Abstract
Context
In natural populations, gene flow often represents a key factor in determining and maintaining genetic diversity. In a worldwide context of habitat fragmentation, assessing the relative contribution of landscape features to gene flow thus appears crucial for sustainable management of species.
Objective
We addressed this issue in Mediterranean mouflon (Ovis gmelini musimon × Ovis sp.) by combining previous knowledge on behavioral ecology with landscape genetics. We also assessed how sex-specific behavioral differences translated in term of functional connectivity in both sexes.
Methods
We relied on 239 individuals genotyped at 16 microsatellite markers. We applied a model optimization approach in a causal modeling framework of landscape genetics to test for the effects on gene flow of habitat types and linear landscape features previously identified as important for movements and habitat selection in both sexes. Five resistance values were alternately assigned to these landscape characteristics leading to a comprehensive set of resistance surfaces.
Results
Isolation by resistance shaped female gene flow, supporting the central role of linear landscape features as behavioral barriers for animal movements. Conversely, no isolation by resistance was detected in males. Although a lack of statistical power cannot be discarded to explain this result, it tended to confirm that males are less influenced by landscape structures during the mating period.
Conclusions
Combining previous knowledge on behavioral ecology with results from landscape genetics was decisive in assessing functional landscape connectivity in both sexes. These results highlighted the need to perform sex-specific studies for management and conservation of dimorphic species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Animal movements are linked to various fundamental biological processes (e.g. foraging, dispersal, mating, Zeller et al. 2012) and determine in turn individual performance, gene flow and population dynamics. At the inter- (e.g. metapopulations, sensu Moilanen and Hanski 1998) as well as at the intra-population scale, maintaining connectivity between demes, habitat patches and individuals makes it possible to avoid strong impacts from stochastic processes (demographic, environmental and genetic) and thus extinction vortices (Crooks and Sanjayan 2006). Among the different elements that can impact and influence individual movements, landscape plays a crucial role (e.g. Harris and Reed 2002; Cozzi et al. 2013; Seidler et al. 2014; Zimmermann et al. 2014). Understanding the potential impact of landscape on animal movements and its consequences on individual performance, gene flow and population dynamics is thus of primary importance for conservation and sustainable management of many species and populations in the current context of habitat loss and fragmentation (Lande 1998; Fahrig 2003).
Landscape impacts on movements depend on several factors (e.g. landscape composition, structure) and among them, a fundamental element is landscape connectivity. Landscape connectivity was originally defined as “the degree to which the landscape facilitates or impedes movement among resource patches” (Taylor et al. 1993). More recently, landscape ecologists have asserted that there are actually two fundamental types of connectivity: structural and functional. Structural connectivity refers only to physical characteristics of landscapes, while functional connectivity considers the behavioral responses of organisms to this physical landscape structure (Taylor et al. 2006; Baguette and Van Dyck 2007). Indeed, physically connected habitats are not necessarily functionally connected and vice versa (e.g. unused corridors, Taylor et al. 2006; Imong et al. 2014). Landscape functional connectivity can vary between species, classes of individuals and individuals, since their behaviors and movements can be drastically different. For instance, in polygynous mammal species, males and females may exhibit very different dispersal movements (natal or reproductive) (Greenwood 1980 but see Clutton-Brock and Lukas 2012) resulting in different functional landscape connectivity values between sexes (e.g. Larroque et al. 2016b). Accordingly, in conservation and management problematics, studies on landscape impacts on movement must consider the functional connectivity of landscape for individual classes (e.g. sex).
Assessing functional landscape connectivity is commonly achieved by identifying “permeable” landscape elements that favor movement and “resistant” elements that impede it (Ray et al. 2002; Beyer et al. 2016; Panzacchi et al. 2016). The costs of moving across different landscape elements are assessed based on biological data to establish resistance surfaces (Zeller et al. 2012, 2016). Among the different biological data types that can be used to estimate the resistance of each landscape element (see Zeller et al. 2012 for a review), genetic data makes it possible to focus on estimating the resistance imposed by the landscape on gene flow. The study of gene flow is of primary concern for populations, since genetic diversity has implications for population adaptability and viability, and hence conservation (Frankham et al. 2004, Segelbacher et al. 2010). Due to the increasing need for landscape connectivity assessment and restoration, the landscape genetics approach, combining population genetics, landscape ecology and spatial statistics (Manel et al. 2003) and enabling the direct measurement of functional connectivity in landscapes, has become a central topic for researchers (Richardson et al. 2016). Unlike classic population genetics, landscape genetics explicitly accounts for landscape resistance to movements and a common approach is to relate genetic distances between individuals, groups or populations to distances that account for landscape resistance to movements (e.g. least-cost distances). Based on this framework, the impacts of landscape elements on gene flow have been illustrated in different species (e.g. Coulon et al. 2004 in roe deer (Capreolus capreolus), Cushman et al. 2006 in black bear (Ursus americanus), Larroque et al. 2016a in European pine marten (Martes martes), Barros et al. 2016 in Egyptian mongoose (Herpestes ichneumon), Olah et al. 2017 in scarlet macaws (Ara macao)).
A commonly used approach for obtaining a final resistance surface best describing resistance of landscape to gene flow is to define several alternative resistance surfaces and then determine which one best explains the spatial pattern of genetic variation observed (“Two-stage empirical approach”, see Zeller et al. 2012). The definition of such alternative resistance surfaces requires defining (i) the landscape characteristics and elements to consider (e.g. those impacting movements) and (ii) the resistance values for each of these landscape elements. The development of GPS monitoring, which enables the recording of fine-scale data on animal movements (Nathan et al. 2008) provides a unique opportunity to shed light on the two previous points by objectively quantifying the most important factors influencing movements (e.g. habitat suitability/selection or step selection studies). Nonetheless, since they do not focus on dispersal only, habitat suitability or preferences are not systematically related to landscape resistance on gene flow (e.g. Wasserman et al. 2010; Reding et al. 2013; Mateo-Sanchez et al. 2015; Roffler et al. 2016; Keeley et al. 2017) and as such might not be the most suitable way to define landscape element resistance values. As an alternative to approaches using a priori information, approaches making no a priori on resistance values are promising. However, testing every possible combination of resistance values for each of the landscape elements considered exponentially increases the number of resistance surfaces to test and concurrently the computation time. So far, and to our knowledge, few studies have used these approaches and tested all possible combinations of resistance values of all environmental parameters (but see Larroque et al. 2016a, b).
Numerous landscape elements, such as linear landscape features (e.g. Kuehn et al. 2007; Hepenstrick et al. 2012; Robinson et al. 2012; Parks et al. 2015; Wilson et al. 2015) or habitat types and topography (e.g. Epps et al. 2007; Perez-Espona et al. 2008; Shirk et al. 2010; Robinson et al. 2012; Parks et al. 2015; Roffler et al. 2016; Creech et al. 2017; Gubili et al. 2017) have been shown to influence gene flow in large herbivores and wild sheep. Elevation has even been found to shape genetic diversity and extinction risks in desert bighorn sheep (Ovis Canadensis nelson, Epps et al. 2004, 2006) supporting the idea that studying impacts of landscape on gene flow in wild sheep could be particularly relevant in this group of species for which multiple conservation (e.g. Corsican mouflon populations, Shackleton & IUCN/SSC Caprinae Specialist Group 1997, Cypriot mouflon, (Ovis orientalis ophion), Valdez 2008) and management purposes exist (e.g. trophy hunting; Harris and Pletscher 2002; Hofer 2002; detrimental effects on native animal and plant species Chapuis et al. 1994; Nogales et al. 2006; Bertolino et al. 2009). Here, we proposed to gain better knowledge on functional connectivity in these species by studying, in a population of Mediterranean mouflon (Ovis gmelini musimon × Ovis sp.), the impacts on gene flow of several landscape characteristics known to influence habitat selection and animal movements. In the studied population, animals have been shown to have marked habitat preferences determined by land cover and topography all year round, with marked differences between sexes (Marchand et al. 2015). Additionally, natural and anthropogenic linear landscape features acted as behavioral barriers and influenced movements and home-range selection of individuals (Marchand et al. 2017a). Accordingly, we evaluated the impacts on genetic differentiation in the study population of the linear landscape features and land-cover classes known to impact habitat selection and movements. We expected sex-specific effects of landscape elements on gene flow, in accordance with the sex-specific space-use observed in this dimorphic species exhibiting strong sexual segregation (Ruckstuhl and Neuhaus 2006; Marchand et al. 2015, 2017a; Bourgoin et al. 2018). More specifically, male gene flow should be much less impacted by landscape features than female, as gene flow is thought to be mostly insured by male reproductive dispersal in this population (reproductive excursions, Marchand et al. unpublished data, Portanier et al. 2017) in which females are philopatric (Dubois et al. 1992, 1994; Dupuis et al. 2002) and natal dispersal would be limited in males (Dubois et al. 1993, 1996; King and Brooks 2003). Furthermore, lower spatial and genetic structures and weaker impacts of linear landscape elements on movements have been observed in males than in females, in particular during the mating period (Dubois et al. 1993, 1996; Marchand et al. 2017a; Portanier et al. 2017) when males exhibit very different behavior as compared to the rest of the year (e.g. hypophagia, Pelletier et al. 2009, increased movements, Karns et al. 2011, Jarnemo 2011) and use coursing as a mating strategy (Bon et al. 1992; Dubois et al. 1993, 1996; Marchand et al. 2015). Since linear landscape features have been shown to have much higher impacts on movements than land cover (Marchand et al. 2017a), we also expected these landscape elements to be the principal driver of gene flow for females.
