Abstract
Parkinson’s disease (PD) is a multi-factorial neurodegenerative disease. Long noncoding RNAs (lncRNAs) have been revealed to be involved in the process of PD. Herein, this study aimed to investigate the potential function and mechanism of JHDM1D-AS1 (JHDM1D antisense 1) in PD process. 1-Methyl-4-phenylpyridinium (MPP +)–induced SK-N-SH cells were used to conduct expression and function analyses. Levels of genes and proteins were examined using real-time reverse transcription PCR (RT-qPCR) and Western blot. Cell viability and apoptosis were determined using CCK-8 assay, flow cytometry, and Western blot, respectively. ELISA analysis was performed for the detection of interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α. The contents of lactate dehydrogenase (LDH), superoxide dismutase (SOD), and malondialdehyde (MDA) were measured using commercial kits. The direct interactions between miR-134-5p and PIK3R3 (Phosphoinositide-3-Kinase Regulatory Subunit 3) or JHDM1D-AS1 were verified by dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. JHDM1D-AS1 expression was decreased by MPP + in SK-N-SH cells in a dose- or time-dependent manner. Functionally, JHDM1D-AS1 overexpression attenuated MPP + -evoked neuronal apoptosis, inflammation, and oxidative stress. Mechanistically, JHDM1D-AS1 competitively bound to miR-134-5p to upregulate the expression of its target PIK3R3. Rescue experiments suggested that miR-134-5p upregulation reversed the inhibitory effects of JHDM1D-AS1 on MPP + -induced neuronal injury. Moreover, inhibition of miR-134-5p protected neurons against MPP + -induced neuronal apoptosis, inflammation, and oxidative stress, which were abolished by PIK3R3 silencing. JHDM1D-AS1 protected against MPP + -induced neuron injury via miR-134-5p/PIK3R3 axis, suggesting the potential involvement of this axis in PD process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parkinson’s disease (PD) is a multi-factorial neurodegenerative disease attributing to the progressive loss of dopaminergic neurons, which leads to tremor, rigidity, and bradykinesia, thus reducing the quality of life (Kalia and Lang 2015; Teive et al. 2016). PD can affect 1–2 per 1000 of the population (Tysnes and Storstein 2017); currently, there are no biomarkers, laboratory tests, or imaging studies to confirm this disease; moreover, current treatment options are largely limited to control the symptoms (Marino et al. 2020). The pathology of PD involves in a wide range of regions of the nervous system, diverse protein aggregates, and neurotransmitters; importantly, there is increasing evidence suggesting that various fundamental cellular processes, including cell apoptosis, inflammation, and oxidative stress, are responsible for PD progression (Abou-Sleiman et al. 2006; Michel et al. 2016; Zhou et al. 2008). Therefore, further investigations underlying the pathology of PD are required for the development of novel therapeutic strategies for PD patients.
Long noncoding RNAs (lncRNAs) are transcripts with a length of approximately 200 nucleotides that have no protein-coding potential (Wang et al. 2011). They have emerged as important regulators in a variety of physiological and pathological processes (Heward and Lindsay 2014; Maass et al. 2014; Quinn and Chang 2016). Accumulating studies indicate that lncRNAs are aberrantly expressed in the blood or tissues of patients with different diseases, including cancers, cardiovascular diseases, and neurodegenerative diseases (Bhan et al. 2017; Haemmig et al. 2017; Wu et al. 2013). In PD, some lncRNAs have also been revealed to involve in the process of this disease. For example, lncRNA MALAT1 induced neuroinflammation and ROS production by recruiting NRF2 in vitro and in PD mouse (Cai et al. 2020). LncRNA H19 was demonstrated to suppress neuronal apoptosis in PD (Zhang et al. 2020). Therefore, targeting lncRNA in PD will be an attractive treatment strategy. JHDM1D-AS1 (JHDM1D antisense 1) is a nutrient starvation-responsive lncRNA. Recently, Liu et al. showed that JHDM1D-AS1 had a prominent neuroprotective effect in spinal cord after brachial plexus injury by alleviating neuronal apoptosis and neuroinflammation (Liu et al. 2020). Moreover, JHDM1D-AS1 was found to be decreased in PD patients (Zhou et al. 2018). Here, we speculated that dysregulation of JHDM1D-AS1 expression might be also involved in the progression of PD.
