Abstract
Covariate analysis in population pharmacokinetics is key for adjusting doses for patients. The main objective of this work was to compare the adequacy of various modeling approaches on covariate clinical relevance decision-making. The full model, stepwise covariate model (SCM) and SCM+ PsN algorithms were compared in a clinical trial simulation of a 383-patient population pharmacokinetic study mixing rich and sparse designs. A one-compartment model with first-order absorption was used. A base model including a body weight effect on CL/F and V/F and a covariate model including 4 additional covariates-parameters relationships were simulated. As for forest plots, ratios between covariates at a specific value and that of a typical individual were calculated with their 90% confidence interval (CI90) using standard errors. Covariates on CL, V and KA were considered relevant if their CI90 fell completely outside the reference area [0.8–1.2]. All approaches provided unbiased covariate ratio estimates. For covariates with a simulated effect, the 3 approaches correctly identify their clinical relevance. However, significant covariates were missed in up to 15% of cases with SCM/SCM+. For covariate with no simulated effects, the full model mainly identified them as non-relevant or with insufficient information while SCM/SCM+ mainly did not select them. SCM/SCM+ assume that non-selected covariates are non-relevant when it could be due to insufficient information, whereas the full model does not make this assumption and is faster. This study must be extended to other methods and completed by a more complex high-dimensional simulation framework.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pharmacokinetics (PK) modeling aims at describing the dynamics of drug concentration over time. One of the main objectives of population PK analysis is to identify and quantify the sources of variability between individuals, which may be due to intrinsic or extrinsic factors, known as covariates. The analysis of covariates is a key step in population PK modeling and more broadly in drug development as it allows the drug dose to be adjusted for patients. For example, specific groups of the population may be under-exposed and therefore not benefit from the expected drug efficacy, or may be over-exposed, and therefore may present an increased risk of toxicity. Model predictions for different values of the covariate can also be performed to investigate the compound PK behavior under new conditions (i.e. interpolations or extrapolations). One frequent example is the prediction of PK in pediatric patients extrapolating from a population PK model built from adult data and taking into account the influence of age-varying covariates such as the body size or organ maturation, as a first insight to guide the dose investigation in pediatric trials [1].
In population PK modeling, the first step is to develop a base model, which is built by focusing mainly on the structural and statistical parts. This model is often covariate-free, but in some cases it may be necessary to include important covariates, like a strong known effect of formulation or body weight, to stabilize it. Once the base model is developed, the set of investigated covariate-parameter relationships need to be defined based on scientific or clinical interest, mechanistic plausibility and prior knowledge related to available covariates. When it is done, covariate model building can be performed using the base model. In pharmacometrics, there are several covariate model building methods which can be divided into 2 main categories: the covariate selection methods, which study the effect of the covariate-parameter relationships selected from the investigated set, and the full modeling methods, which study the effect of all the covariate-parameter relationships of the investigated set. Covariate selection methods comprise the most commonly used stepwise methods [2,3,4] (including stepwise covariate model (SCM) [5, 6] algorithm and its enhanced version SCM+ [7] implemented in Perl-speaks-NONMEM (PsN) [8] and the conditional sampling use for stepwise approach based on correlation tests (COSSAC) algorithm [9] implemented in Monolix) as well as other methods such as the generalized additive model (GAM) method [10], the least absolute shrinkage and selection operator (LASSO) [11], the stochastic approximation for model building algorithm (SAMBA) [12] and machine learning derived methods [13] (including random forest [14], support vector machine (SVM) [14] and genetic algorithm (GA) [15]). Full modeling methods comprise the full fixed effect modeling method [16, 17] (commonly denoted full model) and the full random effect modeling (FREM) method [18]. The health authorities’ recommendations on covariate model building remains fairly broad. In the population PK guidance published in 2020 by the U.S. food and drug administration (FDA) it is stated that “covariate analysis can be performed based on several approaches or their possible combinations (e.g., stepwise covariate analysis, full covariate model approach, the LASSO)” [19], as long as the choice of the method used is justified.
