Abstract
Resveratrol, a phytoalexin present in grapes and other edible foods, has been reported to have beneficial effects against various diseases including cancer. We previously reported that resveratrol and its derivative, caffeic acid-adducted resveratrol, selectively inhibit the three-dimensional (3D) proliferation of a human colorectal cancer cell line, HCT116 with activating KRAS mutation. Herein, we demonstrated that a novel compound, ferulic acid-bound resveratrol, also represses the 3D proliferation of HCT116 cells. We observed that resveratrol conjugated to two ferulic acids represses the 3D proliferation of HCT116 cells more strongly than resveratrol and resveratrol conjugated to one ferulic acid. Resveratrol conjugated to two ferulic acids also inhibited the 3D proliferation of MCF7 human breast cancer cells. We further uncovered that the resveratrol derivative increases the mRNA level of the tumor suppressor p15, a CDK inhibitor that functions as a brake of cell proliferation in HCT116 cells. These results imply that the resveratrol derivative represses 3D proliferation via increasing p15 expression in HCT116 cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Normal colonic epithelium assembles into a three-dimensional (3D) structure in vivo [1], and deregulation of the structure is frequently observed in cancer [2]. This structure can be partially replicated via 3D culture (3DC) in vitro [3]. It was previously reported that HKe3 cells, which were derived from a colorectal cancer cell line, HCT116 via deleting mutant KRAS (constitutively active form), assemble into a normal colonic crypt-like 3D structure in 3DC. On the contrary, HCT116 cells carrying mutant KRAS form an architecture without normal cell polarity and luminal apoptosis, indicating that oncogenic mutation of KRAS is involved in the disruption of cellular polarity and inhibition of apoptosis [3]. Therefore, a 3DC method using these cells could be needed to clarify colorectal tumorigenesis in vivo and identify treatments for colorectal cancer.
Resveratrol is a naturally occurring polyphenol present in many kinds of edible foods such as grapes, peanuts, and blueberry [4], and it is believed to possess efficacy against a number of diseases such as heart disease, neurological disorders, diabetes, obesity, and cancer [5, 6]. Resveratrol effects on the expression of several genes related to cell cycle progression and apoptosis, such as p53, p21, p300/CBP, Apaf1, and BAK, lead to the repression of proliferation and induction of apoptosis in cancer cells [5]. Tsunoda et al. show that resveratrol selectively induces luminal apoptosis and represses the 3D proliferation of HCT116 cells carrying activating KRAS mutation, probably by inhibiting PDE4 activity [7, 8]. We furthermore reported that caffeic acid-bound resveratrol represses the 3D proliferation of HCT116 cells more strongly than resveratrol [9]. The resveratrol derivative exhibited inhibitory effect comparable with 5-fluorouracil, a usual anticancer drug.
Cell cycle progression of mammalian cells is promoted by protein complexes composed of cyclins and cyclin-dependent kinases (CDKs) and repressed by CDK inhibitors (CKIs) [10]. There are seven CKI in mammals, which are divided into two groups: the INK4 family (p15, p16, p18, and p19), which binds to and inhibits CDK4/6, and the CIP/KIP family (p21, p27, and p57), which binds to and inhibits CDK2. Increased CKI expression causes the inactivation of their target cyclin–CDK complexes, leading to the retention of pRB in the hypophosphorylated state, and prevention of cell cycle progression at G1 phase. Disruption of this pathway causes abnormal cell proliferation, leading to cancer development [11]. In this study, we found that a novel compound, in which two ferulic acids were conjugated to resveratrol (UHA025), induces p15 expression and represses the 3D proliferation of HCT116 cells.
Materials and methods
Cell culture
Human colorectal cancer HCT116 cells and breast cancer MCF7 cells were obtained from ATCC (American Type Culture Collection, Frederick, MA, USA). HKe3 cells were established by disrupting at the oncogenic mutant KRAS gene in HCT116 cells [12]. These cells were maintained as previously described [9]. 3DC was performed using a 96-well plate with an ultra-low attachment surface and round bottom (Corning Inc., Corning, NY, USA). Photomicrographs of cell spheroids in 3DC were taken using a CKX41 inverted microscope (Olympus, Tokyo, Japan) and the areas of cell spheroids were measured using ImageJ software.