1-Methyl-4-phenylpyridinium (MPP +) is the active in vivo metabolite of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), which can cause PD-like symptoms by inducing selective degeneration of nigrostriatal dopaminergic (DA) neurons (Risiglione et al. 2020). Due to the difficulty of the acquisition and maintainability of dopaminergic neurons, the neuroblastoma cell line has been widely used as a model for PD (Krishna et al. 2014). Hence, this study used MPP + -stimulated SK-N-SH cells to mimic PD environment in vitro to investigate the potential functions and mechanisms underlying JHDM1D-AS1 in PD, with the goal of suggesting a new therapeutic target for PD treatment.
Materials and Methods
Cell Culture and Treatment
Human SK-N-SH cells were commercially obtained from Cedarlane (Burlington, NC, USA), and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Los Angeles, CA, USA) containing 10% fetal calf serum (Gibco) and 1% penicillin/streptomycin (Gibco) at 37 °C with 5% CO2. Cells from passages 3 to 7 were used for functional experiment.
To mimic the pathological feature of PD in vitro, SK-N-SH cells were challenged with increasing doses (0, 1, 2, 3, or 4 mM) of MPP + (Sigma-Aldrich, St. Louis, MO, USA) for 24 h or exposed to 2 mM MPP + for different times (0 h, 12 h, 24 h, 36 h, or 48 h). Treatment of 2 mM MPP + for 24 h was finally chosen to trigger PD-like neuronal injury in subsequent functional analyses.
Real-time Reverse Transcription PCR (RT-qPCR)
Trizol reagent (Invitrogen, Carlsbad, CA, USA) was applied for the isolation of total RNAs. The generation of complementary DNA (cDNA) was conducted using the PrimeScript RT Reagent Kit (TaKaRa, Otsu, Japan) or PrimeScript miRNA cDNA Synthesis Kit (TaKaRa), and then qPCR was carried out using the SYBR Green (TaKaRa). The relative molecular expression, normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6, was detected by the 2−△△Ct method. The same experiment was repeated three times, and the average was taken. The primer sequences were shown:
JHDM1D-AS1: F 5′-GAAGCGGAGAAGTTTGCTGC-3′, R 5′-AGTTTGGATTCCACGGCACA-3′;
miR-134-5p: F 5′-GCCGAGTGTGACTGGTTGACCA-3′, R 5′-CAGTGCAGGGTCCGAGGTAT-3′;
PIK3R3: F 5′-CTGGAGGGAGGTGATGATGC-3′, R 5′-GTTGAGGCATCTCGGACCAA-3′;
GAPDH: F 5′-CTGGGCTACACTGAGCACC-3′, R 5′- AAGTGGTCGTTGAGGGCAATG-3′;
U6: F 5′-CTCGCTTCGGCAGCACA-3′, R 5′-AACGCTTCACGAATTTGCGT-3′.
Cell Transfection
The pcDNA3.1-JHDM1D-AS1 overexpression vector (JHDM1D-AS1) with empty pcDNA3.1 vector (vector), PIK3R3 specific siRNA (si-PIK3R3, 5′-GGACUU GCUUUAUGGGAAA dTdT-3′) with the nontarget siRNA (si-NC, 5′-AAUUCUC CGAACGUGUCACGU-3′), miR-134-5p mimic (5′- UGUGACUGGUUGACCAGAGGGG-3′), miR-134-5p inhibitor (5′-ACACUGACCAACUGGUCUCCCC-3′), or corresponding control (miRNA NC or inhibitor NC, 5′-AAUCUUGGCCUAACUACUGGCU-3′) were designed and synthesized by Genechem (Shanghai, China). Then, transient transfection in SK-N-SH cells with 40 nM of miRNA mimic, inhibitor, negative controls, or 100 nM of si-PIK3R3, si-NC, or 100 nM of JHDM1D-AS1 or vector was implemented employing Lipofectamine 2000 provided by Invitrogen. After confirmation of the transfection efficiencies, cells were subjected to 2 mM MPP + for 24 h for subsequent analyses.
Cell Counting Kit-8 (CCK-8) Assay
SK-N-SH cells (5000/well) were placed at each well of 96-well plates overnight; then, the optical density (OD) at 450 nm was examined by a spectrophotometric microplate reader after the addition of 10 μL CCK-8 solution (Sigma-Aldrich) in each well for 2 h. All experiments were repeated three times independently.
Flow Cytometry
Briefly, SK-N-SH cells were resuspended in binding buffer to a concentration of 1 × 106/mL, and then incubated with 5 μL FITC annexin V and 10 μL PI (BD Biosciences, Franklin Lakes, NJ, USA). Lastly, the apoptosis rate was measured after 15 min of incubation in the dark using a flow cytometer. Three replicate wells were set in each group, and the experiment was repeated three times.
