Abstract
Infection with the deadly rabies virus (RABV) leads to alteration of cellular gene expression. The RABV, similar to other neurodegenerative diseases may be implicated in neuronal death due to an imbalance in Ca2+ homeostasis. Parvalbumin (PV) and Secretagogin (Scgn), two members of the Calcium-Binding Proteins (CBPs) are useful neuronal markers responsible for calcium regulation and buffering with possible protective roles against infections. This study investigated whether infection with rabies virus causes variance in expression levels of PV and Scgn using the Challenge virus standard (CVS) and Nigerian Street Rabies virus (SRV) strains. Forty-eight, 4-week-old BALB/c mice strains were divided into two test groups and challenged with Rabies virus (RABV) infection and one control group. The presence of RABV antigen was verified by direct fluorescent antibody test (DFAT) and real-time quantitative PCR (qRT-PCR) was used to assess PV and Scgn gene expression. Infection with both virus strains resulted in significant (p < 0.05) increases in expression during early infection. Mid-infection phase caused reduced expression for both genes. However, as infection progressed to the terminal phase, a lower increase in expression was measured. Gene expression and viral load correlation indicated no positive relationship. Neurons with these CBPs may have a greater capacity to buffer calcium and be more resistant to degenerative changes caused by RABV. This implies that, when PV and Scgn expression levels are kept adequately high, the integrity of neurons may be maintained and degeneration caused by RABV infection may be prevented or stopped, hence, these are possible constituents of effective rabies therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most deaths resulting from rabies occur in Africa and Asia (Fooks et al. 2014) and contribute substantially to almost 60,000 human deaths that occur globally per year and 21,476 in Africa (Hampson et al. 2015; World Health Organization (WHO) 2018). Rabies virus (RABV) belongs to the genus Lyssavirus, family Rhabdoviridae, a widespread neurotropic zoonotic viral disease, most commonly spread by the bite of a rabid animal (Kuzmin 2015; Kia et al. 2018; Abdulazeez et al. 2020). The severity of neurologic signs and the fatal outcome that result from infection with rabies are not consistent with the mild inflammation and neuronal degeneration that is seen in the central nervous system (CNS) (Jackson 2007; Singh et al. 2017). By the time prodromal symptoms such as fever, flu-like symptoms and gastrointestinal disturbance begin to show, the virus is already widely disseminated throughout the CNS (Mitrabhakdi et al. 2005; Kuzmin 2015). For adequate infection in the CNS, RABV escapes the host immune response and protects the infected neurons against apoptosis or premature destruction of neurons (Singh et al. 2017).
Many fundamental functions and biological processes are regulated by calcium; its implication and role as a ubiquitous second messenger in metabolism, contraction, cell division and cell growth, among many others, cannot be underemphasized (Verdes et al. 2016). An imbalance in calcium ions (Ca2+) homeostasis can result in the impairment of normal cell functions (Błaszczyk 2016). Its regulation, therefore, is vital to ensure the normal functioning of biological systems. A Ca2+/Gamma-Aminobutyric acid (GABA) mechanism responsible for the stabilization of neuronal activity occurs both at cellular and systemic levels and is highly regulated. A decline in this control will initiate several cascading processes leading to a weakened protective barrier and accumulation of intracellular calcium deposits (Błaszczyk 2016).
A major role in the regulatory mechanism of Ca2+ is played in part by calcium-binding proteins (CBPs). These proteins are effective as buffers against increases in intracellular Ca2+ and also as sensors to promote protein-protein interactions and signaling events (Alpar et al. 2012). Parvalbumin (PV) and Secretagogin (Scgn) belong to these classes of proteins that are responsible for calcium regulation and buffering, and both proteins have been identified in neuronal subpopulations (Tsuboi et al. 2000; Maj et al. 2012). PV is present as the main soluble CBP in fast twitching muscle fibers, Scgn is highly expressed in neuroendocrine cells of the central nervous system (Maj et al. 2012; Garas et al. 2016). Expression patterns of the classical CBPs (Parvalbumin, Calbindin, and Calretinin) have generated broad interest because of the importance of GABAergic neurons in refining the physiological output of neuronal networks (Klausberger and Somogyi 2008). Scgn is plentiful in subsets of neurons engaged in long-distance migration and it is histochemically detectable throughout neuronal morphogenesis (Mulder et al. 2009). When present in neurons otherwise lacking PV, Calbindin, or Calretinin, Scgn may contribute to the refinement of intracellular Ca2+ signaling (Gartner et al. 2001).
Due to the role of calcium homeostasis in dysregulation in neurodegenerative diseases and other pathophysiology (Mattson 2007), there is reason to assume that neurons that contain certain intracellular calcium-binding proteins have a greater capacity to buffer calcium, and therefore would be more resistant to degeneration (Heizmann and Braun 1992). There have also been reports that indicate a possible involvement of the GABAergic system during rabies virus infection (Torres-Fernández et al. 2004; Verdes et al. 2016).