To date, studies in the literature comparing different covariate model building methods have focused mainly on the performance of detecting significant covariates and the accuracy of their estimation. Ribbing et al. (2007) [11] developed a LASSO algorithm in PsN and showed that it outperformed the PsN’s SCM algorithm in predicting covariate models on small datasets by examining the effect of data size, number of covariates and starting model. Sibieude et al. (2021) [14] compared machine learning approaches (random forest and SVM) with PsN’s SCM algorithm and Monolix’s COSSAC algorithm, in terms of covariate detection performance by varying the number of covariates, the correlation and the effect size. Svensson et al. (2022) [7] demonstrated that the number of runs and objective function evaluations were considerably reduced with PsN’s SCM+ algorithm compared to PsN’s SCM algorithm, and that both algorithms performed equally on covariate detection performance (which can be improved by adding a stage-wise filtering stage). Yngman et al. (2022) [20] compared the FREM algorithm newly implemented in PsN with the full model approach and showed that they both performed equally well on covariate effects estimation accuracy using different covariate-parameter relationships, covariate distribution and level of correlation. Finally, Amann et al. (2023) [21] found that using PsN’s FREM algorithm followed by a backward elimination procedure improved covariate identification (higher power), estimates accuracy and precision compared to PsN’s SCM algorithm, particularly for sparse datasets, by varying the number of individuals, the covariate correlation, the covariate effect sizes; however, without backward elimination, FREM produced imprecise estimates for sparse data.
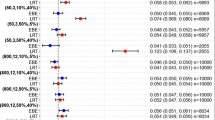
Going as far as quantifying the clinical relevance of covariates is key to perform dose adjustment for patients. In this paper, the term clinical relevance will be used to describe the impact of covariate on the PK only. To our knowledge, no simulation study has been carried out to assess the impact of the different covariates model building methods on covariate clinical relevance using a forest plot based approach. Indeed, to quantify the impact of covariates on drug exposure, health authorities [19, 22] recommend including forest plots [23, 24] in submission packages. These graphs visualize the impact of covariates on parameters of interest, such as primary PK parameters like clearance or volume of distribution, or alternatively on secondary PK parameters such as area under the curve (AUC) or peak concentration (\({C}_{max}\)). For a given value or category of a covariate of interest, the ratio corresponding to a change in parameter value relative to a reference value and its associated 90% confidence interval (CI) are represented. In most cases, the reference value corresponds to the characteristics of the typical individual in the population, however other specific values of interest may also be used. No change from the reference value is materialized by a line at 1 associated with a reference area, often [0.80, 1.20] for primary PK parameters (corresponding to a change of more or less 20% relative to the reference) or [0.80, 1.25] for secondary parameters i.e. AUC or \({C}_{max}\) (coming from bioequivalence testing [19, 25]). To calculate the 90% CI of the ratio, its uncertainty is required i.e. either the standard error (SE), or by using other methods such as samplings in the variance-covariance matrix, bootstrap [26] or sampling-importance resampling (SIR) [27]. The relevance of covariate effects can be inferred from the forest plots depending on the position of the covariate ratio 90% CI relative to the reference line and reference area, as proposed notably in the oncology field to demonstrate treatment effect heterogeneity in different subgroups [28, 29]. Consequently, a precise and accurate estimation of covariate ratios and their associated uncertainty is critical.
The main objective of this work was to compare the appropriateness of different modeling approaches for making decisions on the clinical relevance of covariates. In addition, this work compares the ratio estimates and their corresponding 90% CI. For this study, SCM [5, 6] and SCM+ [7] algorithms implemented in PsN, belonging to the stepwise covariate selection methods, were used and compared to the full model [16, 17], belonging to the full modeling methods.
SCM algorithm consists of a forward iterative loop followed by a backward iterative loop [5, 6]. In the first step of the forward process, each covariate-parameter relationship of the investigated set is univariately added to the starting model i.e. the base model. All the generated models are compared to the starting model and the one that induces the greatest reduction in log-likelihood is selected, if the reduction is significant according to a forward p-value or the algorithm stops. The selected model is defined as the new starting model and the second step begins. The forward process continues step by step until there are no more significant covariate-parameter relationships according to the defined forward p-value. After the forward process, the first step of the backward process begins. Each of the covariate-parameter relationships included in the starting model, i.e. the final forward model, are removed univariately. The model that induces the smallest log-likelihood increase is then selected, if the increase is non-significant according to a backward p-value or the algorithm stops. The selected model considered is defined as the new starting model and the second step begins. The backward process continues step by step until there are no more non-significant covariate-parameter relationships according to the defined backward p-value. The last model obtained corresponds to the final model selected by SCM. SCM+ is an enhanced version of SCM [7]. The set of investigated covariate-parameter relationships is reduced prior to the forward iterative loop. During the first step of the forward process, covariate-parameter relationships that do not induce a significant log-likelihood reduction according to a cutoff p-value are put aside from the set of investigated covariate-parameter relationships. Then, at the end of the forward process, the covariate-parameter relationships put aside are univariately added to the final forward model. If there is any significant log-likelihood reduction according to the defined forward p-value, a forward iterative loop is run again by adding the covariate-parameter relationships put aside from the investigated set and by using the final forward model as the starting model.
The full fixed effect modeling method is completely different from SCM or SCM+, integrating simultaneously all the covariate-parameter relationships of the investigated set into the model [16, 17]. Compared to the stepwise approaches, the full model approach avoids multi-testing issues as it uses the data only one to assess the covariates’ significance.