Western Blot
After isolation of total proteins using RIPA reagent (Beyotime, Beijing, China), protein extracts were separated by 10% polyacrylamide gel and then shifted onto a PVDF membrane (Solarbio, Beijing, China). The membrane was incubated with primary antibodies at 4 °C overnight, and then probed with secondary antibody at room temperature for 2 h. The protein signal was detected using an ECL reagent (Beyotime). The primary antibodies included PCNA (1:5000, ab29, Abcam, Cambridge, MA, USA), Bax (1:1000, ab32503, Abcam), PIK3R3 (Cat #11,889, Cell Signaling Technology, MA, USA), and GAPDH (1:5000, ab181602, Abcam). Experiments were performed three times.
Enzyme-Linked Immunosorbent Assay (ELISA)
The levels of interleukin (IL)-1β, IL-6, and tumor necrosis factor-α (TNF-α) in the culture supernatants of SK-N-SH cells were evaluated by using the commercial the ELISA kits (Solarbio) following the instructions of manufacturer. The same experiment was repeated three times, and the average was taken.
Measurement of Lactate Dehydrogenase (LDH), Malondialdehyde (MDA), or Superoxide Dismutase (SOD)
The contents of LDH, MDA, and SOD were detected using LDH assay kit (Biovision, Wuhan, China), MDA content Assay Kit, or the SOD Assay Kit-WST (Dojindo, Tokyo, Japan) as per the accompanying guidance, respectively. The same experiment was repeated three times, and the average was taken.
Dual-Luciferase Reporter Assay
The 3′UTR of JHDM1D-AS1 or PIK3R3 possessing miR-134-5p binding sites were predicted by using Bioinformatics analysis. Point mutations in binding sites of miR-134-5p were generated by Genema (Shanghai, China). The binding sites were amplified and inserted into the PGL3 Basic vector (Promega, Madison, WI, USA). Next, these constructed reporter plasmids and pRL-TK Renilla vector together miR-134-5p mimic or mimic NC were co-transfected into SK-N-SH cells by using Lipofectamine 2000 (Invitrogen), and the luciferase activity and Renilla activity were determined after 48 h of transfection. Each group was run in triplicate in 24-well plates.
RNA Immunoprecipitation (RIP) Assay
RIP assay was implemented according to the protocol of the EZMagna RIP Kit (Millipore). The lysates of SK-N-SH cells were incubated with magnetic beads conjugated with human Ago2 antibody or negative control IgG. Finally, the precipitated RNAs were purified, and subject to RT-qPCR analysis. Experiments were performed three times.
Statistical Analyses
All data were expressed as mean ± standard deviation. Analysis of variance was applied for multiple group comparisons. Statistical significance was assessed by Student’s t-test for independent two groups or Mann–Whitney U-test for non-parametric independent two-group. P < 0.05 suggested significant differences.
Results
JHDM1D-AS1 Expression Is Decreased in MPP + -Induced SK-N-SH Cells
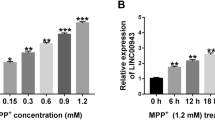
First, the expression profile of JHDM1D-AS1 in MPP + -stimulated SK-N-SH cells was investigated. As shown in Fig. 1A, the expression of JHDM1D-AS1 in SK-N-SH cells was found to be decreased with the increasing doses of MPP + (0, 1, 2, 3, or 4 mM) for 24 h. Moreover, SK-N-SH cells were exposed to 2 mM MPP + for different times (0, 12, 24, 36, or 48 h); it was discovered that JHDM1D-AS1 expression was time-dependently downregulated in SK-N-SH cells (Fig. 1B). Therefore, we demonstrated that JHDM1D-AS1 expression was significantly affected by MPP + treatment in SK-N-SH cells, suggesting the potential involvement of JHDM1D-AS1 in the pathology of PD.