Reagents
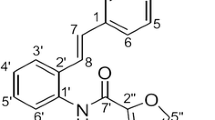
Resveratrol (3,4′,5-trihydroxy-trans-stilbene) and ferulic acid were purchased from Sigma (St. Louis, MO, USA). Ferulic acid-adducted resveratrol formulations (named UHA023, UHA024, and UHA025) were prepared by UHA Mikakuto Co., Ltd. (Osaka, Japan) as previously described (UHA023 [13], UHA024 [14], UHA025 [15]).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
qRT-PCR was performed as previously described [16]. Briefly, the extraction of RNA was performed using an RNeasy Plus Kit (Qiagen, Hilden, Germany). Complementary DNA was synthesized from RNA using a SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA). qPCR was performed using a QuantiTect SYBR Green PCR Master Mix (Qiagen) and an Mx3000P Real-Time Q-PCR System (Agilent Technologies, Santa Clara, CA, USA). The sequence of qPCR primers for human p15, p18, p19, p21, p27, p57, and GAPDH is described in [17].
Results
We recently reported that resveratrol and its derivative, caffeic acid-adducted resveratrol, significantly inhibit the 3D proliferation of HCT116 cells [7,8,9]. In this study, we furthermore searched resveratrol derivatives that repress the proliferation of HCT116 cells in 3DC. HCT116 cells were treated with dimethyl sulfoxide (DMSO, control), resveratrol, or resveratrol derivatives at 25 µM for 7 days. We found that the novel resveratrol derivatives UHA023, UHA024, and UHA025 (Fig. 1a) have stronger inhibitory effects on HCT116 cell spheroid growth in 3DC than resveratrol (Fig. 1b, c). These compounds were synthesized by conjugating resveratrol to one (UHA023 and UHA024) or two ferulic acids (UHA025). We next determined the IC50 for each of these compounds in 3DC. The IC50 was > 20 µM for both UHA023 (Fig. 2a, b) and UHA024 (Fig. 2c, d), compared with 5.86 µM ± 0.82 for UHA025 (Fig. 2e, f). We previously reported an IC50 of 2.39 µM for 5-fluorouracil (5-Fu), a usual anticancer drug used in colon cancer therapy [9]. These results indicate that UHA025 represses the 3D proliferation of HCT116 cells more strongly than UHA023 and UHA024 (Table 1) and a similar inhibitory effect as 5-Fu.
The ferulic acid-adducted resveratrol derivatives UHA023, UHA024, and UHA025 inhibit the three-dimensional proliferation of HCT116 cells. a Chemical structures of resveratrol, ferulic acid, UHA023, UHA024, and UHA025. b 100 HCT116 cells were seeded and treated with dimethyl sulfoxide (Control; Ctr), resveratrol, UHA023, UHA024, or UHA025 at 25 µM on days 0, 2, and 4 in three-dimensional culture. The spheroids were observed on day 7. Scale bar = 500 μm. c The area of the spheroids on day 7 was normalized as the relative ratio to control cells treated with dimethyl sulfoxide. Data represent the means and standard deviations of four spheroids. N.D. not detected
Resveratrol conjugated to two ferulic acids (UHA025) represses the 3D proliferation of HCT116 cells more strongly than resveratrol adducted to one ferulic acid (UHA023 and UHA024). a, c, e 100 HCT116 cells were seeded and treated with UHA023 (a), UHA024 (b), or UHA025 (c) at the indicated concentrations on days 0, 2, and 4 in three-dimensional culture. The pictures of spheroids were taken on the indicated days. Scale bar = 500 μm. b, d, f The size of spheroids was measured on days 2, 4, and 7. The area of the spheroids was normalized as the relative ratio to control cells treated with dimethyl sulfoxide on day 2. Data are presented as means and standard deviations of four spheroids
UHA025 is synthesized by binding two ferulic acids to resveratrol. It has been reported that ferulic acid has the inhibitory effect on the proliferation of human colon cancer Caco-2 cells in 2D culture [18]. Therefore, we examined whether ferulic acid also represses the proliferation of HCT116 cells in 3DC. HCT116 cells were treated with resveratrol or ferulic acid at a concentration of 10 µM, either alone or in combination, for 5 days. Although UHA025 significantly decreased the size of HCT116 cell spheroids compared with the findings in control cells, resveratrol, ferulic acid, or both had no effect on the size of HCT116 cell spheroids (Fig. 3a, b). These results represent that chemical binding of ferulic acid to resveratrol is required for the inhibitory effect of UHA025 on the 3D proliferation of HCT116 cells.