Although advances have been made in the field, the mechanism of rabies pathogenesis – the concise processes by which the virus induces the disease or causes functional alterations in rabies virus-infected neurons is still not completely understood (Schutsky et al. 2014). RABV can offset the balance of the Ca2+/GABA mechanism by redirecting cellular resources towards its viral gene expression by promoting immune evasion (Rivas et al. 2016).
The mechanisms that allow the RABV virus to invade and partly hide from the host’s immune defenses, sometimes for extended periods, before overwhelming the host are fascinating (Madhu et al. 2016), though not fully understood. Improved understanding of the mechanisms underlying rabies neuropathogenesis in animal models is necessary for the development of new therapeutic approaches.
Recent novelty study on Scgn, a calcium-binding protein, has been carried out (Mulder et al. 2009; Zahola et al. 2019), but its implication in rabies infection has never been established. Showing differing expression patterns of Scgn and also PV in RABV infected animal model may be a requisite step to determine effective therapy.
Materials and methods
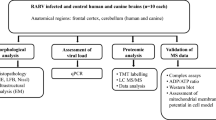
Animals and virus strains
Female BALB/c mice (approximately 3–4 weeks old) were purchased from and housed in Department of Veterinary Public Health and Preventive Medicine animal facility, ABU, Zaria, under pathogen-free conditions with access to food and water ad libitum. All animal experiments described in this study were performed following guidelines of The Nigerian council on Animal Care and ethical approval was granted by Ahmadu Bello University Committee for Animal Health. The challenge virus standard (CVS-11) - a mouse-brain adapted fixed RABV strain and Nigeria street RABV (SRV), isolated from a clinical specimen (rabid dog brain) were obtained from National Veterinary Research Institute (NVRI), Vom and used in this study. All the Researchers involved were vaccinated before the commencement of the experimentations.
Virus infection and tissue extraction
Virus stocks were prepared using a sample of macerated infected dog brain which was diluted to 10% in PBS containing 2% normal equine serum and antibiotics (200 μl/ml of penicillin and 4 mg/ml of streptomycin) (Torres-Fernández et al. 2004). Three-day-old suckling mice were inoculated with either CVS-11 or SRV dilution of this preparation, the mice were euthanized when they became moribund and the brains harvested. A 10% (w/v) suspension was prepared by homogenizing the brains in PBS and antibiotics. The homogenates were centrifuged and the supernatant collected. Animals in the test groups were injected in the left hind limb with 30 μl of this infecting suspension (approximately 1 × 106 infectious particles of RABV). 2 Groups of 18 mice each were inoculated intramuscularly with the viral inoculum for CVS-11 strain (CVS group) and SRV strain (SRV group) respectively. A third group was inoculated under the same conditions with 30 μl of diluting solution for viral inoculum without the virus. All groups of mice were monitored daily for clinical findings. Mice were euthanized and tissue samples collected at 3, 6, 9, 12, 15, and 18 days post-infection (p.i). The harvested brains were suspended in RNA later and stored at −20 °C.
Direct fluorescent antibody test (DFAT)
Impression smears of the samples were made on glass slides to detect the presence of rabies antigen in harvested tissues from all the groups. This was done following the protocol as described by WHO (Rupprecht et al. 2018). The smeared slides were air-dried at room temperature before fixing in cold acetone for 30 mins at −20 °C. The slides were then air-dried for 5 mins and the rabies conjugate (Fujirebio Diagnostics, Inc., Malvern, Pennsylvania, 19,355, USA) was applied to the smear. This was incubated for 30 mins at 37 °C in a humid chamber, after which excess conjugate was washed 3 times with phosphate-buffered saline (PBS) (pH 7.4) and allowed to air dry. Control slides were also prepared in the same way. The slides were viewed under an epifluorescence microscope, the presence of brilliant apple-green fluorescence or greenish-yellow fluorescence against a dark regarded as positive.
RNA extraction and complementary (cDNA) synthesis
Total RNA was isolated from mouse brain tissue using a Norgen® Extraction kit (Norgen Biotek Corp., Ontario, Canada) as per the manufacturer’s instructions. RNA purity and concentration were determined using a Nanodrop spectrophotometer (ND-1000) done by measuring its absorbance at 260 nm (A260). cDNA was synthesized from the extracted total RNA using a SensiFAST™ cDNA synthesis kit (Bioline Reagents Ltd., UK) according to the manufacturer’s instruction: a master mix containing 5 μl of RNA template (70 ng), 1 μl of reverse transcriptase, 4 μl of a 5x TransAmp buffer (containing a blend of random hexamers and oligo dT) and 10 μl of RNase free water to a total volume of 20 μl was prepared. The PCR cycling conditions were: 25 °C/10 mins (Primer annealing), 42 °C/15 mins (reverse transcription), 85 °C/5 mins (inactivation), 4 °C (hold), for 35 cycles. The products were stored at −20 °C until further processing.