JHDM1D-AS1 expression is decreased in MPP + -induced SK-N-SH cells. A RT-qPCR analysis of JHDM1D-AS1 expression level in SK-N-SH cells 24 h following MPP + exposure (0, 1, 2, 3, or 4 mM). B RT-qPCR analysis of JHDM1D-AS1 expression level in SK-N-SH cells exposed to 2 mM MPP + for 0, 12, 24, 36, or 48 h. *P < 0.05
JHDM1D-AS1 Overexpression Restrains MPP + -Induced Neuronal Apoptosis, Inflammation, and Oxidative Stress
Next, JHDM1D-AS1 was overexpressed in SK-N-SH cells, followed by the treatment of 2 mM MPP + for 24 h to investigate the functions of JHDM1D-AS1 in MPP + -induced neuronal injury. The results of RT-qPCR suggested that JHDM1D-AS1 expression in MPP + -challenged SK-N-SH cells was significantly upregulated by the introduction of JHDM1D-AS1 overexpression vector (Fig. 2A). Functionally, upregulation of JHDM1D-AS1 reduced MPP + -induced inhibition of cell proliferation (Fig. 2B) and enhancement of cell apoptosis (Fig. 2C) in SK-N-SH cells. Moreover, Western blot analysis showed that the protein level of PCNA was increased while Bax was decreased in MPP + -induced SK-N-SH cells after elevation of JHDM1D-AS1 (Fig. 2D, E). Besides that, MPP + led to the elevation of inflammatory cytokines IL-1β, IL-6, and TNF-α in SK-N-SH cells, which was reversed by JHDM1D-AS1 upregulation (Fig. 2F–H). Meanwhile, JHDM1D-AS1 upregulation abolished MPP + -triggered increase of LDH and MDA production, as well as decrease of SOD content in SK-N-SH cells (Fig. 2I–K). Altogether, JHDM1D-AS1 ameliorated MPP + -induced neuronal dysfunction.
JHDM1D-AS1 overexpression restrains MPP + -induced neuronal apoptosis, inflammation, and oxidative stress. A–K SK-N-SH cells were transfected with JHDM1D-AS1 or vector, followed by the treatment of 2 mM MPP + for 24 h. A RT-qPCR analysis of JHDM1D-AS1 expression level in SK-N-SH cells. B CCK-8 for cell viability. C Flow cytometer for cell apoptosis. D, E Western blot analysis of the protein levels of PCNA and Bax. F–H ELISA analysis for IL-1β, IL-6, and TNF-α levels. I–K Measurement of LDH, MDA, and SOD levels in cells using commercial kits. *P < 0.05
MiR-134-5p Is a Target of JHDM1D-AS1
The potential target miRNAs of JHDM1D-AS1 were then predicted by using starBase online tool; 19 miRNAs were found that might be the target of JHDM1D-AS1; among them, miR-134-5p, miR-129-5p, miR-421, miR-9, and miR-101-3p were reported to be related to PD. When we overexpressed JHDM1D-AS1 in SK-N-SH cells, it was found that JHDM1D-AS1 overexpression significantly reduced miR-134-5p expression (Fig. S2). Therefore, miR-134-5p was selected for subsequent analysis. JHDM1D-AS1 contains a putative binding site on miR-134-5p (Fig. 3A). After confirming the elevation efficiency of miR-134-5p mimic (Fig. 3B), it was proved that miR-134-5p mimic overtly reduced the luciferase activity of the wild-type JHDM1D-AS1 vector in SK-N-SH cells, while SK-N-SH cells co-transfected with mutated one with miR-134-5p mimic did not exhibit a significant difference (Fig. 3C). Moreover, RIP assay showed JHDM1D-AS1 and miR-134-5p were significantly pulled down by Ago2 antibody compared with the negative control IgG (Fig. 3D), further verifying the binding between miR-134-5p and JHDM1D-AS1. The expression of miR-134-5p was increased by MPP + treatment (Fig. 3E), which was the opposite of that of JHDM1D-AS1 expression trend. Besides that, JHDM1D-AS1 overexpression reduced miR-134-5p expression level in SK-N-SH cells (Fig. 3F). Therefore, we confirmed that JHDM1D-AS1 targeted miR-134-5p and suppressed its expression.