Based on 16 microsatellite loci, we tested a comprehensive set of resistance maps by using five possible resistance values for each of the nine landscape elements known to impact mouflon movements and representing natural or anthropogenic linear features, resource and/or refuge areas. We then selected for each sex the resistance surface best describing the landscape genetic resistance using a model optimization framework modified from the univariate optimization procedure of Shirk et al. (2010) and Larroque et al. (2016a, b). Then, to determine if there was a spatial genetic structure and to disentangle between isolation-by-distance (IBD, when only geographic distance plays a role) and isolation-by-resistance (IBR, when landscape resistance impacts gene flow, McRae 2006) patterns of genetic variation, we used a causal modeling framework (Cushman et al. 2006, 2013), assessing if least-cost distances were correlated to levels of genetic differentiation between individuals.
Material and methods
Study population, data collection and species
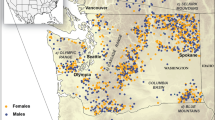
Data were collected in the low mountain Caroux-Espinouse massif (43°38′N, 2°58′E, 17,000 ha, 130-1,124 m asl, southern France, Fig. 1a). The studied Mediterranean mouflon population grew from the release of nineteen individuals between 1956 and 1960 (Garel et al. 2005; Portanier et al. 2017) and inhabits a National Hunting and Wildlife Reserve (1658 ha, 532-1,124 m above sea level; hereafter called “reserve”). Habitat is characterized by deep valleys indenting plateaus and creating a mosaic of ridges and talwegs (Marchand et al. 2017a, Fig. 1c, d). Vegetation is an irregular mosaic of deciduous and coniferous forests with open areas dominated by rocky areas and broom moorlands (Marchand et al. 2015, Table 1, Fig. 1b). Within the reserve, hunting is forbidden and recreational activities are restricted to hiking on a few main trails (Marchand et al. 2014a) but in surrounding unprotected areas, both sexes are harvested each year since 1973 (Garel et al. 2007).
Description of the Caroux-Espinouse massif where landscape resistance was investigated for Mediterranean mouflon. a Geographic location of the Caroux-Espinouse massif in France. b Land-cover classes considered in landscape genetics analyses. Grass-rich areas were classified in two classes according to elevation and slope (on plateaus with slope < 10°, elevation > 900 m or on slopes with slope > 10°, elevation < 900 m). Rocky areas were classified according to slopes (gently-sloped rocky areas if slope was < 30° and steeply-sloped rocky areas if slope was > 30°). c Linear landscape features considered in landscape genetics analyses. Anthropogenic linear features included roads, tracks, hiking trails, while natural linear features included ridges, talwegs, forest edges. d Digital elevation model represented with spatial locations of individuals as defined in the main text
Since 1974, individuals have been annually captured in the reserve. Animals are marked and biometric measurements and hair sampling are performed. Marked animals are visually monitored year-round and spatially located. Since 2003, some individuals have also been equipped with GPS collars (see Marchand et al. 2014b, 2015 for more details). All captures, handling, sampling and collaring were done according to the appropriate national laws for animal welfare and procedures were approved by the pertinent administration.
In the study population, philopatric (Dubois et al. 1992, 1994; Dupuis et al. 2002) Mediterranean mouflon ewes are sexually mature from 1.5 years of age and monotocous (twinning rate is < 3%; Garel et al. 2005). Although probably sexually mature at 2 years old (see Festa-Bianchet 2012 in bighorn sheep (Ovis canadensis)), only rams three or more years old have been observed involved in rutting activities (Bon et al. 1992, 1995). Mature males and females segregate most of the year but less during the rut as males join females to mate (Bon and Campan 1989; Bon et al. 1992; Dubois et al. 1993; Le Pendu et al. 1996; Cransac et al. 1998; Bourgoin et al. 2018). The mating system is thought to be polygynous with a few large males achieving most of the mating (see Geist 1971 for Dall’s sheep (Ovis dalli), see Jarman 1983, Hogg 1987 for bighorn sheep).
As other wild sheep species, Mediterranean mouflon are known to be poor dispersers with expected small dispersal distances, especially for females (Geist 1971; Dubois et al. 1994; Gross et al. 2000; Worley et al. 2004; Marchand et al. unpublished data). Males are nevertheless more mobile than females, with both natal dispersers and philopatric individuals observed (unknown proportions, see Dubois et al. 1993, 1996; King and Brooks 2003). Additionally, rams have been shown to perform excursions (temporary movements outside an established home range) during the rutting period (October–December, Bon et al. 1992; Marchand et al. unpublished data) that are thought to insure gene flows (Portanier et al. 2017). Both faithful to their rutting site and unfaithful males have been observed (Dubois et al. 1993, 1996; Dupuis et al. 2002; Martins et al. 2002).
Mediterranean mouflon is a wild grazer ungulate species (Cransac et al. 1997; Cazau et al. 2011; Marchand et al. 2013). Sex-specific trade-offs between habitats providing food and cover have been found in this population (Marchand et al. 2015). Typically, safe habitats (rocky or cover areas) are favored over food-rich habitats during rutting/hunting (September–February) by both sexes, and also during lambing (March–June, Bon et al. 1993) by females. Females switched to the best foraging (grass-rich areas on plateaus) habitats in summer when lambs were weaned. Additionally, linear landscape features have been found to be even more determinants for animal movements than land cover, with mouflon avoiding crossing both anthropogenic (i.e. roads, tracks and hiking trails) and natural landscape features (i.e. ridges, talwegs and forest edges, Marchand et al. 2017a). No seasonal migration has been reported in this population (mean overlap of home ranges through the year for males: 89.8 ± 2.5% and for females: 88.4 ± 4.5%; Marchand et al. 2014b and unpublished data).
Microsatellites genotyping
We used hair samples from 262 individuals trapped between 2010 and 2014, i.e. within a time period inferior to generation time of mouflon (4.21 years, Hamel et al. 2016), which should limit temporal genetic structure. Genotyping was performed by the Antagene laboratory (Limonest, France, www.antagene.com) following the procedure presented in Portanier et al. (2017). Each DNA sample was typed at 16 microsatellites markers (see Portanier et al. 2017). Analyses were performed on individuals of both sexes expected to have a fixed home range outside the rutting period, i.e. females two or more years old and males four or more years old (e.g. Dubois et al. 1992, 1993; Dupuis et al. 2002). Information about genetic diversity and Hardy–Weinberg equilibrium in the population are available in Portanier et al. (2017).
Among the genotyped samples, two outliers and five pairs of twins were identified using a correspondence analysis (CA) and the matching option in GenAlex v.6.501 (Peakall and Smouse 2006, 2012), respectively. Outliers, and one of the two twins for each pair, were randomly deleted to prevent bias in subsequent analyses. Among the remaining individuals, we considered in the following analyses 239 individuals (77 males and 162 females) with fixed home ranges, at least 13 successfully genotyped genetic markers and for which spatial locations were available. Comparison of the observed genotypes with the distribution of randomized genotypes generated with the program MICROCHECKER v.2.2.3 (Van Oosterhout et al. 2004) revealed that there were no null alleles in the data set.
Landscape genetics analyses
We used a causal modeling framework (Legendre and Troussellier 1988; Legendre 1993; Cushman et al. 2006, 2013) to distinguish between isolation by distance (IBD) and isolation by resistance (IBR) as spatial drivers of genetic differentiation in our population.
Genetic distances
Since it was especially designed to estimate genetic differentiation among individuals in continuous populations and was shown to perform well in landscape genetics studies (Shirk et al. 2017), the âr (Rousset 2000) pairwise genetic distance was calculated using SPAGeDI 1.5 software (Hardy and Vekemans 2002).
Spatial locations of mouflon
We considered genotyped individuals for which at least five spatial locations were available. GPS spatial locations (85 females and 32 males: from 5 to 8570 locations) or visual resighting (104 females and 45 males without GPS collars: from 5 to 73 locations) were averaged to assign each individual at the centroid of all its spatial locations (Fig. 1d). This centroid value was used as spatial location for each individual.