Quantitative real time PCR
The cDNA synthesis reaction product was used as template and Quantitative real-time Polymerase Chain Reaction (qRT-PCR) measurement to determine gene expression was carried out with 5x HOT FIREPol Evagreen® qPCR Mix Plus (Solis Bio-Dyne, Tartu, Estonia) using a BIOER Line-Gene system (Hangzhou, China). qPCR was performed in reaction mixtures containing 5 μl of 5x HOT FIREPol Evagreen qPCR mix plus, 0.6 μl each of forward and reverse primers (Table 1), 2 μl of template cDNA, 16.8 μl of nuclease-free PCR-grade water, to a final volume of 25 μl. The Real-time PCR cycling conditions were: (i) 95 °C for 12 mins (Initial activation) (ii) 40 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 20 s. All reactions included negative controls containing the amplification master mix and dH2O. The Ct (cycle threshold) values obtained from the PCR data and expression values were calculated according to the using 2-ΔΔCt method of relative quantification. The results were normalized to mouse Phosphoglycerate Kinase 1 (PGK-1), which had minimal variation in all normal and infected rabies samples that were tested.
Viral load determination
To determine the viral load in infected brain tissues, a nested PCR was carried out using the Solis BioDyne 5x FIREPol Master Mix (Ready to Load) (Solis Bio-Dyne, Tartu, Estonia) PCR synthesis kit and cDNA samples of virus standards for CVS-11 and SRV strains of the rabies virus. The cDNA, which was obtained by amplifying the N-gene of the virus strain, was then quantified and serial dilutions were prepared from this standard of known concentration. Using a second set of primers specific to inner regions of the amplicons (Nested), qPCR was carried out on each serial dilution, and Ct values were obtained. A standard curve of Ct against Log10 quantity was plotted for each dilution. qPCR was carried out using the second primer set (nested primers) to generate Ct values for the test samples. Quantity of viral material was extrapolated from the standard curve using the generated Ct values. Primer sets are shown in Table 2.
Statistical analysis
Correlation analysis was done using Pearson Correlation analysis to determine the relationship between viral load for each measured infection day of the study and fold change expression of Parvalbumin and Secretagogin genes (Pvalb and Scgn) for the same day.
Each experiment was repeated three times to address biological variability. Numerical results were presented as Mean ± Standard deviation (mean ± S.D.). Variability between groups was measured with Independent Sample t-tests using the Statistical Package for Social Sciences Software (SPSS) version 20.0 (SPSS Inc., Chicago, Illinois, USA), p values less than 0.05 were considered statistically significant. Correlation analysis was measured with Statistical Package for Social Sciences Software (SPSS) version 20.0 and p values <0.05 were considered significant.
Results
Qualitative detection of rabies viral antigen was positive for both viral strains (Table 3), CVS infected group exhibited a higher infectivity rate in the sample populations (89%) compared to the SRV group (83%). An increase in viral concentration was seen overtime through the progressive increase in fluorescent intensity and antigen distribution. Viral antigen was not detected on day 3 p.i for SRV infected group but was detected in 33% of the CVS infected group. We wanted to mimic the natural method of viral entry, hence our reason for choosing the intramuscular inoculation route. Clinical signs of rabies (ruffled fur, increased agitation, paralysis) were observed throughout the infection period, but occurring earlier in the CVS group (Table 3).
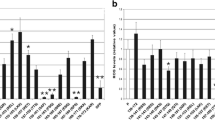
As shown in Fig. 1, parvalbumin expression show CVS infected groups with a significant (p < 0.05) increase in expression (9.44, 10.46; Day 3, 6 p.i) during early infection. Progression of infection caused a significant (p < 0.05) reduction in expression on days 9 and 12 p.i (about 7.77-fold change) and as infection reached the terminal stage, we saw an increase in expression, although not significant at p < 0.05 compared with controls.
Parvalbumin fold change in expression in CVS-11 and SRV infected mice, relative to PGK-1. The brains from uninfected controls as well as from both CVS-11 and SRV infected mice were collected at days 3, 6, 9, 12, 15, and 18 p.i, and RNA was isolated for qPCR as detailed in Materials and Methods. The relative fold changes are expressed in the graph, values marked * are statistically significant (p < 0.05) from controls. PGK-1: Phosphoglycerate kinase-1, CVS-11: Challenge virus standard, SRV: Street rabies virus
The SRV infected group showed a significant (p < 0.05) 5.96-fold upregulation at day 3 and a significant (p < 0.05) 0.35-fold down-regulation of Parvalbumin gene expression at day 9 compared with controls (Fig. 1). Expression was upregulated towards the terminal stage with a significant (p < 0.05) rise in fold change values to 2.23 at day 18 p.i.
Secretagogin expression in CVS-11 and SRV infected samples (Fig. 2) show CVS infected samples demonstrating an increased expression (9.03 and 4.81-fold upregulation for days 3 and 6 p.i) during early infection which was significant (p < 0.05) only at day 6 p.i when compared with controls. Mid-infection showed a significant (p < 0.05) 0.3-fold downregulation of Scgn and as infection reached the terminal stage, expression began to rise and a 1.66-fold upregulation was observed. SRV infected samples showed an upregulation of expression (7.82) which was significant (p < 0.05) on day 6. Mid-infection showed a significant (p < 0.05) downregulation of gene expression (0.35) which gradually began to rise again as infection progressed towards the terminal stage, with day 18 showing a 6.77-fold upregulation.