MiR-134-5p is a target of JHDM1D-AS1. A The putative binding site between JHDM1D-AS1 and miR-134-5p. B RT-qPCR analysis of miR-134-5p expression in SK-N-SH cells transfected with miR-134-5p mimic or mimic NC. C Dual-luciferase reporter assay for the luciferase activity of wild and mutated JHDM1D-AS1 reporter after miR-134-5p overexpression in SK-N-SH cells. D Anti-Ago2 RIP assay was used in SK-N-SH cells to determine JHDM1D-AS1 and miR-134-5p RNA enrichment in immunoprecipitate complexes. E RT-qPCR analysis of miR-134-5p expression in SK-N-SH cells treated with 2 mM MPP + for 24 h. F RT-qPCR analysis of miR-134-5p expression in SK-N-SH cells transfected with JHDM1D-AS1 or vector. *P < 0.05
JHDM1D-AS1 Restrains MPP + -Induced Neuronal Apoptosis, Inflammation, and Oxidative Stress Through miR-134-5p
Thereafter, whether miR-134-5p mediated the effects of JHDM1D-AS1 on MPP + -induced neuronal injury was explored. The results of RT-qPCR suggested that introduction of miR-134-5p mimic rescued JHDM1D-AS1 overexpression–induced decrease in miR-134-5p expression in MPP + -challenged SK-N-SH cells (Fig. 4A). After that, function analysis showed that miR-134-5p upregulation reversed JHDM1D-AS1 restoration-evoked enhancement in proliferation and inhibition in apoptosis in MPP + -challenged SK-N-SH cells (Fig. 4B–E). MiR-134-5p upregulation caused the elevation of inflammatory cytokines IL-1β, IL-6, and TNF-α in JHDM1D-AS1-overexpressed SK-N-SH cells in the presence of MPP + (Fig. 4F–H). Moreover, the reduction of LDH and MDA production, as well as elevation of SOD content mediated by JHDM1D-AS1 upregulation, was also abolished by miR-134-5p mimic in MPP + -challenged SK-N-SH cells (Fig. 4I–K). Taken together, JHDM1D-AS1/miR-134-5p axis was responsible for MPP + -induced neuronal injury.
JHDM1D-AS1 restrains MPP + -induced neuronal apoptosis, inflammation, and oxidative stress through miR-134-5p. A–K SK-N-SH cells were transfected with vector, JHDM1D-AS1, JHDM1D-AS1 + miRNA NC, or JHDM1D-AS1 + miR-134-5p mimic, followed by the treatment of 2 mM MPP + for 24 h. A RT-qPCR analysis of miR-134-5p expression level in SK-N-SH cells. B CCK-8 for cell viability. C Flow cytometer for cell apoptosis. D, E Western blot analysis of the protein levels of PCNA and Bax. F–H ELISA analysis for IL-1β, IL-6, and TNF-α levels. I–K Measurement of LDH, MDA, and SOD levels in cells using commercial kits. *P < 0.05
PIK3R3 Is a Target of miR-134-5p
Based on the prediction of starBase online tool and previous studies, PIK3R3, NUCKS1, VPS13D, TRIM11, and DDX3Y were predicted that might be the target of miR-134-5p. Then, it was proved that miR-134-5p inhibitor led to a marked increase of PIK3R3 (Fig. S3); therefore, we selected PIK3R3 as the subsequent analysis target. The potential binding site of miR-134-5p on PIK3R3 is shown in Fig. 5A. The results of dual-luciferase reporter assay showed that miR-134-5p overexpression reduced the luciferase activity of the wild-type PIK3R3 vector in SK-N-SH cells, while there were no notable changes in the luciferase activity of the mutated PIK3R3 vector after miR-134-5p overexpression (Fig. 5B). Further RIP assay indicated that miR-134-5p and PIK3R3 were preferentially pulled down by anti-Ago2 pellet relative to the IgG immunoprecipitates in SK-N-SH cells (Fig. 5C). PIK3R3 was observed to be decreased in MPP + -challenged SK-N-SH cells (Fig. 5D). Moreover, it was observed that miR-134-5p inhibitor significantly reduced miR-134-5p expression in SK-N-SH cells (Fig. 5E); furthermore, Western blot analysis showed that miR-134-5p downregulation led to an increase of PIK3R3 expression in SK-N-SH cells (Fig. 5F). All these results confirmed that PIK3R3 was a target of miR-134-5p, and was negatively regulated by miR-134-5p. Besides that, Western blot analysis also exhibited that JHDM1D-AS1 elevated PIK3R3 expression in SK-N-SH cells, which was reversed by miR-134-5p mimic (Fig. 5G), suggesting that JHDM1D-AS1/miR-134-5p axis could regulate PIK3R3 expression.