IBD model
The IBD model predicts that genetic distance between individuals increases with increasing geographic (Euclidean) distance. In the IBD model, pairwise Euclidean geographic distances (straight line, hereafter EuD) were calculated. These distance were topographically corrected (i.e. taking into account elevation changes between pixels, digital elevation model with a 25 m resolution, BD ALTI©, French National Institute of Geography, www.ign.fr, see Fig. 1d) following the method reported in Larroque et al. (2016a).
IBR models definition and optimization
The IBR model predicts that genetic distance between individuals increases with increasing resistance of the landscape between them. In IBR models, the resistance distance was calculated using least-cost pathways between each pair of individuals.
Seven land-cover classes and two types of linear structures (natural and anthropogenic) known to impact habitat selection, home range selection and movements of mouflon were considered in the different resistance surfaces modeled (Fig. 1b, c, Table 1). These habitat categories explicitly took into account slope and elevation parameters (Fig. 1b, Table 1, see Marchand et al. 2015, 2017a for details). Habitat map was derived from a SPOT satellite image and field validation in a 25 × 25 m grid (Tronchot 2008), and the digital elevation model was used to derive slope (see Marchand et al. 2015). Each pixel was characterized by the dominant habitat type (Fig. 1b). Linear landscape features were derived in accordance with Marchand et al. (2017a). Natural linear landscape features referred to ridges, talwegs and forest edges and were extracted from the digital elevation model and the BD FORÊT© (French National Institute of Geography, http://www.ign.fr). Anthropogenic linear landscape features referred to roads, tracks and hiking trails and were extracted from the BD CARTO© (French National Institute of Geography, http://www.ign.fr). A 15 m buffer was applied to linear landscape features to reinforce their size and avoid raster breaks. In order to avoid artificial map boundary effects that might affect directional choice of the least-cost algorithm and bias resistance values estimations (see Koen et al. 2010), we buffered 2 km around individual locations to define study area limits (Fig. 1d). In our case, two kilometers made it possible to obtain a circular area of 1250 ha around each individual, which is largely representative of the expected maximum area of a mouflon home range (1000 ha, see Marchand 2013). In the final map, all habitat classes were represented in similar proportions (< 10%) with the exception of deciduous forests and natural linear landscape features, which are largely more represented (24 and 23% respectively, see Fig. 1b, Online Resource 1). Missing data in the final map (see Online Resource 1) are mostly located in the external range of the study area and are due to the fact that the precise habitat map had lower extent than extents obtained after buffering of 2 km (see Fig. 1b, d).
Five varying resistance values were alternatively assigned to each of the nine landscape elements leading to 1,953,125 different possible combinations of resistance values (scenarios hereafter). Values were chosen to illustrate the case where the landscape element was not resistant to movements (resistance of 1), weakly and moderately resistant (25 and 50 respectively) and highly and totally resistant (75 and 100 respectively). In order to identify which scenario best described genetic data, we calculated least-cost distance matrices (LCD) measuring the cumulative cost (as recommended by Etherington and Holland 2013) under each scenario using the costDistance function in gdistance R package (van Etten 2017).
For each sex, we then evaluated each of the 1,953,125 scenarios based on the correlation between genetic distances âr (G) and the log transformed LCD using partial mantel tests, partialling out the log transformed Euclidean distance (G ~ log(LCD)i|log(EuD) for the ith scenario). In accordance with the model optimization procedures of Shirk et al. (2010) and Larroque et al. (2016a, b), the top scenario was identified by a unimodal peak of support in partial Mantel r values (LCDtop). When no unimodal peak of support was identified, we selected significant (two-tailed p values) scenarios having a positive Mantel r and calculated the Relative Support index (RS, Cushman et al. 2013). RS is the difference in Mantel r of the models (i) G ~ log(LCD)i|log(EuD) and (ii) G ~ log(EuD)|log(LCD)i and was introduced to prevent the high type I error rates of Mantel tests. We ranked models according to their RS and selected the scenario having the highest RS (LCDtop hereafter). In keeping with the recommendations of Cushman et al. (2013) to counterbalance the high type I error rates and to decrease the risks of finding spurious support for IBD null models while IBR is the true process, we used an alpha level of 0.01 to evaluate the significance of Mantel tests. It is especially recommended when highly correlated alternative hypotheses are confronted, like IBD and IBR are in the present study (see Online Resource 2), because Mantel-based methods are less reliable (Cushman et al. 2013; Zeller et al. 2016).
According to this lower discriminant power of causal modeling when alternative scenarios are highly correlated, we assessed the sensitivity of our results to the number of selected scenarios by calculating averaged scenarios across the first n models (see Table S3, Online Resource 3). Averaging resistance values allowed us to construct an averaged resistance surface based on the n first models and to calculate least-cost distances under this averaged model (LCDav).
All Mantel and partial Mantel tests were performed using the ecodist package in R (Goslee and Urban 2007) and 9999 permutations.
Principal driver of the genetic differentiation
Once we identified the top or the averaged scenarios among all the alternative resistance models, we performed diagnostic tests of causal modeling (Cushman et al. 2006, 2013) to distinguish between the three possible drivers of genetic differentiation in each sex (IBD, IBR and no structure). We first tested for correlations between genetic and Euclidean and resistance distances alone using simple Mantel tests (G ~ log(EuD) and G ~ log(LCD), tests “t1” and “t2” hereafter, respectively). Causal modeling then predicts that for IBR to be recognized as the principal driver of genetic differentiation, partial Mantel tests between genetic and resistance distances must be significant for G ~ log(LCD)|log(EuD) and non-significant for G ~ log(EuD)|log(LCD) (tests “t3” and “t4” hereafter, respectively).
Results
IBR model optimization and driver of the genetic differentiation
No IBD patterns were detected (tests t1, Table 2) for both sexes indicating that genetic differentiation is not determined by Euclidean distances. No unimodal peak of support for scenarios was observed in Mantel r values, neither for females nor for males (Online Resource 4). In females, we calculated the RS for the 4906 scenarios having a significant positive Mantel r (p < 0.01, mean Mantel r = 0.08, mean RS = 0.15) and selected the best one to perform causal modeling. We also performed causal modeling on the averaged scenarios (LCDav, see Table S4, Online Resource 3) and obtained qualitatively the same results indicating that results were robust. As expected, IBR was identified as the principal driver of the genetic structure since the test t3 was significant and had positive relative support (RS = 0.16), while the test t4 was non-significant (Table 2). As we expected, model optimization indicated very high resistance values of linear landscape features (natural as well as anthropogenic), coniferous forests and grass rich areas on slopes (Fig. 2a). These resistance values mean that these landscape elements are between 75 and 100 times more resistant than any landscape element having a resistance of 1. Deciduous forests and broom moorlands had relatively low resistance values while gently- and steeply-sloped rocky areas and grass-rich areas on plateaus did not oppose any resistance to female gene flow (Fig. 2a). In addition, the resistance values for steeply-sloped rocky areas and linear landscape features changed only slightly between the first top scenario and averaged values over 4906 scenarios (difference < 10%, see Fig. 2b) indicating the marked robustness of these results as compared to other habitat types and supporting the expected preeminent role of linear landscape features on female gene flow.
Resistance values obtained in the best scenario for each of the landscape elements considered in landscape genetics analyses for females (a). A resistance value of 100 refers to totally resistant landscape element while a value of 1 refers to totally permeable landscape elements. b Variation of resistance values between the first top scenario (highest RS) and the resistance values obtained after averaging across 4906 best scenarios for females Mediterranean mouflon. Variation was calculated as the absolute value of the difference: v1 − vav
In males, only one scenario had a significant positive Mantel r (p < 0.01, Mantel r = 0.14, RS = 0.26) and accordingly no scenario averaging was performed. However, no conclusion can be drawn from resistance values observed (see Online Resource 3, Table S5) in the top scenario since the conditions to conclude an effect of landscape on gene flow were not met (t3 and t4 were both non-significant, Table 2). Although relative support is positive (RS = 0.26), these results supported the expectation that landscape features had much lower effects on gene flow in males than in females.
Discussion
Landscape genetics revealed sex-specific effects of landscape on gene flow in the studied Mediterranean mouflon population. In females, the significant IBR detected revealed that the role of the landscape on genetic differentiation was mostly related to the high resistance of linear landscape features and unsafe habitats (e.g. coniferous forests) to gene flow, and the higher permeability of the habitats perceived as safe (e.g. deciduous forests and rocky areas). These results are thus in accordance with previous studies on movements and habitat selection (Marchand et al. 2015, 2017a). In contrast, no IBR was detected in males, suggesting that landscape features had limited impact on gene flow, and confirming previous results showing that landscape characteristics had less influence on movements of males during the rutting period and consequently on population spatial genetic structure (Dubois et al. 1993, 1996; Marchand et al. 2017a; Portanier et al. 2017). Combining previous knowledge on movements and habitat selection with the results from this landscape genetics approach was hence decisive in characterizing functional connectivity of both sexes in the studied population.