Ferulic acid, a component of UHA025, has no influence on the three-dimensional proliferation of HCT116 cells. a 600 HCT116 cells were seeded and treated with dimethyl sulfoxide (Control; Ctr), ferulic acid, resveratrol, UHA025, or combinations of these agents at 10 µM and cultured for 5 days. The pictures of spheroids were taken on day 5. Scale bar = 500 μm. b The area of the spheroids on day 5 was normalized as the relative ratio to control cells treated with dimethyl sulfoxide. Data are presented as means and standard deviations of four spheroids. The significance test was performed using the two-tailed Student’s t test. ***P < 0.001, n.s. not significant
Recently, we reported that resveratrol, as well as its derivative caffeic acid-adducted resveratrol, selectively represses the 3D proliferation of HCT116 cells, but not HKe3 cells, which were established by deleting the oncogenic mutant KRAS gene, leaving only one wild-type KRAS gene in HCT116 cells [8, 9]. This finding suggested that resveratrol and caffeic acid-adducted resveratrol specifically target an oncogenic KRAS-mediated signaling. We, therefore, examined the effect of UHA025 on the 3D proliferation of HKe3 cells. HCT116 and HKe3 cells were treated with UHA025, 5-Fu, or DMSO at 10 µM for 6 days. 5-Fu and UHA025 significantly decreased the size of both HCT116 (Fig. 4a, b) and HKe3 (Fig. 4c, d) cell spheroids in 3DC compared with the findings for control cells. However, the inhibitory effect of UHA025 in HKe3 cells was lower than that in HCT116 cells, suggesting that UHA025 partially inhibits an oncogenic KRAS-mediated signaling pathway. Furthermore, we examined the inhibitory effect of UHA025 on the 3D proliferation of MCF-7 human breast cancer cells. UHA025 decreased the size of MCF cell spheroids in 3DC in a concentration-dependent manner (IC50 = 8.61 ± 0.73 µM) (Fig. 4e, f). These results indicate that UHA025 represses the 3D proliferation of several types of cancer cells.
UHA025 inhibits the three-dimensional (3D) proliferation of HKe3 and MCF-7 cells. a, c 600 HCT116 cells (a) and 1800 HKe3 cells (c) were seeded and treated with dimethyl sulfoxide (Control; Ctr), 5-fluorouracil (5-Fu), or UHA023 at 10 µM on day 0. The pictures of spheroids were taken on day 6. Scale bar = 500 μm. b, d The area of the spheroids on day 6 was normalized as the relative ratio to control cells treated with dimethyl sulfoxide. Data are presented as means and standard deviations of four spheroids. The significance test was performed using the two-tailed Student’s t test. ***P < 0.001, **P < 0.01. e 100 MCF-7 cells were seeded and treated with UHA025 at the indicated concentrations on days 0, 2, and 4. The pictures of spheroids were taken on the indicated days. Scale bar = 500 μm. f The size of spheroids were measured on days 2, 4, and 7. The area of the spheroids was normalized as the relative ratio to control cells treated with dimethyl sulfoxide on day 2. Data are presented as means and standard deviations of four spheroids
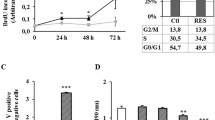
We finally attempted to reveal the mechanism by which UHA025 inhibits the 3D proliferation of HCT116 cells. We examined whether UHA025 effects on the expression of CKIs function as brakes of cell proliferation via inhibiting CDK [10]. qRT-PCR assays illustrated that UHA025 significantly increased the amount of p15 mRNA, but not those of other CKIs (Fig. 5). The expression of p16 gene was not detected in HCT116 cells. These results suggest that UHA025 induces p15 transcription, leading to the inhibition of 3D proliferation of HCT116 cells.