PIK3R3 is a target of miR-134-5p. A The putative binding site between PIK3R3 and miR-134-5p. B Dual-luciferase reporter assay for the luciferase activity of wild and mutated PIK3R3 reporter after miR-134-5p overexpression in SK-N-SH cells. C Anti-Ago2 RIP assay was used in SK-N-SH cells to determine PIK3R3 and miR-134-5p RNA enrichment in immunoprecipitate complexes. D Western blot analysis of PIK3R3 expression in SK-N-SH cells treated with 2 mM MPP + for 24 h. E The interference efficiency of miR-134-5p inhibitor or inhibitor NC in SK-N-SH using RT-qPCR. F Western blot analysis of PIK3R3 expression in SK-N-SH cells transfected with miR-134-5p inhibitor or inhibitor NC. G Western blot analysis of PIK3R3 expression in SK-N-SH cells transfected with vector, JHDM1D-AS1, JHDM1D-AS1 + miRNA NC, or JHDM1D-AS1 + miR-134-5p mimic. *P < 0.05
Inhibition of miR-134-5p Suppresses MPP + -Induced Neuronal Apoptosis, Inflammation, and Oxidative Stress via PIK3R3
To investigate whether miR-134-5p/PIK3R3 axis was involved in MPP + -induced neuronal injury, three siRNAs targeting PIK3R3 were designed and the knockdown efficiencies have been validated by qRT-PCR with the decrease of PIK3R3 expression level in SK-N-SH cells (Fig. S1), and si-PIK3R3#1 (si-PIK3R3) was selected for subsequent analysis due to the highest decrease of PIK3R3 expression after si-PIK3R3#1 transfection. The interference efficiency of si-PIK3R3 (si-PIK3R3#1) was also confirmed by using Western blot (Fig. 6A). Then, SK-N-SH cells were co-transfection with miR-134-5p inhibitor and PIK3R3 siRNA, followed by MPP + treatment for 24 h. Further function analyses indicated that inhibition of miR-134-5p promoted cell proliferation and suppressed apoptosis in MPP + -challenged SK-N-SH cells, which were reversed by PIK3R3 knockdown (Fig. 6B–E). Moreover, miR-134-5p inhibition reduced the release of IL-1β, IL-6, and TNF-α in MPP + -challenged SK-N-SH cells, while PIK3R3 knockdown abolished these effects (Fig. 6F–H). Besides that, PIK3R3 knockdown attenuated miR-134-5p inhibition-evoked suppression of oxidative stress in SK-N-SH cells under MPP + treatment, evidenced by the elevation of LDH and MDA production, as well as decrease of SOD content (Fig. 6I–K). Collectively, miR-134-5p/PIK3R3 axis was engaged in MPP + -induced neuronal injury.
Inhibition of miR-134-5p suppresses MPP + -induced neuronal apoptosis, inflammation, and oxidative stress PIK3R3. A Western blot analysis of PIK3R3 expression in SK-N-SH cells transfected with si-NC or si-PIK3R3. B–K SK-N-SH cells were co-transfection with miR-134-5p inhibitor and PIK3R3 siRNA, followed by MPP + treatment for 24 h. B CCK-8 for cell viability. C Flow cytometer for cell apoptosis. D, E Western blot analysis of the protein levels of PCNA and Bax. F–H ELISA analysis for IL-1β, IL-6, and TNF-α levels. I–K Measurement of LDH, MDA, and SOD levels in cells using commercial kits. *P < 0.05
Discussion
Recently, the fundamental role of lncRNAs in central nervous system is increasingly emerging, and the dysregulation of lncRNAs can result in neuronal death and is functionally and mechanistically associated with neurobiological processes related to learning, memory, and disorders (Dexter and Jenner 2013; Riva et al. 2016; Salta and De Strooper 2012). JHDM1D-AS1 is a functional lncRNA; previous studies discovered that JHDM1D-AS1 performed oncogenic role in several cancers, such as gastric (Wu et al. 2021), lung (Yao et al. 2019), and pancreatic (Kondo et al. 2017) cancers. However, the effects of JHDM1D-AS1 on the process of PD remain vague. In this study, JHDM1D-AS1 expression was demonstrated to be decreased in MPP + -induced SK-N-SH cells, which was consistent with previous findings (Zhou et al., 2018). Furthermore, we proved that overexpression of JHDM1D-AS1 reduced MPP + -mediated apoptosis, inflammation, and oxidative stress in SK-N-SH cells, suggesting the neuroprotective effect of JHDM1D-AS1.