Landscape impacts on female movements and gene flow
In numerous animal species (e.g. Ehrlich 1961 in insects, Harris and Reed 2002 in non-migratory birds, Frantz et al. 2010 in badgers (Meles meles), Zimmermann et al. 2014 in large carnivores), linear landscape features (natural and anthropogenic) have been shown to impact movements. In the study population, these linear features are crossed less than expected all year round, although they can easily be (i.e. are behavioral barriers instead of physical barriers), with consequences on the design of individual home range (Marchand et al. 2017a). In addition, in Mediterranean mouflon as in numerous ungulate species, habitat selection is driven by a pervasive food/cover trade-off (Festa-Bianchet 1988; Dussault et al. 2005; Ciuti et al. 2009; Mabille et al. 2012; Marchand et al. 2015). During the rutting period, habitats perceived as unsafe (plateaus) are thus avoided by females while habitats perceived as safe and/or providing resources are selected (i.e. deciduous forests or rocky areas on slopes providing high visibility and low accessibility for predators, see Marchand et al. 2015). Here, we showed that these behavioral characteristics translated into consequences on gene flow and spatial genetic structure in females. Indeed, both natural and anthropogenic linear landscape features had very high resistance values, with a very strong impact on female gene flow and movements, both here and in studies of habitat selection. Furthermore, resistance values for linear features were highly consistent across scenarios, highlighting the preeminent role of such features for female movements. Similarly, unsafe habitats such as coniferous forests appeared as totally resistant while deciduous forests and rocky areas appeared as weakly and not resistant to gene flow, respectively, in the present study. Resistance values observed for grass rich areas on plateaus were nevertheless unexpected since this habitat type is avoided during the rutting period (see Marchand et al. 2015). The low resistance value observed for steeply-sloped rocky areas was almost the same across all scenarios, suggesting that this landscape feature is among the principal drivers of gene flow in females. In accordance with observations made in other wild sheep species (see Epps et al. 2007; Roffler et al. 2016; Creech et al. 2017), land cover and topography also determined how and through which habitat types female gene flow occurred in the Mediterranean mouflon population under study.
In other large herbivores species, linear landscape features have also been shown to impact movements (e.g. bison (Bison bison), Bruggeman et al. 2006, 2007, pronghorn (Antilocapra Americana), Seidler et al. 2014, red deer (Cervus elaphus), Prokopenko et al. 2017). However, most studies focused on physical barriers. While ridges imposed directional dispersal movements (individuals follow the ridges, Long et al. 2010) in male white-tailed deer (Odocoileus virginianus), to our knowledge no other study investigated the effects of such natural behavioral barriers on gene flow in large herbivores. Similarly, although anthropogenic linear landscape features have been studied much more and shown to impact gene flow in numerous ungulate species (e.g. red deer, Frantz et al. 2012, roe deer (Capreolus capreolus), Breyne et al. 2014, bighorn sheep, Creech et al. 2017), most studies referring to such features have considered highways or wide, frequently traveled roads representing physical barriers (e.g. Epps et al. 2005; Wilson et al. 2015). It was only recently shown that even unpaved roads with little traffic could impact the presence probability and space use of animals (e.g. red deer and wild boars (Sus scrofa), D’Amico et al. 2016, Mt. Graham red squirrels (Tamiasciurus hudsonicus grahamensis), Chen and Koprowski 2016). Anthropogenic linear features considered in our study were composed of one small remote road, tracks and hiking trails with relatively low human use. They nevertheless imposed a very high resistance to female gene flow, providing evidence that the behavioral limitations imposed by these elements impacted movements during the mating season.
One mechanism involved in avoidance of linear landscape features can be an increased perceived risk near these features (“landscape of fear”, Laundré et al. 2001). Indeed, females favored safe environments during the rutting period, and linear landscape features could appear as unsafe due to vegetation gaps (gap avoidance, e.g. D’Amico et al. 2016; Chen and Koprowski 2016). In addition, despite a low rate of human activity (0.40 tourist groups per hour, Martinetto et al. 1998) and the absence of hunting in the protected area (but occurring outside), behavioral responses to human disturbances and hunting have been observed in the protected area during the rutting period (Benoist et al. 2013; Marchand et al. 2014a). Human activities could thus participate in the landscape of fear (e.g. increased vigilance in moose (Alces alces) along roads, Ciuti et al. 2012) and females might avoid crossing linear landscape elements to decrease contacts with humans.
Marchand et al. (2017a) also hypothesized that linear landscape features could represent visual landmarks in cognitive maps driving animal navigation and, showed their role in the definition of the boundaries of individual home range. Since females are highly philopatric and less mobile than males (Dubois et al. 1992; Dupuis et al. 2002), the cognitive map may be another mechanism in accordance with our expectations, explaining linear landscape features avoidance. It is worth noting that in numerous ungulate species, including Mediterranean mouflon, females exhibit a marked socio-spatial structure and live in spatially separated groups (e.g. Geist 1971; Festa-Bianchet 1991; Garel et al. 2007; Portanier et al. 2017) for which boundaries can rely on linear landscape features (e.g. Laurian et al. 2008; Bartzke et al. 2015). These groups can be differentiated in terms of behaviors, body mass and fitness (e.g. Coulson et al. 1999; Pettorelli et al. 2001, 2003; Focardi et al. 2002; Garel et al. 2007), sanitary status (e.g. Altizer et al. 2003; Marchand et al. 2017b), or genetics (e.g. Nussey et al. 2005; Podgórski et al. 2014; Noble et al. 2016; Portanier et al. 2017), highlighting that behavioral separations imposed by linear landscape features between groups can have implications for populations in numerous aspects and reinforce spatial genetic structure.
Behavioral ecology and landscape genetics are complementary for males
Regarding males, no impact of landscape was detected on gene flow. A lack of statistical power due to a lower sample size for males than for females cannot be discarded to explain this result. Indeed, performing a power analysis (i.e. applying landscape genetics analyses on five randomly selected female sub-samples of 70 individuals), we evidenced that IBR signal is not systematically detected when considering male’s sample size (see Online Resource 5). On the other hand, the absence of a landscape genetic signal was expected in males and had a strong biological support. Indeed, in numerous ungulate species, males exhibit seasonal movement patterns (e.g. Pépin et al. 2009; Unterthiner et al. 2012; White et al. 2012) with increased movements during the mating period (e.g. Karns et al. 2011; Jarnemo 2011). In Mediterranean mouflon, during the rutting period, males are more mobile and prone to move toward unfamiliar areas than females (Dubois et al. 1993, 1996; Dupuis et al. 2002; Marchand et al. 2017a). They are also thought to insure most of the gene flow through reproductive excursions (Portanier et al. 2017; Marchand et al. unpublished data) leading to lower levels of spatial genetic structure (Portanier et al. 2017). Accordingly, the absence of landscape effect on gene flow in males was biologically meaningful and tended to confirm that landscape had less impact on male reproductive movements than on females.
Although habitat preferences have been highlighted for males during autumn, they are the same as female preferences (Marchand et al. 2015) and authors hypothesized that habitat selection by males during the rutting period is the consequence of female choices since Ovis species and Mediterranean mouflon use coursing as mating strategies, with males following females (Hogg 1984; Hogg and Forbes 1997; Bon et al. 1992; Coltman et al. 2002). In addition, Marchand et al. (2017a) demonstrated impacts of some linear landscape features on male movements during the rutting period, but they did not account for excursions outside home ranges since they only considered movement steps included in the 95% home range. These excursions might constitute reproductive excursions and resistance of linear landscape features measured within home ranges might therefore overestimate the genetic resistance of these landscape elements. Recent studies have indeed shown that the behavioral state (e.g. foraging, dispersing) is an important factor to take into account when studying landscape connectivity to avoid potentially erroneous conclusions about isolation of patches (Abrahms et al. 2017; Blazquez-Cabrera et al. 2016; Maiorano et al. 2017). For instance, focusing on ‘inside home range’ locations instead of on ‘outside home range’ locations (dispersal and exploratory movements) could lead to underestimating landscape functional connectivity (Blazquez-Cabrera et al. 2016). Our results might illustrate this case since the linear landscape features having an impact on male movements seemed to have no impact on male gene flow. Genetic connectivity and connectivity linked to ‘inside home range’ movements (e.g. when foraging) can be different since habitat selection aims at maximizing individuals’ fitness on their home range while reproductive dispersal movements achieve a different goal and lead individuals to modify their behavior (Larroque 2015). Mating may thus be a sufficient motivation to overcome landscape constraints to movements.