For both SCM/SCM+ and the full model, it has been suggested [4, 17] that correlated covariates should be avoided, as they may hinder the identification of covariates that are truly influential.
A clinical trial simulation study was conducted inspired by a real population PK analysis of emicizumab [30], a humanized bispecific monoclonal antibody administered subcutaneously to patients with hemophilia A [31,32,33]. Hemophilia A is a rare genetic blood disorder affecting mainly men and caused by a mutation of the F8 gene located on the X chromosome. Patients with this disease suffer lifelong bleeding due to a deficiency in plasma clotting factor VIII (FVIII) and may develop anti-FVIII antibodies (FVIII-inhibitors) when treated with intravenous injections of FVIII.
This paper outlined the simulation study design, detailing how covariate analysis was performed using the full model, SCM and SCM+ algorithms implemented in PsN. The methodology for evaluating estimates accuracy and the validity of their associated uncertainty was detailed. The covariate clinical relevance was evaluated using a CI-based definition as proposed in the literature [28, 29]. By combining the results of ratios calculated for multiple percentiles or classes, this newly proposed decision-making rule allows reaching a single conclusion on the clinical relevance of a covariate-parameter relationship. The accuracy of covariate estimates and ratio estimates with the evaluation of their corresponding uncertainties were presented, as well as the effectiveness of the full model, PsN’s SCM and SCM+ algorithms to assess the covariate clinical relevance.
Materials and Methods
Model and Notations
Let \({y}_{ij}\) be the response of individual \(i \in \{1, ...,N\}\) at time \({t}_{ij}\) with \(j \in \{1, ..., {n}_{i}\}\) and \({n}_{i}\) the total number of observations for individual \(i\):
where \(f\) is the non-linear structural model depending on \({\phi }_{i}\), the vector of individual parameters for subject \(i\). \({\varepsilon }_{ij}\sim N(0,1)\) refers to the measurement error for the individual \(i\), at the time \({t}_{ij}\), with \(a\) the additive and \(b\) the proportional term of the residual unexplained variability.
The vector of individual parameters is function of \(\mu\) the fixed effect vector, \({\eta }_{i}\sim N(0,\Omega )\) the random-effect vector of individual \(i\) with \(\Omega\) the variance covariance matrix, \({C}_{i}\) the covariate values vector of individual \(i\), and \(\beta\) the covariate parameters vector. Parameters were log-normally distributed to ensure positiveness. Therefore, \({\phi }_{i}\) can be written as follows:
with \(g\) the function describing the covariate-parameter relationship which can have any shape but is usually linear, exponential, power or piecewise linear. For instance, in the case of a continuous covariate (\({c}_{1}\)) described using a power function and a binary covariate covariate (\({c}_{2}\), equal to \(REF\), the reference category, or \(NREF\), the other category not being the reference) described using a linear function acting on the same parameter with index \(p\), the vector of individual parameters for subject \(i\) can be detailed as follows:
where \({\mu }_{p}\) is the fixed effect for the \(p\)th parameter and \({\eta }_{p,i}\), the random effect of individual \(i\), for the \(p\)th parameter. \({c}_{1,i}\) and \({c}_{2,i}\) are the covariate values of the individual \(i\), with \(\overline{{c }_{1}}\) the median value of the continuous covariate \({c}_{1}\) in the studied population. \({1}_{{c}_{2,i}=NREF}\) denotes the indicatrice function which is equal to 1 when \({c}_{2,i}=NREF\) and 0 when \({c}_{2,i}=REF\). \({\beta }_{p,{c}_{1}}\) and \({\beta }_{p,{\text{NREF}}}\) are the covariate parameters on the \(p\)th parameter of \({c}_{1}\) and the \(NREF\) category relative to the REF category of \({c}_{2}\), respectively.
The final vector of parameters to estimate with their standard error (SE) is therefore \(\theta =\left\{\mu ,\beta ,\Omega ,a,b\right\}\).