LncRNAs can act as miRNA sponges, which have been identified as competing endogenous RNAs (Chen et al. 2019; Chen and Wang 2019). In the present study, a binding between JHDM1D-AS1 and miR-134-5p was identified, and JHDM1D-AS1 targetedly suppressed miR-134-5p expression. A precious study showed that miR-134-5p contributed to plasticity deficit in Aβ(1–42)-induced model of Alzheimer’s disease (Baby et al. 2020). In PD, Feng et al. suggested that miR-134-5p reversed the neuroprotective effects of circDLGAP4 in PD (Feng et al. 2020). In the current work, we revealed an increased level of miR-134-5p in MPP + -stimulated SK-N-SH cells, functionally, knockdown of miR-134-5p repressed neuronal apoptosis, inflammation and oxidative stress that caused by MPP + treatment. Moreover, miR-134-5p upregulation attenuated the neuroprotective effects of JHDM1D-AS1.
PIK3R3, also named p55PIK, is one of the class IA regulatory subunits of PI3Ks; it has been reported to be abnormally overexpressed in several types of cancers, and acted as an oncogene to promote cancer progression by regulating various biological behaviors (Wang et al. 2013; Wang et al. 2012; Yu et al. 2015). However, PIK3R3 was observed to be decreased in neurotoxin paraquat-stimulated human dopaminergic SH-SY5Y cells, which led to apoptosis induction (Zhou et al. 2014). Moreover, Zhang et al. showed that lncRNA H19 elevated PIK3R3 expression via miR-585-3p to enhance MPP + -mediated neuronal apoptosis (Zhang et al. 2020). In the current work, we verified that miR-134-5p directly targeted PIK3R3; moreover, JHDM1D-AS1 could regulate PIK3R3 expression through sponging miR-134-5p. Therefore, a JHDM1D-AS1/miR-134-5p/PIK3R3 axis was identified. Additionally, we also observed that PIK3R3 knockdown resulted in a reduction of the neuroprotective effect mediated by miR-134-5p in the presence MPP + .
In conclusion, this work firstly demonstrated that JHDM1D-AS1 restrained MPP + -mediated neuronal apoptosis, inflammation, and oxidative stress via elevating PIK3R3 through miR-134-5p (Fig. 7), indicating a novel insight into the pathogenesis of PD. However, our research has some limitations. The data presented are based on a limited number of cells in vitro; more evidence was needed to investigate the regulatory function of JHDM1D-AS1/miR-134-5p/PIK3R3 axis in vivo.
Availability of Data and Materials
Please contact the correspondence author for the data request.
References
Abou-Sleiman PM, Muqit MM, Wood NW (2006) Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci 7:207–219
Baby N, Alagappan N, Dheen ST, Sajikumar S (2020) MicroRNA-134–5p inhibition rescues long-term plasticity and synaptic tagging/capture in an Aβ(1–42)-induced model of Alzheimer’s disease. Aging Cell 19: e13046.
Bhan A, Soleimani M, Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77:3965–3981
Cai LJ, Tu L, Huang XM, Huang J, Qiu N, Xie GH, Liao JX, Du W, Zhang YY, Tian JY (2020) LncRNA MALAT1 facilitates inflammasome activation via epigenetic suppression of Nrf2 in Parkinson’s disease. Mol Brain 13:130
Chen J, Lin Y, Jia Y, Xu T, Wu F, Jin Y (2019) LncRNA HAND2-AS1 exerts anti-oncogenic effects on ovarian cancer via restoration of BCL2L11 as a sponge of microRNA-340-5p. J Cell Physiol 234:23421–23436
Chen S, Wang J (2019) HAND2-AS1 inhibits invasion and metastasis of cervical cancer cells via microRNA-330-5p-mediated LDOC1. Cancer Cell Int 19:353
Dexter DT, Jenner P (2013) Parkinson disease: from pathology to molecular disease mechanisms. Free Radic Biol Med 62:132–144
Feng Z, Zhang L, Wang S, Hong Q (2020) Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun 522:388–394
Haemmig S, Simion V, Yang D, Deng Y, Feinberg MW (2017) Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr Opin Cardiol 32:776–783
Heward JA, Lindsay MA (2014) Long non-coding RNAs in the regulation of the immune response. Trends Immunol 35:408–419
Kalia LV, Lang AE (2015) Parkinson’s disease. Lancet 386:896–912
Kondo A, Nonaka A, Shimamura T, Yamamoto S, Yoshida T, Kodama T, Aburatani H, Osawa T (2017) Long noncoding RNA JHDM1D-AS1 promotes tumor growth by regulating angiogenesis in response to nutrient starvation. Mol Cell Biol 37:
Krishna A, Biryukov M, Trefois C, Antony PM, Hussong R, Lin J, Heinäniemi M, Glusman G, Köglsberger S, Boyd O, Van Den Berg BH, Linke D, Huang D, Wang K, Hood L, Tholey A, Schneider R, Galas DJ, Balling R, May P (2014) Systems genomics evaluation of the SH-SY5Y neuroblastoma cell line as a model for Parkinson’s disease. BMC Genomics 15:1154
Liu LP, Zhang J, Pu B, Li WQ, Wang YS (2020). Upregulation of JHDM1D-AS1 alleviates neuroinflammation and neuronal injury via targeting miR-101–3p-DUSP1 in spinal cord after brachial plexus injury. Int Immunopharmacol 89: 106962.