UHA025 significantly increases the mRNA levels of p15. 2000 HCT116 cells were seeded and treated with dimethyl sulfoxide (Control; Ctr) or UHA025 at 5 µM on day 0 and then cultured for 6 days in 3D. Quantitative reverse transcription-polymerase chain reaction was performed to detect the mRNA expression of CDK inhibitors p15, p18, p19, p21, p27, and p57. Data were normalized as the relative ratio to control cells treated with dimethyl sulfoxide. Means and standard deviations were calculated from the data for three independent experiments. The significance test was performed using the two-tailed Student’s t test. *P < 0.05, **P < 0.01, n.s. not significant
Discussion
In this research, we showed that a novel compound, UHA025, has marked inhibitory effects on the 3D proliferation of HCT116 and MCF-7 cells. Recently, we reported that resveratrol and its derivative, caffeic acid-conjugated resveratrol, selectively inhibit the 3D proliferation of HCT116 cells with activating KRAS mutation, but not HKe3 cells lacking KRAS mutation, indicating that these compounds specifically inhibit an oncogenic KRAS-mediated signaling [8, 9]. We found that UHA025 (IC50 = 5.86 µM) has a stronger inhibitory effect on HCT116 cell spheroid growth in 3DC than the caffeic acid-adducted resveratrol (IC50 = 9.52 µM) [9]. The inhibitory effect of UHA025 on the 3D proliferation of HKe3 cells was lower than that of HCT116 cells, even though UHA025 inhibited the 3D proliferation of both HCT116 and HKe3 cells. This result suggests that UHA025 partially represses an oncogenic KRAS-mediated signaling as well as resveratrol and its derivative, caffeic acid-conjugated resveratrol.
Cell proliferation is strictly regulated by cell cycle regulators such as cyclin/CDK complexes, which function as accelerators, and CKIs, which function as brakes [10]. In this study, we found that UHA025 increases the mRNA levels of the CKI p15 in HCT116 cells. However, the detailed molecular mechanism by which UHA025 induces p15 expression remains to be determined. p15 expression is mainly regulated at the transcriptional level. Several studies have shown that the transcription factor Smads directly binds to the p15 promoter and activates p15 transcription in response to TGF-beta [19,20,21]. We have also reported that a long non-coding RNA, ANRIL, binds to polycomb protein complexes, which are transcriptional repressors, and recruits them on the p15 locus, leading to the transcriptional repression of p15 [22]. Taken together, UHA025 may activate p15 transcription through these aforementioned factors. It will be important to determine the detailed molecular mechanism of p15 induction by UHA025. We also found that UHA025 does not have an effect on the expression level of p15 mRNA in MCF7 cells (data not shown), suggesting that the action mechanism of UHA025 in 3D proliferation differs among cell types and UHA025 is also involved in the regulation of genes other than p15 gene.
As UHA025 (IC50 = 5.86 µM) displayed similar inhibitory effects on the 3D proliferation of HCT116 cells as 5-Fu (IC50 = 2.39 µM), a usual anticancer drug, this novel compound could be a potential anticancer drug candidate.