Secretagogin fold change in expression in CVS-11 and SRV infected mice, relative to PGK-1. The brains from uninfected controls as well as from both CVS-11 and SRV infected mice were collected at days 3, 6, 9, 12, 15, and 18 p.i, and RNA was isolated for qPCR as detailed in Materials and Methods. The relative fold changes are expressed in the graph, values marked * are statistically significant (p < 0.05) from controls. PGK-1: Phosphoglycerate kinase-1, CVS-11: Challenge virus standard, SRV: Street rabies virus
In the comparative expression of Parvalbumin and Secretagogin for both viral strains, PV and Scgn showed very similar expression levels on day 3 of infection for CVS-infected mice. Other measured days showed a higher PV expression (Fig. 1) than Scgn (Fig. 2). Although a fold downregulation of expression was observed in Secretagogin during mid-infection, CVS-infected samples maintained an upregulation of PV expression at that stage. Gene expression in SRV infected mice showed higher expression in PV during early and mid-infection stages (Fig. 1) while Scgn expression levels were higher at the terminal stage (Fig. 2). Downregulation of expression was seen for both genes on day 6 and day 18 p.i, Scgn showed a 6.77-fold upregulation (Fig. 2) compared to the 2.2-fold seen in PV (Fig. 1).
Relationship between gene expression and viral load in infected samples
To determine if the viral load of infection affects gene expression, we used nested RT-PCR to assess mRNA levels of RABV nucleotide protein as a measure of virus replication. Table 4 shows the viral load quantification in the test samples. Pearson’s correlation coefficient was used as a simple measure of the linear variation between viral RNA load and gene expression. As shown in Table 5, PV expression showed a moderate negative correlation for CVS (r = −0.696) and a weak negative correlation (r = −0.379) for SRV, both correlation values were statistically non-significant (p < 0.05). For Scgn, the association between fold change expression and viral load showed also, a weak negative correlation (r = −0.352) for CVS and a weak positive correlation (r = 0.290) for SRV. Both values presented were not statistically significant (p < 0.05). The values are shown in Table 5.
Discussion
More prevalent in the developing world than in industrialized countries, rabies is a major health hazard affecting a vast population (White and Lloyd 2012). RABV infects almost exclusively the neurons, with neuro-invasiveness and neurotropism being the major defining characteristics of the pathogenesis of a classical rabies infection (Gnanadurai et al. 2015). While there is some understanding of the roles of viral proteins in rabies pathogenesis, the contribution of host factors to RABV transcription replication and axonal trans-synaptic spread remains unknown. Infection and replication of viruses in cells of vertebrates result in the alteration of expression of cellular genes and affects the regulation of cellular mRNA translation, localization, and degradation (White and Lloyd 2012). Changes in host gene expression could be a cellular antivirus response, a virus-induced response, or a nonspecific response that neither promotes nor prevents virus infection (Saha and Rangarajan 2003). The outcome of RABV infection is determined by the convergence of several different virus–host interactions. In this study, we examined Pvalb and Scgn expression after infection with Challenge Virus Standard (CVS-11) or Nigeria street Rabies virus (SRV) strains of rabies virus. Our study showed that rabies infection in mice elicited alterations in gene expression of our interest genes.
Both genes showed differential expression levels through the course of infection. This is consistent with findings that report that rabies infection elicits various cellular responses in infected neurons, resulting in an increase or decrease of various genes and proteins (Prosniak et al. 2001; Miao et al. 2017; Abdulazeez et al. 2020). The expression of PV and Scgn were similar in trend through the infection period with early infection showing notable increase. The onset of clinical signs (gradual paralysis of hind limbs, hunched back) observed from day 8pi resulted in decreased expression values: 1.04 and 0.53 (CVS and SRV) for PV and 0.35 and 0.35 (CVS and SRV) for SG. This was about a 5–7 fold reduction in expression from the early infection stage. As infection progressed to the terminal phase, we saw expression levels begin to rise again.
Because cellular degeneration is accompanied by impaired Ca2+ homeostasis, CBPs which play a major role in Ca2+ homeostasis have been postulated to have a protective role (Heizmann and Braun 1992). Several studies have shown that an increase in viral propagation causes an alteration in host proteins; Shraddha and Abhay (2015) report total protein content reduction with progression of infection, and Torres-Fernández et al. (2004) reports an increase in some of the proteins that regulate Calcium homeostasis during rabies infection. Abdulazeez et al. (2020) reports an early infection increase and a late infection stage decrease of interferon-gamma mRNA levels. Our findings report upregulation of PV and Scgn during early infection and a downregulation during mid-infection. Consistent with our findings during mid-infection are studies that have shown neuronal proteins involved in synapse, neurotransmission, apoptosis, and metabolism being downregulated (Zandi et al. 2009; Kluge et al. 2013). Calcium is a major player in neurotransmission and cellular degeneration is accompanied by impaired Ca2+ homeostasis, CBPs which play a major role in Ca2+ homeostasis have been postulated to have a protective role (Heizmann and Braun 1992).
