (2014) The prevalence of Parkinson’s disease: a systematic review and meta-analysis. Mov Disord 29(13):1583–1590. https://doi.org/10.1002/mds.25945
Prommahom A, Dharmasaroja P (2021) Effects of eEF1A2 knockdown on autophagy in an MPP(+)-induced cellular model of Parkinson’s disease. Neurosci Res 164(55–69). https://doi.org/10.1016/j.neures.2020.03.013
Puspita L, Chung SY, Shim JW (2017) Oxidative Stress and Cellular Pathologies in Parkinson’s Disease 10(1):53. https://doi.org/10.1186/s13041-017-0340-9
Qin X, Zhang X, Li P et al (2021) MicroRNA-185 activates PI3K/AKT signalling pathway to alleviate dopaminergic neuron damage via targeting IGF1 in Parkinson’s disease. J Drug Target 1–9. https://doi.org/10.1080/1061186x.2021.1886300
Ravanidis S, Bougea A (2021) Differentially Expressed Circular RNAs in Peripheral Blood Mononuclear Cells of Patients with Parkinson’s Disease 36(5):1170–1179. https://doi.org/10.1002/mds.28467
Raza C, Anjum R, Shakeel NUA (2019) Parkinson’s disease: mechanisms, translational models and management strategies. Life Sci 226(77–90). https://doi.org/10.1016/j.lfs.2019.03.057
Reich SG, Savitt JM (2019) Parkinson’s disease. Med Clin North Am 103(2):337–350. https://doi.org/10.1016/j.mcna.2018.10.014
Rybak-Wolf A, Stottmeister C, Glažar P et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870–885. https://doi.org/10.1016/j.molcel.2015.03.027
Salzman J (2016) Circular RNA expression: its potential regulation and function. Trends Genet 32(5):309–316. https://doi.org/10.1016/j.tig.2016.03.002
Sveinbjornsdottir S (2016) The clinical symptoms of Parkinson’s disease. J Neurochem 139 Suppl 1(318–324). https://doi.org/10.1111/jnc.13691
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17(5):272–283. https://doi.org/10.1038/nrg.2016.20
Trist BG, Hare DJ, Double KL (2019) Oxidative Stress in the Aging Substantia Nigra and the Etiology of Parkinson’s Disease 18(6):e13031. https://doi.org/10.1111/acel.13031
Wang W, Lv R, Zhang J et al (2021) circSAMD4A participates in the apoptosis and autophagy of dopaminergic neurons via the miR‑29c‑3p‑mediated AMPK/mTOR pathway in Parkinson’s disease. Mol Med Rep 24(1). https://doi.org/10.3892/mmr.2021.12179
Wen Z, Zhang J, Tang P et al (2018) Overexpression of miR-185 inhibits autophagy and apoptosis of dopaminergic neurons by regulating the AMPK/mTOR signaling pathway in Parkinson’s disease. Mol Med Rep 17(1):131–137. https://doi.org/10.3892/mmr.2017.7897
Winge MCG, Marinkovich MP (2019) Epidermal activation of the small GTPase Rac1 in psoriasis pathogenesis. Small GTPases 10(3):163–168. https://doi.org/10.1080/21541248.2016.1273861
Xie H, Hu H, Chang M et al (2016) Identification of chaperones in a MPP(+)-induced and ATRA/TPA-differentiated SH-SY5Y cell PD model. Am J Transl Res 8(12):5659–5671
Zhang Z, Yang T, Xiao J (2018) Circular RNAs: promising biomarkers for human diseases. EBioMedicine 34(267–274. https://doi.org/10.1016/j.ebiom.2018.07.036
Zhong L, Ju K, Chen A et al (2021) Circulating CircRNAs panel acts as a biomarker for the early diagnosis and severity of Parkinson’s disease. Front Aging Neurosci 13(684289). https://doi.org/10.3389/fnagi.2021.684289
Zhu J, Wang S, Liang Y et al (2018) Inhibition of microRNA-505 suppressed MPP+ -induced cytotoxicity of SHSY5Y cells in an in vitro Parkinson’s disease model. Eur J Pharmacol 835(11–18). https://doi.org/10.1016/j.ejphar.2018.07.023
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of The First Affiliated Hospital of Jinzhou Medical University. Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
1. Hsa_circ_0004381 knockdown suppressed MPP+-induced neuronal cell damage.
2. Hsa_circ_0004381 directly bound with miR-185-5p.
3. RAC1 was a target of miR-185-5p.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, H., Wang, C. & Zhang, X. Circular RNA hsa_circ_0004381 Promotes Neuronal Injury in Parkinson’s Disease Cell Model by miR-185-5p/RAC1 Axis. Neurotox Res 40, 1007–1019 (2022). https://doi.org/10.1007/s12640-022-00525-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-022-00525-3