Maass PG, Luft FC, Bähring S (2014) Long non-coding RNA in health and disease. J Mol Med (berl) 92:337–346
Marino BLB, De Souza LR, Sousa KPA, Ferreira JV, Padilha EC, Da Silva C, Taft CA, Hage-Melim LIS (2020) Parkinson’s disease: a review from pathophysiology to treatment. Mini Rev Med Chem 20:754–767
Michel PP, Hirsch EC, Hunot S (2016) Understanding dopaminergic cell death pathways in Parkinson disease. Neuron 90:675–691
Quinn JJ, Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17:47–62
Risiglione P, Leggio L, Cubisino SaM, Reina S, Paternò G, Marchetti B, Magrì A, Iraci N, Messina A (2020) High-resolution respirometry reveals MPP(+) mitochondrial toxicity mechanism in a cellular model of Parkinson’s disease. Int J Mol Sci 21:
Riva P, Ratti A, Venturin M (2016) The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res 13:1219–1231
Salta E, De Strooper B (2012) Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol 11:189–200
Teive HA, Bertucci DCF, Munhoz RP (2016) Unusual motor and non-motor symptoms and signs in the early stage of Parkinson’s disease. Arq Neuropsiquiatr 74:781–784
Tysnes OB, Storstein A (2017) Epidemiology of Parkinson’s disease. J Neural Transm (vienna) 124:901–905
Wang G, Chen C, Yang R, Cao X, Lai S, Luo X, Feng Y, Xia X, Gong J, Hu J (2013) p55PIK-PI3K stimulates angiogenesis in colorectal cancer cell by activating NF-κB pathway. Angiogenesis 16:561–573
Wang G, Deng Y, Cao X, Lai S, Tong Y, Luo X, Feng Y, Xia X, Gong J, Hu J (2012) Blocking p55PIK signaling inhibits proliferation and induces differentiation of leukemia cells. Cell Death Differ 19:1870–1879
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472:120–124
Wu M, Liu Y, Pu YS, Ma Y, Wang JH, Liu EQ (2021) JHDM1D-AS1 aggravates the development of gastric cancer through miR-450a-2–3p-PRAF2 axis. Life Sci 265: 118805.
Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A (2013) Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull 97:69–80
Yao G, Chen K, Qin Y, Niu Y, Zhang X, Xu S, Zhang C, Feng M, Wang K (2019) Long non-coding RNA JHDM1D-AS1 interacts with DHX15 protein to enhance non-small-cell lung cancer growth and metastasis. Mol Ther Nucleic Acids 18:831–840
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F, Sun L, Zhang Y, Cui Y, Zhang F, Li J, He X, Yao M (2015) MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 34:413–423
Zhang Y, Xia Q, Lin J (2020) LncRNA H19 attenuates apoptosis in MPTP-induced Parkinson’s disease through regulating miR-585-3p/PIK3R3. Neurochem Res 45:1700–1710
Zhou C, Huang Y, Przedborski S (2008) Oxidative stress in Parkinson’s disease: a mechanism of pathogenic and therapeutic significance. Ann N Y Acad Sci 1147:93–104
Zhou Y, Gu C, Li J, Zhu L, Huang G, Dai J, Huang H (2018) Aberrantly expressed long noncoding RNAs and genes in Parkinson’s disease. Neuropsychiatr Dis Treat 14:3219–3229
Zhou Y, Li F, Tian X, Wang B, Ding M, Pang H (2014) Changes in phosphatidylinositol 3-kinase 55 kDa gamma expression and subcellular localization may be caspase 6 dependent in paraquat-induced SH-SY5Y apoptosis. Hum Exp Toxicol 33:761–771
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, C., Zhang, H. & Li, J. LncRNA JHDM1D-AS1 Suppresses MPP + -Induced Neuronal Injury in Parkinson’s Disease via miR-134-5p/PIK3R3 Axis. Neurotox Res 39, 1771–1781 (2021). https://doi.org/10.1007/s12640-021-00437-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-021-00437-8