The upregulation observed during early infection may have been a result of antiviral response to infection. Ito et al. (2016) indicate that activation of the interferon system leads to establishment of antiviral response and helps shape effective adaptive response. We hypothesize that the increase in gene expression observed during both early and terminal infection stages were adaptive responses by the host to limit neuronal death. Zhang et al. (2016) reported adaptive immunity for attenuated rabies virus strain leading to subtle clinically observable signs and viral clearance from the CNS. RABV expresses proteins that can inhibit interferon pathways and can form inhibitory interactions with cellular factors (Ojedapo et al. 2020). This could cause the downregulation of gene expression observed during mid-infection.
Functional studies in neurons revealed that parvalbumin significantly buffers calcium influx signals, causing a decrease in the overall calcium elevations and slowing down the rate of calcium increases in these cells (Chard et al. 1993). Also, Secretagogin may contribute to the refinement of intracellular calcium signaling and in cases where it is co-expressed with any of the classical CBPs, a substantial enhanced calcium buffering capacity could be obtained (Mulder et al. 2009). CBPs have been shown to exert neuroprotective effects by reducing the magnitude of cytosolic calcium elevations (Schwaller 2009). Various researches have been done on PV implication in rabies infection (Torres-Fernández et al. 2004; Rengifo et al. 2016). This is the first that attempts to establish how the progression of the disease affects the expression of Secretagogin.
Despite major intrinsic differences in the etiology of neurodegenerative disorders, deregulated calcium homeostasis has emerged as a common underlying mechanism of neuronal loss in Alzheimer’s Disease (AD), Parkinson’s diseases (PD), amyotrophic lateral sclerosis (ALS), among others (Popugaeva et al. 2017; Duda et al. 2016; Sirabella et al. 2018). In addition to direct effects on neuronal survival, altered Ca2+ homeostasis may also contribute to the initiation or progression of the neurodegenerative process by enhancing neuronal vulnerability to metabolic and other stressors (Toescu and Verkhratsky 2004; Toescu and Vreugdenhil 2010). Palop and Mucke (2010), Zallo et al. (2018) both suggest that dysfunction of PV cells and inhibitory functions contribute to Alzheimer’s Disease-related functional impairments. Involvement of Scgn in neuronal survival in Alzheimer’s Disease has also been reported (Attems et al. 2007, 2008; Zahola et al. 2019). The down-regulation of these genes observed during mid-infection could be the cause of the dysregulation prominent with rabies infection, due to a decreased ability for these proteins to buffer calcium effectively.
The results we obtained indicate an imbalance of calcium regulation during infection and suggest that CBPs may be of importance in regulating calcium homeostasis during infection because of the varying changes in mRNA expression levels of the genes throughout infection period.
Furthermore, we also examined the association between gene expression levels and the viral load of infection. Even though the negative correlation sign indicates an inverse relationship between the viral load and the gene expression, the weak values we obtained indicate that this relationship is almost negligible. Various studies have tried to correlate expression levels of genes with the viral load of infection (de Boer et al. 2007; Ragin et al. 2007). The results from these studies report that variation in viral load was not reflected in the expression of the genes. It also showed that mRNA transcripts were highly variable and did not show a correlation to the physical status of the virus. Overall, changes in global RNA expression reflect responses to viral replication rather than a mechanism that might explain viral control.
Conclusion
Collectively, our study reveals that Scgn expression is affected by infection of CVS-11 and SRV and that both PV and Scgn play an important role in the pathogenesis of rabies. The altered expression of SG and PV may reflect cellular dysfunction associated with rabies thus, maintaining them might serve to preserve Calcium and hence cellular integrity of the neurons. Relevant signaling pathways, varying immune host responses, and pathogeny still provide vast areas to conduct intensive investigations regarding rabies pathogenesis.