References
O’Brien LE, Zegers MM, Mostov KE (2002) Building epithelial architecture: insights from three-dimensional culture models. Nat Rev Mol Cell Biol 3:531–537. https://doi.org/10.1038/nrm859
Griffith LG, Swartz MA (2006) Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol 7:211–224. https://doi.org/10.1038/nrm1858
Tsunoda T, Takashima Y, Fujimoto T, Koyanagi M, Yoshida Y, Doi K, Tanaka Y, Kuroki M, Sasazuki T, Shirasawa S (2010) Three-dimensionally specific inhibition of DNA repair-related genes by activated KRAS in colon crypt model. Neoplasia 12(5):397–404
Mukherjee S, Dudley JI, Das DK (2010) Dose-dependency of resveratrol in providing health benefits. Dose Response 8:478–500
Signorelli P, Ghidoni R (2005) Resveratrol as an anticancer nutrient: molecular basis, open questions and promises. J Nutr Biochem 16:449–466. https://doi.org/10.1016/j.jnutbio.2005.01.017
Carrizzo A, Forte M, Damato A, Trimarco V, Salzano F, Bartolo M, Maciag A, Puca AA, Vecchione C (2013) Antioxidant effects of resveratrol in cardiovascular, cerebral and metabolic diseases. Food Chem Toxicol 61:215–226. https://doi.org/10.1016/j.fct.2013.07.021
Tsunoda T, Ishikura S, Doi K, Matsuzaki H, Iwaihara Y, Shirasawa S (2014) Resveratrol induces luminal apoptosis of human colorectal cancer HCT116 cells in three-dimensional culture. Anticancer Res 34(8):4551–4555
Tsunoda T, Ishikura S, Doi K, Iwaihara Y, Hidesima H, Luo H, Hirose Y, Shirasawa S (2015) Establishment of a three-dimensional floating cell culture system for screening drugs targeting KRAS-mediated signaling molecules. Anticancer Res 35(8):4453–4459
Okamoto H, Matsukawa T, Doi S, Tsunoda T, Sawata Y, Naemura M, Ohnuki K, Shirasawa S, Kotake Y (2018) A novel resveratrol derivative selectively inhibits the proliferation of colorectal cancer cells with KRAS mutation. Mol Cell Biochem 442(1–2):39–45
Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13:1501–1512
Sherr CJ (1996) Cancer cell cycles. Science 274:1672–1677
Shirasawa S, Furuse M, Yokoyama N, Sasazuki T (1993) Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 260:85–88
Doi S, Kishi A, Matsukawa T, Nojima M, Matsui T, Yamada Y, Yamada I (2015) A novel resveratrol derivative. Japan. Patent 5703887
Kishi A, Matsukawa T, Doi S, Nojima M, Matsui T, Yamada Y, Yamada I (2015) A novel resveratrol derivative. Japan. Patent 5673091
Doi S, Matsukawa T, Kishi A, Nojima M, Matsui T, Yamada Y, Yamada I (2015) A novel resveratrol derivative. Japan. Patent 5728972
Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y (2007) pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev 21:49–54. https://doi.org/10.1101/gad.1499407
Kotake Y, Goto T, Naemura M, Inoue Y, Okamoto H, Tahara K (2017) Long noncoding RNA PANDA positively regulates proliferation of osteosarcoma cells. Anticancer Res 37:81–85. https://doi.org/10.21873/anticanres.11292
Janicke B, Hegardt C, Krogh M, Onning G, Akesson B, Cirenajwis HM, Oredsson SM (2011) The antiproliferative effect of dietary fiber phenolic compounds ferulic acid and p-coumaric acid on the cell cycle of Caco-2 cells. Nutr Cancer 63:611–622. https://doi.org/10.1080/01635581.2011.538486
Hannon GJ, Beach D (1994) p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371:257–261. https://doi.org/10.1038/371257a0
Rich JN, Zhang M, Datto MB, Bigner DD, Wang XF (1999) Transforming growth factor-beta-mediated p15(INK4B) induction and growth inhibition in astrocytes is SMAD3-dependent and a pathway prominently altered in human glioma cell lines. J Biol Chem 274:35053–35058
Feng XH, Lin X, Derynck R (2000) Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J 19:5178–5193. https://doi.org/10.1093/emboj/19.19.5178
Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30:1956–1962. https://doi.org/10.1038/onc.2010.568
Acknowledgements
We thank Keishi Tamura and Haruna Okamoto for their helpful discussions and technical assistance. This work was supported by JSPS KAKENHI Grant Number 17K07184 (to YK) and the Naito Foundation (to YK). We thank Joe Barber Jr., PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The resveratrol derivatives (UHA023, UHA024, and UHA025) used in this study were provided by UHA Mikakuto Co., Ltd. Taiji Matsukawa and Satoshi Doi are employees of UHA Mikakuto Co., Ltd. The authors have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sawata, Y., Matsukawa, T., Doi, S. et al. A novel compound, ferulic acid-bound resveratrol, induces the tumor suppressor gene p15 and inhibits the three-dimensional proliferation of colorectal cancer cells. Mol Cell Biochem 462, 25–31 (2019). https://doi.org/10.1007/s11010-019-03606-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-019-03606-8