Alternatively, landscape effects on male gene flow might not yet be strong enough to lead to a detectable IBR pattern. Landguth et al. (2010) evidenced that when too few generations elapsed, landscape effects on genetic differentiation are not detectable by Mantel tests. In the Caroux-Espinouse population, only 14 generations elapsed since introduction (Garel et al. 2005). Due to higher philopatry and spatial stability (Dubois et al. 1992, 1994; Martins et al. 2002; Marchand et al. 2017a), landscape effects were expected to be stronger for females than for males, which might also explain why IBR pattern was already detectable for females while not for males.
Conclusions and perspectives
In the current context of global changes, investigating processes (e.g. gene flow) and not only patterns (e.g. spatial structure) is necessary to better predict consequences of global changes on populations and species (Plard et al. 2015; Moran et al. 2016). As illustrated here, combining previous knowledge of movements and habitat selection with the results from a landscape genetics approach focusing on gene flow was decisive to better understand functional connectivity in the studied population. Spatial approaches allowed us to focus on landscape elements known to be important for Mediterranean mouflon movements and habitat selection and the landscape genetics approach allowed us to accurately describe landscape functional connectivity.
Our findings showed that males and females can present very different resistance surfaces, highlighting the necessity of performing sex-specific landscape connectivity studies, especially in polygynous mammals exhibiting sex-biased dispersal. Our results also showed strong impacts of behavioral barriers on gene flow and spatial genetic structure and illustrated the importance of also accounting for such types of barriers when studying functional connectivity. More specifically, more studies on impacts of natural linear landscape features acting as behavioral barriers are needed. In the current context of habitat loss and fragmentation (Fahrig 2003) reducing wildlife corridors and isolating populations (Crooks and Sanjayan 2006), our results can serve to better understand spatial genetic patterns of other mouflon populations, especially endangered ones (e.g. Corsican mouflon Ovis gmelini musimon var. corsicana).
Some landscape factors constantly appeared as a constraint to gene flow in wild sheep landscape genetic studies (e.g. slope, Epps et al. 2007; Roffler et al. 2016; Creech et al. 2017, present study), but landscape resistance in one study area does not systematically correspond to what happens in another study area (e.g. Larroque et al. 2016a, Short-Bull et al. 2011). More research is thus needed to improve our understanding of landscape impacts on wild sheep gene flow. Another important point here is that, although natal dispersal of males is expected to be limited (Dubois et al. 1993, 1996; King and Brooks 2003), it can occur, and more studies on natal dispersal will be needed to fully understand gene flow in Mediterranean mouflon. In addition, using the least-cost path approach we assumed that individuals knew ahead which path will be optimal (Balkenhol et al. 2016). This hypothesis is not unrealistic for Mediterranean mouflon given their abilities to recognize familiar areas and move away from linear landscape elements before being in direct contact with (see Marchand et al. 2017a). Circuit-theory has nevertheless been applied in landscape genetics to release this assumption and could represent an interesting complementary approach to the present study. Furthermore, although being the most used (Zeller et al. 2016), landscape genetics Mantel based methods have been criticized due to inflated type I error (e.g. Legendre et al. 2015) and low discrimination power between highly correlated alternative hypotheses (Zeller et al. 2016). Here we used Mantel r and relative support (see Cushman et al. 2013) and performed scenarios’ averaging to avoid these biases. While no clear consensus has yet been reached about the best method in landscape genetics (Balkenhol et al. 2016), alternative methods to Mantel tests are promising (see Shirk et al. 2018) and applying such approaches in Mediterranean mouflon would allow to confirm our results.
Finally, landscape functional connectivity encompasses several components among which genetic connectivity but also for instance demographic or behavioral connectivity (see Lowe and Allendorf 2010). The data type which should be used to estimate landscape functional connectivity depends on the question that is addressed. For example, genetic connectivity can overlook most of the host movements potentially leading to parasite transmission across the landscape since it will not only be determined by reproductive movements but instead by several movement types (e.g. migratory movements, Conner and Miller 2004). Management strategies should thus account for all functional connectivity components, and several data types enabling the measurement of different connectivity components are thus needed to fully understand impacts of landscape on populations.
Data availability
Microsatellite and spatial data sets will be available from the Dryad Digital Repository.
References
Abrahms B, Sawyer SC, Jordan NR, McNutt JW, Wilson AM, Brashares JS (2017) Does wildlife resource selection accurately inform corridor conservation? J Appl Ecol 54:412–422
Altizer S, Nunn CL, Thrall PH, Gittleman JL, Antonovics J, Cunningham AA, Dobson AP, Ezenwa V, Jones KE, Pedersen AB, Poss M, Pulliam JRC (2003) Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu Rev Ecol Evol Syst 34:517–547
Baguette M, Van Dyck H (2007) Landscape connectivity and animal behavior: functional grain as a key determinant for dispersal. Landscape Ecol 22:1117–1129
Balkenhol N, Cushman SA, Storfer AT, Waits LP (2016) Landscape genetics: concepts, methods, applications. Wiley-Blackwell, West Sussex, UK
Barros T, Cushman SA, Carvalho J, Fonseca C (2016) Mediterranean scrubland and elevation drive gene flow of a Mediterranean carnivore, the Egyptian mongoose Herpestes ichneumon (Herpestidae). Biol J Linn Soc 120:195–209
Bartzke GS, May R, Solberg EJ, Rolandsen CM, Røskaft E (2015) Differential barrier and corridor effects of power lines, roads and rivers on moose (Alces alces) movements. Ecosphere 6:art67
Baudière A (1970) Recherches phytogéographiques sur la bordure méridionale du Massif Central français (Les Monts de l’Espinouse). Thèse de doctorat, Université de Montpellier, Montpellier, France
Benoist S, Garel M, Cugnasse JM, Blanchard P (2013) Human disturbances, habitat characteristics and social environment generate sex-specific responses in vigilance of Mediterranean mouflon. PLoS ONE 8:e82960
Bertolino S, Di Montezemolo NC, Bassano B (2009) Food-niche relationships within a guild of alpine ungulates including an introduced species. J Zool 277:63–69
Beyer HL, Gurarie E, Börger L, Panzacchi M, Basille M, Herfindal I, Van Moorter B, R. Lele S, Matthiopoulos J (2016) “You shall not pass!”: quantifying barrier permeability and proximity avoidance by animals. J Anim Ecol 85:43–53
Blazquez-Cabrera S, Gastón A, Beier P, Garrote G, Simón MÁ, Saura S (2016) Influence of separating home range and dispersal movements on characterizing corridors and effective distances. Landscape Ecol 31:2355–2366
Bon R, Campan R (1989) Social tendencies of the Corsican Mouflon Ovis ammon musimon in the Caroux-Espinouse massif (South of France). Behav Process 19:57–78
Bon R, Dardaillon M, Estevez I (1993) Mating and lambing periods as related to age of female mouflon. J Mammal 74:752–757
Bon R, Gonzalez G, Bosch MD, Cugnasse JM (1992) Ram rut-involvement in a hunted population of mouflons. Acta Theriol 37:63–71
Bon R, Recarte JM, Gonzalez G, Cugnasse JM (1995) Courtship and behavioral maturation of male mouflons. Acta Theriol 40:283–294
Bourgoin G, Marchand P, Hewison AJM, Ruckstuhl KE, Garel M (2018) Social behaviour as a predominant driver of sexual, age-dependent and reproductive segregation in Mediterranean mouflon. Anim Behav 136:87–100
Breyne P, Mergeay J, Casaer J (2014) Roe deer population structure in a highly fragmented landscape. Eur J Wildl Res 60:909–917
Bruggeman JE, Garrott RA, Bjornlie D, White PJ, Watson FGR, Borkowski JJ (2006) Temporal variability in winter travel patterns of Yellowstone bison: the effects of road grooming. Ecol Appl 16:1539–1554