Limitations
Using absolute quantification and employing a range of plasmids for quantification of the viral gene would be a better approach and would give better insight and more clarity to the work.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Abdulazeez M, Kia GSN, Abarshi MM, Muhammad A, Ojedapo CE, Atawodi JC, Dantong D, Kwaga JKP (2020) Induction of rabies virus infection in mice brain may up and down regulate type II interferon gamma via epigenetic modifications. Metab Brain Dis 35:819–827. https://doi.org/10.1007/s11011-020-00553-y
Alpar A, Attems J, Mulder J, Hokfelt T, Harkany T (2012) The renaissance of Ca2+binding proteins in the nervous system: secretagogin takes center stage. Cell Signal 24(2):378–387. https://doi.org/10.1016/j.cellsig.2011.09.028
Attems J, Preusser M, Grosinger-Quass M, Wagner L, Lintner F, Jellinger K (2008) Calcium-binding protein secretagogin-expressing neurones in the human hippocampus are largely resistant to neurodegeneration in Alzheimer’s disease. Neuropath Appl Neuro 34(1):23–32. https://doi.org/10.1111/j.1365-2990.2007.00854.x
Attems J, Quass M, Gartner W, Nabokikh A, Wagner L, Steurer S et al (2007) Immunoreactivity of calcium binding protein secretagogin in the human hippocampus is restricted to pyramidal neurons. Exp Gerontol 42(3):215–222. https://doi.org/10.1016/j.exger.2006.09.018
Błaszczyk JW (2016) Parkinson’s disease and Neurodegeneration: GABA-collapse hypothesis. Front Neurosci 10:269. https://doi.org/10.3389/fnins.2016.00269
Chard PS, Bleakman D, Christakos S, Fullmer CS, Miller RJ (1993) Calcium buffering properties of calbindin D28k and parvalbumin in rat sensory neurons. J Physiol 472(1):341–357. https://doi.org/10.1113/jphysiol.1993.sp019950
de Boer MA, Jordanova ES, Kenter GG, Peters AA, Corver WE, Trimbos JB, Fleuren GJ (2007) High human papillomavirus oncogene mRNA expression and not viral DNA load is associated with poor prognosis in cervical cancer patients. Clin Cancer Res 13(1):132–138. https://doi.org/10.1158/1078-0432.CCR-06-1568
Duda J, Pötschke C, Liss B (2016) Converging roles of ion channels, calcium, metabolic stress, and activity pattern of Substantia nigra dopaminergic neurons in health and Parkinson's disease. J Neurochem 139:156–178. https://doi.org/10.1111/jnc.13572
Fooks AR, Banyard AC, Horton DL, Johnson N, McElhinney LM, Jackson AC (2014) Current status of rabies and prospects for elimination. Lancet 384(9951):1389–1399. https://doi.org/10.1016/S0140-6736(13)62707-5
Garas FN, Shah RS, Kormann E, Doig NM, Vinciati F, Nakamura KC, Sharott A (2016) Secretagogin expression delineates functionally-specialized populations of striatal parvalbumin-containing interneurons. Elife 5(16088). https://doi.org/10.7554/eLife.16088.001
Gartner W, Lang W, Leutmetze F, Domanovits H, Waldhäusl W, Wagner L (2001) Cerebral expression and serum detectability of secretagogin, a recently cloned EF-hand Ca2+−binding protein. Cereb Cortex 11(12):1161–1169. https://doi.org/10.1093/cercor/11.12.1161
Gnanadurai CW, Yang Y, Huang Y, Li Z, Leyson CM, Cooper TL, Fu ZF (2015) Differential host immune responses after infection with wild-type or lab-attenuated rabies viruses in dogs. PLoS Negl Trop Dis 9(8):1–15. https://doi.org/10.1371/journal.pntd.0004023
Hampson K, Coudeville L, Lembo T, Sambo M, Kieffer A, Attlan M, Barrat J, Blanton JD, Briggs DJ, Cleaveland S, Costa P, Freuling CM, Hiby E, Knopf L, Leanes F, Meslin FX, Metlin A, Miranda ME, Müller T, Nel LH, Recuenco S, Rupprecht CE, Schumacher C, Taylor L, Vigilato MAN, Zinsstag J, Dushoff J, on behalf of the Global Alliance for Rabies Control Partners for Rabies Prevention (2015) Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis 9(4):e0003709. https://doi.org/10.1371/journal.pntd.0003709
Heizmann CW, Braun K (1992) Changes in Ca(2+)-binding proteins in human neurodegenerative disorders. Trends Neurosci 15(7):259–264. https://doi.org/10.1016/0166-2236(92)90067-I
Ito N, Moseley GW, Sugiyama M (2016) The importance of immune evasion in the pathogenesis of rabies virus. J Vet Med Sci 78(7):1089–1098. https://doi.org/10.1292/jvms.16-0092
Jackson AC (2007) Pathogenesis. In: Jackson AC, Wunner WH (eds) Rabies, 2nd edn. Elsevier Academic Press, London, pp 341–381. https://doi.org/10.1016/B978-012369366-2/50010-5
Kia GSN, Huang Y, Zhou M, Zhou Z, Gnanadurai CW, Leysona CM, Umoh JU, Kazeem HM, Ehizibolo DO, Kwaga JKP, Nwosu CI, Fu ZF (2018) Molecular characterization of a rabies virus isolated from trade dogs in plateau state, Nigeria. Sokoto J Vet Sci 16:54–62. https://doi.org/10.4314/sokjvs.v16i2.8
Klausberger T, Somogyi P (2008) Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321(5885):53–57. https://doi.org/10.1126/science.1149381
Kluge M, Schmidt D, Uhr M, Steiger A (2013) Ghrelin suppresses nocturnal secretion of luteinizing hormone (LH) and thyroid stimulating hormone (TSH) in patients with major depression. J Psychiatr Res 47(9):1236–1239. https://doi.org/10.1016/j.jpsychires.2013.05.010
Kuzmin IV (2015) Virus isolation in animals: the mouse inoculation test. In: Rupprecht C, Nagarajan T (eds) Current laboratory techniques in rabies diagnosis. Research and Prevention. Academic Press, CA, pp 13–23. https://doi.org/10.1016/B978-0-12-801919-1.00002-6
Madhu BP, Singh KP, Saminathan M, Singh R, Shivasharanappa N, Sharma AK, Malik YS, Dhama K, Manjunatha V (2016) Role of nitric oxide in the regulation of immune responses during rabies virus infection in mice. Virus Disease 27(4):387–399. https://doi.org/10.1007/s13337-016-0343-7
Maj M, Milenkovic I, Bauer J, Berggard T, Veit M, Ilhan-Mutlu A et al (2012) Novel insights into the distribution and functional aspects of the calcium binding protein secretagogin from studies on rat brain and primary neuronal cell culture. Front Mol Neurosci 5:84. https://doi.org/10.3389/fnmol.2012.00084
Mattson MP (2007) Calcium and neurodegeneration. Aging Cell 6(3):337–350. https://doi.org/10.1111/j.1474-9726.2007.00275.x
Miao FM, Zhang SF, Wang SC, Liu Y, Zhang F, Hu RL (2017) Comparison of immune responses to attenuated rabies virus and street virus in mouse brain. Arch Virol 162(1):247–257. https://doi.org/10.1007/s00705-016-3081-7
Mitrabhakdi E, Shuangshoti S, Wannakrairot P (2005) Difference in neuropathogenetic mechanisms in human furious and paralytic rabies. J Neurol Sci 238(1–2):3–10. https://doi.org/10.1016/j.jns.2005.05.004
Mulder J, Zilberter M, Spence L, Tortoriello G, Uhlen M, Yanagawa Y, Aujard F, Hokfelt T, Harkany T (2009) Secretagogin is a Ca2+− binding protein specifying subpopulations of telencephalic neurons. Proc Natl Acad Sci 106(52):22492–22497. https://doi.org/10.1073/pnas.0912484106
Ojedapo CE, Muhammad A, Kia GS, Abarshi MM, Abdulazeez M, Atawodi JC, Kwaga JK (2020) Rabies virus induction in mice upregulates B7-H1 via epigenetic modifications. Virus Diseases 31:1–9. https://doi.org/10.1007/s13337-020-00588-w
Palop JJ, Mucke L (2010) Amyloid-β–induced neuronal dysfunction in Alzheimer's disease: from synapses toward neural networks. Nat Neurosci 13(7):812–818. https://doi.org/10.1038/nn.2583
Popugaeva E, Pchitskaya E, Bezprozvanny I (2017) Dysregulation of neuronal calcium homeostasis in Alzheimer's disease–a therapeutic opportunity. Biochem Biophys Res Commun 483(4):998–1004. https://doi.org/10.1016/j.bbrc.2016.09.053
Prosniak M, Hooper DC, Dietzschold B, Koprowski H (2001) Effect of rabies virus infection on gene expression in mouse brain. Proc Natl Acad Sci 98(5):2758–2763. https://doi.org/10.1073/pnas.051630298
Ragin CCR, Modugno F, Gollin SM (2007) The epidemiology and risk factors of head and neck cancer: a focus on human papillomavirus. J Dent Res 86(2):104–114 10.1177%2F154405910708600202
Rengifo AC, Umbarila VJ, Garzón MJ, Torres-Fernández O (2016) Differential effect of the route of inoculation of rabies virus on NeuN Immunoreactivity in the cerebral cortex of mice. Int J Morphol 34(4):1362–1368. https://doi.org/10.4067/S0717-95022016000400031
Rivas HG, Schmaling SK, Gaglia MM (2016) Shutoff of host gene expression in influenza a virus and herpesviruses: similar mechanisms and common themes. Viruses 8(4):102. https://doi.org/10.3390/v8040102
Rupprecht CE, Fooks AR, Abela-Ridder B (eds) (2018) Laboratory techniques in rabies, vol 1, 5th edn. World Health Organization, Geneva
Saha S, Rangarajan PN (2003) Common host genes are activated in mouse brain by Japanese encephalitis and rabies viruses. J Gen Virol 84(7):1729–1735. https://doi.org/10.1099/vir.0.18826-0
Schutsky K, Portocarrero C, Hooper DC, Dietzschold B, Faber M (2014) Limited brain metabolism changes differentiate between the progression and clearance of rabies virus. PLoS One 9(4):e87180. https://doi.org/10.1371/journal.pone.0087180
Schwaller B (2009) The continuing disappearance of “pure” Ca2+ buffers. Cell Mol Life Sci 66(2):275–300. https://doi.org/10.1007/s00018-008-8564-6