Bruggeman JE, Garrott RA, White PJ, Watson FGR, Wallen R (2007) Covariates affecting spatial variability in bison travel behavior in Yellowstone National Park. Ecol Appl 17:1411–1423
Bull RAS, Cushman SA, Mace R, Chilton T, Kendall KC (2011) Why replication is important in landscape genetics: American black bear in the Rocky Mountains. Mol Ecol 20:1092–1107
Cazau M, Garel M, Maillard D (2011) Responses of heather moorland and Mediterranean mouflon foraging to prescribed-burning and cutting. J Wildl Manage 75:967–972
Chapuis JL, Boussès P, Barnaud G (1994) Alien mammals, impact and management in the French subantarctic islands. Biol Conserv 67:97–104
Chen HL, Koprowski JL (2016) Barrier effects of roads on an endangered forest obligate: influences of traffic, road edges, and gaps. Biol Conserv 199:33–40
Ciuti S, Northrup JM, Muhly TB, Simi S, Musiani M, Pitt JA, Boyce MS (2012) Effects of humans on behaviour of wildlife exceed those of natural predators in a landscape of fear. PLoS ONE 7:e50611
Ciuti S, Pipia A, Grignolio S, Ghiandai F, Apollonio M (2009) Space use, habitat selection and activity patterns of female sardinian mouflon (Ovis orientalis musimon) during the lambing season. Eur J Wildl Res 55:589–595
Clutton-Brock TH, Lukas D (2012) The evolution of social philopatry and dispersal in female mammals. Mol Ecol 21:472–492
Coltman DW, Festa-Bianchet M, Jorgenson JT, Strobeck C (2002) Age-dependent sexual selection in bighorn rams. Proc R Soc B Biol Sci 269:165–172
Conner MM, Miller MW (2004) Movement patterns and spatial epidemiology of a prion disease in mule deer population units. Ecol Appl 14:1870–1881
Coulon A, Cosson JF, Angibault JM, Cargnelutti B, Galan M, Morellet N, Petit E, Aulagnier S, Hewisson AJM (2004) Landscape connectivity influences gene flow in a roe deer population inhabiting a fragmented landscape: an individual–based approach. Mol Ecol 13:2841–2850
Coulson T, Albon S, Pilkington J, Clutton-Brock T (1999) Small scale spatial dynamics in fluctuating ungulate population. J Anim Ecol 68:658–671
Cozzi G, Broekhuis F, Mcnutt JW, Schmid B (2013) Comparison of the effects of artificial and natural barriers on large African carnivores: implications for interspecific relationships and connectivity. J Anim Ecol 82:707–715
Cransac N, Gerard JF, Maublanc ML, Pépin D (1998) An example of segregation between age and sex classes only weakly related to habitat use in mouflon sheep (Ovis gmelini). J Zool 244:371–378
Cransac N, Valet G, Cugnasse JM, Rech J (1997) Seasonal diet of mouflon (Ovis gmelini): comparison of population sub-units and sex-age classes. La Terre la Vie 52:21–36
Creech TG, Epps CW, Landguth EL, Wehausen JD, Crowhurst RS, Holton B, Monello RJ (2017) Simulating the spread of selection-driven genotypes using landscape resistance models for desert bighorn sheep. PLoS ONE 12:e0176960
Crooks KR, Sanjayan M (2006) Connectivity conservation: maintaining connections for nature. In: Crooks KR, Sanjayan M (eds) Connectivity conservation. Cambridge University Press, Cambridge, pp 1–28
Cugnasse JM (1990) Inventaire faunistique du massif du Caroux et des monts de l’Espinouse (Hérault):(mammifères, oiseaux, reptiles et batraciens). Office national de la chasse, Station d’étude sur la faune des milieux méditerranéens, Paris, France
Cushman SA, McKelvey KS, Hayden J, Schwartz M (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486–499
Cushman SA, Wasserman TN, Landguth EL, Shirk AJ (2013) Re-evaluating causal modeling with mantel tests in landscape genetics. Diversity 5:51–72
D’Amico M, Périquet S, Román J, Revilla E (2016) Road avoidance responses determine the impact of heterogeneous road networks at a regional scale. J Appl Ecol 53:181–190
Dubois M, Bon R, Cransac N, Maublanc ML (1994) Dispersal patterns of Corsican mouflon ewes: importance of age and proximate influences. Appl Anim Behav Sci 42:29–40
Dubois M, Gerard J, Maublanc ML (1992) Seasonal movements of females Corsican mouflon (Ovis ammon) in a Mediterranean mountain range, southern France. Behav Process 26:155–166
Dubois M, Khazraïe K, Guilhem C, Maublanc ML, Le Pendu Y (1996) Philopatry in mouflon rams during the rutting season: psycho-ethological determinism and functional consequences. Behav Process 35:93–100
Dubois M, Quenette P-Y, Bideau E (1993) Seasonal range use by European mouflon rams in medium altitude mountains. Acta Theriol 38:185–198
Dupuis J, Badia J, Maublanc ML, Bon R (2002) Survival and spatial fidelity of mouflon (Ovis gmelini): a bayesian analysis of an age-dependent capture-recapture model. J Agric Biol Environ Stat 7:277–298
Dussault C, Ouellet J, Courtois R, Huot J, Breton L, Jolicoeur H (2005) Linking moose habitat selection to limiting factors. Ecography 28:619–628
Ehrlich PR (1961) Intrinsic barriers to dispersal in checkerspot butterfly. Science 134:108–109
Epps CW, McCullough DR, Wehausen JD, Bleich VC, Rechel JL (2004) Effects of climate change on population persistence of desert-dwelling mountain sheep in California. Conserv Biol 18:102–113
Epps CW, Palsbøll PJ, Wehausen JD, Roderick GK, McCullough DR (2006) Elevation and connectivity define genetic refugia for mountain sheep as climate warms. Mol Ecol 15:4295–4302
Epps CW, Palsboll PJ, Wehausen JD, Roderick GK, Ramey RR, McCullough DR (2005) Highways block gene flow and cause a rapid decline in genetic diversity of desert bighorn sheep. Ecol Lett 8:1029–1038
Epps CW, Wehausen JD, Bleich VC, Torres SG, Brashares JS (2007) Optimizing dispersal and corridor models using landscape genetics. J Appl Ecol 44:714–724
Etherington TR, Penelope Holland E (2013) Least-cost path length versus accumulated-cost as connectivity measures. Landscape Ecol 28:1223–1229
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Festa-Bianchet M (1988) Seasonal range selection in bighorn sheep: conflicts between forage quality, forage quantity, and predator avoidance. Oecologia 75:580–586
Festa-Bianchet M (1991) The social system of bighorn sheep: grouping patterns, kinship and female dominance rank. Anim Behav 42:71–82
Festa-Bianchet M (2012) The cost of trying: weak interspecific correlations among life-history components in male ungulates. Can J Zool 90:1072–1085
Focardi S, Raganella Pelliccioni E, Petrucco R, Toso S (2002) Spatial patterns and density dependence in the dynamics of a roe deer (Capreolus capreolus) population in central Italy. Oecologia 130:411–419
Frankham R, Ballou JD, Briscoe DA (2004) A primer of conservation genetics. Cambridge University Press, New York
Frantz AC, Bertouille S, Eloy MC, Licoppe A, Chaumont F, Flamand MC (2012) Comparative landscape genetic analyses show a Belgian motorway to be a gene flow barrier for red deer (Cervus elaphus), but not wild boars (Sus scrofa). Mol Ecol 21:3445–3457
Frantz AC, Pope LC, Etherington TR, Wilson GJ, Burke T (2010) Using isolation-by-distance-based approaches to assess the barrier effect of linear landscape elements on badger (Meles meles) dispersal. Mol Ecol 19:1663–1674
Garel M, Cugnasse JM, Loison A, Gaillard JM, Vuiton C, Maillard D (2005) Monitoring the abundance of mouflon in South France. Eur J Wildl Res 51:69–76
Garel M, Cugnasse JM, Maillard D, Gaillard JM, Hewison AJM, Dubray D (2007) Selective harvesting and habitat loss produce long-term life history changes in a mouflon population. Ecol Appl 17:1607–1618
Geist V (1971) Mountain sheep: a study in behavior and evolution. University of Chicago Press, Chicago, Illinois
Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19
Greenwood PJ (1980) Mating systems, philopatry and dispersal in birds and mammals. Anim Behav 28:1140–1162
Gross JE, Singer FJ, Moses ME (2000) Effects of disease, dispersal and area on bighorn sheep restoration. Restor Ecol 8:25–37
Gubili C, Mariani S, Weckworth BV, Galpern P, McDevitt AD, Hebblewhite M, Nickel B, Musiani M (2017) Environmental and anthropogenic drivers of connectivity patterns: a basis for prioritizing conservation efforts for threatened populations. Evol Appl 10:199–211
Hamel S, Gaillard JM, Yoccoz NG, Albon S, Côté SD, Craine JM, Festa-Bianchet M, Garel M, Lee P, Moss C, Nussey DH, Pelletier F, Stien A, Tveraa T (2016) Cohort variation in individual body mass dissipates with age in large herbivores. Ecol Monogr 86:517–543
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Harris RB, Pletscher DH (2002) Incentives toward conservation of argali Ovis ammon: a case study of trophy hunting in Western China. Oryx 36:373–381