Shraddha M, Abhay C (2015) Rabies virus severely influences host protein during terminal stages of infection. Int J Curr Microbiol Appl Sci 4(10):129–137
Singh R, Singh KP, Cherian S, Saminathan M, Kapoor S, Manjunatha Reddy GB, Panda S, Dhama K (2017) Rabies–epidemiology, pathogenesis, public health concerns and advances in diagnosis and control: a comprehensive review. Veterinary Q 37(1):212–251. https://doi.org/10.1080/01652176.2017.1343516
Sirabella R, Valsecchi V, Anzilotti S, Cuomo O, Vinciguerra A, Cepparulo P, Brancaccio P, Guida N, Blondeau N, Canzoniero LMT, Franco C, Amoroso S, Annunziato L, Pignataro G (2018) Ionic homeostasis maintenance in ALS: focus on new therapeutic targets. Front Neurosci 12:510. https://doi.org/10.3389/fnins.2018.00510
Toescu EC, Verkhratsky A (2004) Ca2+ and mitochondria as substrates for deficits in synaptic plasticity in normal brain ageing. J Cell Mol Med 8(2):181–190. https://doi.org/10.1111/j.1582-4934.2004.tb00273.x
Toescu EC, Vreugdenhil M (2010) Calcium and normal brain ageing. Cell Calcium 47(2):158–164. https://doi.org/10.1016/j.ceca.2009.11.013
Torres-Fernández O, Yepes GE, Gómez JE, Pimienta HJ (2004) Effect of rabies virus infection on the expression of parvalbumin, calbindin and calretinin in mouse cerebral cortex. Biomedica 24(1):63–78 Retrieved January 24, 2021, from http://www.scielo.org.co/scielo.php?script=sci_arttext&pid=S0120-41572004000100009&lng=en&tlng=en
Tsuboi K, Kimber TA, Shults CW (2000) Calretinin-containing axons and neurons are resistant to an intrastriatal 6-hydroxydopamine lesion. Brain Res 866(1–2):55–64. https://doi.org/10.1016/S0006-8993(00)02219-8
Verdes JM, de Sant'Ana FJF, Sabalsagaray MJ, Okada K, Calliari A, Moraña JA, de Barros CSL (2016) Calbindin D28k distribution in neurons and reactive gliosis in cerebellar cortex of natural rabies virus-infected cattle. J Vet Diagn Investig 28(4):361–368. https://doi.org/10.1177/1040638716644485
White JP, Lloyd RE (2012) Regulation of stress granules in virus systems. Trends Microbiol 20(4):175–183. https://doi.org/10.1016/j.tim.2012.02.001
World Health Organization (2018) WHO expert consultation on rabies: third report, vol 1012. World Health Organization
Zahola P, Hanics J, Pintér A, Máté Z, Gáspárdy A, Hevesi Z, Echevarria D, Adori C, Barde S, Törőcsik B, Erdélyi F, Szabó G, Wagner L, Kovacs GG, Hökfelt T, Harkany T, Alpár A (2019) Secretagogin expression in the vertebrate brainstem with focus on the noradrenergic system and implications for Alzheimer’s disease. Brain Struct Funct 224(6):2061–2078. https://doi.org/10.1007/s00429-019-01886-w
Zallo F, Gardenal E, Verkhratsky A, Rodríguez JJ (2018) Loss of calretinin and parvalbumin positive interneurones in the hippocampal CA1 of aged Alzheimer’s disease mice. Neurosci Lett 681:19–25. https://doi.org/10.1016/j.neulet.2018.05.027
Zandi F, Eslami N, Soheili M, Fayaz A, Gholami A, Vaziri B (2009) Proteomics analysis of BHK-21 cells infected with a fixed strain of rabies virus. Proteomics 9(9):2399–2407. https://doi.org/10.1002/pmic.200701007
Zhang D, He F, Bi S, Guo H, Zhang B, Wu F, Liang J, Yang Y, Tian Q, Ju C, Fan H, Chen J, Guo X, Luo Y (2016) Genome-wide transcriptional profiling reveals two distinct outcomes in central nervous system infections of rabies virus. Front Microbiol 7:751. https://doi.org/10.3389/fmicb.2016.00751
Acknowledgments
The authors are grateful to the Africa Center of Excellence for Neglected Tropical Diseases and Forensic Biotechnology, (ACENTDFB), Ahmadu Bello University (ABU) Zaria, for funding this unique Project. The National Veterinary Research Institute (NVRI) for the provision of the Rabies viruses used in this study.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study, conception, and design. Material preparation, data collection, and analysis were performed by Brenda Kanu, George C. Korie, Grace S.N. Kia, and Ishaya S. Tekki. Supervision was performed by Grace S.N. Kia and Idowu A. Aimola. The first draft of the manuscript was written by Brenda Kanu and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Compliance with ethical standards.
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kanu, B., Kia, G.S.N., Aimola, I.A. et al. Rabies virus infection is associated with alterations in the expression of parvalbumin and secretagogin in mice brain. Metab Brain Dis 36, 1267–1275 (2021). https://doi.org/10.1007/s11011-021-00717-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-021-00717-4