Harris RJ, Reed JM (2002) Behavioral barriers to non-migratory movements of birds. Ann Zool Fenn 39:275–290
Hepenstrick D, Thiel D, Holderegger R, Gugerli F (2012) Genetic discontinuities in roe deer (Capreolus capreolus) coincide with fenced transportation infrastructure. Basic Appl Ecol 13:631–638
Hofer D (2002) The lion’s share of the hunt: Trophy hunting and conservation: A review of the legal eurasian tourist hunting market and trophy trade under cites. Technical report, TRAFFIC Europe, Brussels, Belgium
Hogg JT (1984) Mating in bighorn sheep: multiple creative male strategies. Science 225:526–529
Hogg JT (1987) Intrasexual competition and mate choice in rocky mountain bighorn sheep. Ethology 75:119–144
Hogg JT, Forbes SH (1997) Mating in bighorn sheep: frequent male reproduction via a high-risk “unconventional” tactic. Behav Ecol Sociobiol 41:33–48
Imong I, Robbins MM, Mundry R, Bergl R, Kühl HS (2014) Informing conservation management about structural versus functional connectivity: a case-study of Cross River gorillas. Am J Primatol 76:978–988
Jarman P (1983) Mating system and sexual dimorphism in large, terrestrial, mammalian herbivores. Biol Rev 58:485–520
Jarnemo A (2011) Male red deer (Cervus elaphus) dispersal during the breeding season. J Ethol 29:329–336
Karns GR, Lancia RA, DePerno CS, Conner MC (2011) Investigation of adult male white-tailed deer excursions outside their home range. Southeast Nat 10:39–52
Keeley ATH, Beier P, Keeley BW, Fagan ME (2017) Habitat suitability is a poor proxy for landscape connectivity during dispersal and mating movements. Landsc Urban Plan 161:90–102
King R, Brooks SP (2003) Survival and spatial fidelity of mouflons: the effect of location, age, and sex. J Agric Biol Environ Stat 8:486–513
Koen EL, Garroway CJ, Wilson PJ, Bowman J (2010) The effect of map boundary on estimates of landscape resistance to animal movement. PLoS ONE 5:1–8
Kuehn R, Hindenlang KE, Holzgang O, Senn J, Stoeckle B, Sperisen C (2007) Genetic effect of transportation infrastructure on roe deer populations (Capreolus capreolus). J Hered 98:13–22
Lande R (1998) Anthropogenic, ecological and genetic factors in extinction and conservation. Res Popul Ecol 40:259–269
Landguth EL, Cushman SA, Schwartz MK, McKelvey KS, Murhpy M, Luikart G (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191
Larroque J (2015) Same looks, different ecology: ecological and genetic insights on two syntopic mustelids species, the European Pine marten (Martes martes) and the Stone marten (Martes foina). Thèse de doctorat, Université de Lyon, Lyon, France
Larroque J, Ruette S, Vandel J-M, Devillard S (2016a) Divergent landscape effects on genetic differentiation in two populations of the European pine marten (Martes martes). Landscape Ecol 31:517–531
Larroque J, Ruette S, Vandel JM, Queney G, Devillard S (2016b) Age and sex-dependent effects of landscape cover and trapping on the spatial genetic structure of the stone marten (Martes foina). Conserv Genet 17:1293–1306
Laundré JW, Hernández L, Altendorf KB (2001) Wolves, elk, and bison: reestablishing the “landscape of fear” in Yellowstone National Park, U.S.A. Can J Zool 79:1401–1409
Laurian C, Dussault C, Ouellet J-P, Courtois R, Poulin M, Breton L (2008) Behavior of moose relative to a road network. J Wildl Manag 72:1550–1557
Le Pendu Y, Maublanc ML, Briedermann L, Dubois M (1996) Spatial structure and activity in groups of Mediterranean mouflon (Ovis gmelini). A comparative study. Appl Anim Behav Sci 46:201–216
Legendre P (1993) Spatial autocorrelation: trouble or new paradigm? Ecology 74:1659–1673
Legendre P, Fortin MJ, Borcard D (2015) Should the Mantel test be used in spatial analysis? Methods Ecol Evol 6:1239–1247
Legendre P, Troussellier M (1988) Aquatic heterotrophic bacteria: modeling in the presence of spatial autocorrelation. Limnol Oceanogr 33:1055–1067
Long ES, Diefenbach DR, Wallingford BD, Rosenberry CS (2010) Influence of roads, rivers, and mountains on natal dispersal of white-tailed deer. J Wildl Manag 74:1242–1249
Lowe WH, Allendorf FW (2010) What can genetics tell us about population connectivity? Mol Ecol 19:3038–3051
Mabille G, Dussault C, Ouellet JP, Laurian C (2012) Linking trade-offs in habitat selection with the occurrence of functional responses for moose living in two nearby study areas. Oecologia 170:965–977
Maiorano L, Boitani L, Chiaverini L, Ciucci P (2017) Uncertainties in the identification of potential dispersal corridors: the importance of behaviour, sex, and algorithm. Basic Appl Ecol 21:66–75
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Marchand P (2013) Déterminants spatio-temporels de la sélection de l’habitat chez le mouflon méditerranéen Ovis gmelini musimon × Ovis sp. Thèse de doctorat, Université de Grenoble, Grenoble, France
Marchand P, Freycon P, Herbaux J, Game Y, Toïgo C, Gilot-fromont E, Rossi S, Hars J (2017a) Sociospatial structure explains marked variation in brucellosis seroprevalence in an Alpine ibex population. Sci Rep 7:1–12
Marchand P, Garel M, Bourgoin G, Dubray D, Maillard D, Loison A (2014a) Impacts of tourism and hunting on a large herbivore’s spatio-temporal behavior in and around a French protected area. Biol Conserv 177:1–11
Marchand P, Garel M, Bourgoin G, Dubray D, Maillard D, Loison A (2015) Coupling scale-specific habitat selection and activity reveals sex-specific food/cover trade-offs in a large herbivore. Anim Behav 102:169–187
Marchand P, Garel M, Bourgoin G, Duparc A, Dubray D, Maillard D, Loison A (2017b) Combining familiarity and landscape features helps break down the barriers between movements and home ranges in a non-territorial large herbivore. J Anim Ecol 86:371–383
Marchand P, Garel M, Bourgoin G, Michel P, Maillard D, Loison A (2014b) Habitat-related variation in carcass mass of a large herbivore revealed by combining hunting and GPS data. J Wildl Manag 78:657–670
Marchand P, Redjadj C, Garel M, Cugnasse JM, Maillard D, Loison A (2013) Are mouflon Ovis gmelini musimon really grazers? A review of variation in diet composition. Mamm Rev 43:275–291
Martinetto K, Cugnasse J, Gilbert Y (1998) La cohabitation du mouflon Méditerranéen (Ovis gmelini musimon x Ovis sp.) et des touristes dans le massif du Caroux-Espinouse (Hérault). Gibier Faune Sauvage 15:905–919
Martins AG, Netto NT, Aulagnier S, Borges A, Dubois M, Vicente L, Gerard J-F, Maublanc ML (2002) Population subdivision among mouflon sheep (Ovis gmelini) ewes and ranging behaviour of rams during the rut. J Zool 258:27–37
Mateo-Sánchez MC, Balkenhol N, Cushman SA, Pérez T, Domínguez A, Saura S (2015) A comparative framework to infer landscape effects on population genetic structure: are habitat suitability models effective in explaining gene flow? Landscape Ecol 30:1405–1420
McRae BH (2006) Isolation by resistance. Evolution 60:1551–1561
Moilanen A, Hanski I (1998) Metapopulation dynamics: effects of habitat quality and landscape structure. Ecology 79:2503–2515
Moran EV, Hartig F, Bell DM (2016) Intraspecific trait variation across scales: implications for understanding global change responses. Glob Chang Biol 22:137–150
Nathan R, Getz WM, Revilla E, Holyoak M, Kadmon R, Saltz D, Smouse PE (2008) A movement ecology paradigm for unifying organismal movement research. PNAS 105:19052–19059
Noble CW, Bono JM, Pigage HK, Hale DW, Pigage JC (2016) Fine-scale genetic structure in female mule deer (Odocoileus hemionus). West North American Nat 76:417–426
Nogales M, Rodriguez-Luengo JL, Marrero P (2006) Ecological effects and distribution of invasive non-native on the Canary Islands. Mamm Rev 36:49–65
Nussey DH, Coltman DW, Coulson T, Kruuk LEB, Donald A, Morris SJ, Clutton-Brock TH, Pemberton J (2005) Rapidly declining fine-scale spatial genetic structure in female red deer. Mol Ecol 14:3395–3405
Olah G, Smith AL, Asner GP, Brightsmith DJ, Heinsohn RG, Peakall R (2017) Exploring dispersal barriers using landscape genetic resistance modelling in scarlet macaws of the Peruvian Amazon. Landscape Ecol 32:445–456
Panzacchi M, Van Moorter B, Strand O, Saerens M, Kivimäki I, St. Clair CC, Herfindal I, Boitani L (2016) Predicting the continuum between corridors and barriers to animal movements using Step Selection Functions and Randomized Shortest Paths. J Anim Ecol 85:32–42
Parks LC, Wallin DO, Cushman SA, McRae BH (2015) Landscape-level analysis of mountain goat population connectivity in Washington and southern British Columbia. Conserv Genet 16:1195–1207
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pelletier F, Mainguy J, Côté SD (2009) Rut-induced hypophagia in male bighorn sheep and mountain goats: foraging under time budget constraints. Ethology 115:141–151
Pépin D, Morellet N, Goulard M (2009) Seasonal and daily walking activity patterns of free-ranging adult red deer (Cervus elaphus) at the individual level. Eur J Wildl Res 55:479–486
Perez-Espona S, Perez-Barberia FJ, Mcleod JE, Jiggins CD, Gordon IJ, Pemberton JM (2008) Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol Ecol 17:981–996
Pettorelli N, Dray S, Gaillard JM, Chessel D, Duncan P, Illius A, Guillon N, Klein F, Van Laere G (2003) Spatial variation in springtime food resources influences the winter body mass of roe deer fawns. Oecologia 137:363–369
Pettorelli N, Gaillard JM, Duncan P, Ouellet JP, Van Laere G (2001) Population density and small-scale variation in habitat quality affect phenotypic quality in roe deer. Oecologia 128:400–405
Plard F, Gaillard J, Coulson T, Delorme D, Warnant C, Michallet J, Tuljapurkar S, Krishnakumar S, Bonenfant C, Lyon D, Lyon F-, De Biom L (2015) Quantifying the influence of measured and unmeasured individual differences on demography. J Anim Ecol 84:1434–1445
Podgórski T, Scandura M, Jedrzejewska B (2014) Next of kin next door—philopatry and socio-genetic population structure in wild boar. J Zool 294:190–197
Portanier E, Garel M, Devillard S, Marchand P, Andru J, Maillard D, Bourgoin G (2017) Introduction history overrides social factors in explaining genetic structure of females in Mediterranean mouflon. Ecol Evol 7:1–12
Prokopenko CM, Boyce MS, Avgar T (2017) Characterizing wildlife behavioural responses to roads using integrated step selection analysis. J Appl Ecol 54:470–479
Ray N, Lehmann A, Joly P (2002) Modeling spatial distribution of amphibian populations: a GIS approach based on habitat matrix permeability. Biodivers Conserv 11:2143–2165
Reding DM, Cushman SA, Gosselink TE, Clark WR (2013) Linking movement behavior and fine-scale genetic structure to model landscape connectivity for bobcats (Lynx rufus). Landscape Ecol 28:471–486
Richardson JL, Brady SP, Wang IJ, Spear SF (2016) Navigating the pitfalls and promise of landscape genetics. Mol Ecol 25:849–863
Robinson SJ, Samuel MD, Lopez DL, Shelton P (2012) The walk is never random: subtle landscape effects shape gene flow in a continuous white-tailed deer population in the Midwestern United States. Mol Ecol 21:4190–4205
Roffler GH, Schwartz MK, Pilgrim KL, Talbot SL, Sage GK, Adams LG, Luikart G (2016) Identification of landscape features influencing gene flow: how useful are habitat selection models? Evol Appl 9:805–817
Rousset F (2000) Genetic differentiation between individuals. J Evol Biol 13:58–62
Ruckstuhl KE, Neuhaus P (2006) Sexual segregation in vertebrates - ecology of the two sexes. Cambridge University Press, Cambridge
Segelbacher G, Cushman SA, Epperson BK, Fortin MJ, Francois O, Hardy OJ, Holderegger R, Taberlet P, Waits LP, Manel S (2010) Applications of landscape genetics in conservation biology: concepts and challenges. Conserv Genet 11:375–385
Seidler RG, Long RA, Berger J, Bergen S, Beckmann JP (2014) Identifying impediments to long-distance mammal migrations. Conserv Biol 29:99–109
Shackleton, DM, IUCN/SSC Caprinae Specialist Group (1997) Wild sheep and goats and their relatives: status survey and conservation action plan for Caprinae. IUCN, Gland, Switzerland and Cambridge
Shirk AJ, Landguth EL, Cushman SA (2017) A comparison of individual-based genetic distance metrics for landscape genetics. Mol Ecol Resour 17:1308–1317
Shirk AJ, Landguth EL, Cushman SA (2018) A comparison of regression methods for model selection in individual-based landscape genetic analysis. Mol Ecol Resour 18:55–67
Shirk AJ, Wallin DO, Cushman SA, Rice CG, Warheit KI (2010) Inferring landscape effects on gene flow: a new model selection framework. Mol Ecol 19:3603–3619
Taylor PD, Fahrig L, Henein K, Merriam G (1993) Connectivity is a vital element of landscape structure. Oikos 68:571–573
Taylor P, Fahrig L, With K (2006) Landscape connectivity: a return to the basics. In: Crooks KR, Sanjayan M (eds) Connectivity conservation. Cambridge University Press, Cambridge, pp 29–43
Tronchot M (2008) Cartographie des habitats du massif du Caroux-Espinouse (Hérault) à partir de photos aeriennes de 2005. Technical report. Groupement d’Interêt Environnemental et Cynégétique du Caroux-Espinouse - Office National de la Chasse et de la Faune Sauvage, Paris, France
Unterthiner S, Ferretti F, Rossi L, Lovari S (2012) Sexual and seasonal differences of space use in Alpine chamois. Ethol Ecol Evol 24:257–274
Valdez R (2008) Ovis orientalis. The IUCN Red List of Threatened Species 2008: e.T15739A5076068
van Etten J (2017) R Package gdistance: distances and routes on geographical grids. J Stat Softw 76:1–21
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wasserman TN, Cushman SA, Schwartz MK, Wallin DO (2010) Spatial scaling and multi-model inference in landscape genetics: Martes americana in northern Idaho. Landscape Ecol 25:1601–1612
White KS, Gregovich DP, Barten NL, Scott R (2012) Moose population ecology and habitat use along the Juneau Access road corridor, Alaska. Final wildlife research report. ADF&G/DWC/WRR-2012-03. Alaska Department of Fish and Game, Juneau, AK. USA
Wilson RE, Farley SD, McDonough TJ, Talbot SL, Barboza PS (2015) A genetic discontinuity in moose (Alces alces) in Alaska corresponds with fenced transportation infrastructure. Conserv Genet 16:791–800
Worley K, Strobeck C, Arthur A, Carey J, Schwantje H, Veitch A, Coltman DW (2004) Population genetic structure of North American thinhorn sheep (Ovis dalli). Mol Ecol 13:2545–2556
Zeller KA, Creech TG, Millette KL, Crowhurst RS, Long RA, Wagner HH, Balkenhol N, Landguth EL (2016) Using simulations to evaluate Mantel-based methods for assessing landscape resistance to gene flow. Ecol Evol 6:4115–4128
Zeller KA, McGarigal K, Whiteley AR (2012) Estimating landscape resistance to movement: a review. Landscape Ecol 27:777–797
Zimmermann B, Nelson L, Wabakken P, Sand H, Liberg O (2014) Behavioral responses of wolves to roads: scale-dependent ambivalence. Behav Ecol 25:1353–1364
Acknowledgements
We warmly thank all the professionals from the Office National de la Chasse et de la Faune Sauvage (Service Départemental 34, Jeanne Duhayer and Christian Itty) and numerous trainees for their technical support in trapping, tagging, sampling, and monitoring VHF and GPS-collared mouflons. We would like to thank Dominique Dubray for his involvement in the deployment of GPS collars, all the technical personnel of LBBE for helping with laboratory steps of the study, and the Antagene laboratory (Limonest, France, www.antagene.com) for DNA extraction and genotyping. We would also like to thank Elodie Bonneau and Julie Andru for the first laboratory and genetic analyses, Patrick James and the James lab members for helpful comments on the first draft of the manuscript, and Denise Mirat for checking the English. We also gratefully acknowledge IN2P3 and the CC LBBE/PRABI for providing computer resources, and the bioinformatics team of LBBE for their advice on computational optimization of scripts. Finally, we thank two anonymous referees for helpful comments on the manuscript. This research project and E. Portanier’s collaboration were funded by the Office National de la Chasse et de la Faune Sauvage, Vetagro Sup and LBBE.
Funding
This study was funded by the Office National de la Chasse et de la Faune Sauvage, VetagroSup and Laboratoire de Biométrie et de Biologie Evolutive (Grant Numbers 2013/20/6171 and 2015/14/6171).
Author information
Authors and Affiliations
Contributions
EP, MG, PM, DM, GB and SD conceptualized and designed the research. EP, JL, MG, PM and SD conducted data analyses. All authors contributed in interpreting the results and writing the paper.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Portanier, E., Larroque, J., Garel, M. et al. Landscape genetics matches with behavioral ecology and brings new insight on the functional connectivity in Mediterranean mouflon. Landscape Ecol 33, 1069–1085 (2018). https://doi.org/10.1007/s10980-018-0650-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-018-0650-z