Simulation Study
Population PK Models
A population PK model has been developed by Retout et al. (2020) [30] to characterize the PK of emicizumab in adult and pediatric patients (> 1 year of age) with hemophilia A using a database of \(N=389\) individuals. Patients were included in 5 phase I/II and phase III clinical studies (see Table S1.1 in the Supplementary File 1). The PK sampling scheme was either rich (phase I/II or run in phase of one of the phase III study) or sparse (phase III studies with mostly through samples) and 3 dosing regimens were investigated: an administration every week, every 2 weeks or every 4 weeks. In Retout et al. (2020) [30], from 383 evaluable PK profiles, it was found that a 1-compartment model with a first-order absorption and a linear elimination best described the emicizumab PK data. This model had 3 PK parameters: the apparent clearance (CL/F), the apparent volume of distribution (V/F) and the absorption rate (KA), where F denotes the bioavailability. A correlation was observed between the 2 parameters CL/F and V/F as well as between CL/F and KA. A total of 3 continuous covariates were included in the final model, i.e. the body weight (BW) on V/F and CL/F, the albumin (ALB) on CL/F and the age (AGE) over 30 years old on F. In addition, 1 categorical covariate was included in the final model, i.e. the race (RACE) on V/F. This covariate was defined in 4 categories: White (WHT) the reference, Black (BLK), Asian (ASN) and Other (OTH). Only the BLK category showed an effect.
For the simulation study, 2 models were defined derived from this population PK model: a base model and a covariate model. No correlation between parameters were considered for these 2 models (diagonal variance-covariance matrix).
The base model was similar to the one reported in Retout et al. (2020) [30], including a BW effect on CL/F and V/F using a power function:
For the covariate model, it was very similar to the final population PK model reported in Retout et al. (2020) [30], including the same 4 covariates i.e. BW, ALB, AGE and RACE. However, the AGE effect was modeled on CL/F and V/F instead of on F, using a power function for simplification purposes:
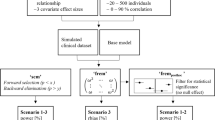
Simulation Settings
A total of \(S=200\) datasets of \(N=383\) patients were simulated with the base model and the covariate model. The simulation design was similar to the real data in terms of dosing regimens, number and collection times of PK samples and patients associated covariates (Tale S1.1 in the Supplementary File 1). Individual PK parameters, errors and individual emicizumab PK concentrations were simulated. The values used to simulate the parameters are shown in Table 1 and were obtained by fitting the base model and the covariate model to the real data. A dataset simulated with the base and a dataset simulated with the covariate model (i.e. dataset n°1, the first of the 200 simulated datasets) are displayed in Fig. S1.1 in the Supplementary File 1.
Covariate Analysis
The set of investigated covariate-parameter relationships are described in Table 2. There were 5 continuous covariates, i.e. BW, AGE, ALB, aspartate aminotransferase (AST), bilirubin (BILI), and 2 categorical covariates, i.e. patient status (STAT) with 2 categories: non-inhibitor (NINH, the reference) and FVIII inhibitor (INH) and the RACE with 4 categories: WHT (the reference), BLK, ASN and OTH. The reference category was defined as the most frequent one. The covariate distributions and correlations of the real database of \(N=389\) patients with hemophilia A are provided in Table S1.2, Figs. S1.2, and S1.3 in the Supplementary File 1. All covariates were non-normally distributed.
The full model, SCM and SCM+ were applied to each of the datasets simulated with the base model and the covariate model. To provide a reference model, the simulated model (i.e. the base model or the covariate model) was fitted to the simulated datasets.
Estimation and Implementation
The entire analysis was run using NONMEM version 7.4.3 on a High Performance Computing (HPC) environment. The HPC had 66 compute nodes using each 2 x Intel® Xeon® Platinum 9242 Processor (48 cores per processor) @ 2.3 Ghz, 96 cores and 768 GB DDR4-2933 of RAM memory.
The datasets were simulated with PsN version 5.3.0 using the sse tool. A total of 24 cores were allocated and the seed was fixed.
For the parameter estimations, each run was launched with 24 cores, 6 threads using PsN version 5.3.2. The first-order conditional estimation with interaction (FOCEi) algorithm was used. SE were derived from the covariance matrix computed as \({R}^{-1}S{R}^{-1}\), with \(R\) and \(S\) the Hessian and the Cross-Product Gradient matrix, respectively.
The reference model was launched using the execute PsN tool. The full model was launched using the parallel retries PsN tool. Each fit was launched with 5 retries and the best fit was kept. SCM and SCM+ were launched using the scm and scmplus PsN tools, respectively. The forward p-value was set to 0.05 (PsN default value), the backward p-value was set to 0.01 (which is assumed to be one of the most commonly used [4, 5, 11, 14] but is not the PsN default value); and for SCM+, the cutoff p-value was set to 0.05 (PsN default value). For the specific case of categorical covariate with more than 2 categories i.e. the RACE, the overall effect was tested with SCM and SCM+. The effect of the 4 categories i.e. WHT (the reference), BLK, ASN and OTH were evaluated simultaneously. Thus, if the RACE was found to be significant and was therefore selected, the effect of the BLK, ASN and OTH race relative to the WHT (the reference) were all estimated. When a covariate was not selected by SCM or SCM+, the covariate parameter estimate and its SE were set to 0. In addition, the covariate ratio estimate was set to 1 with a CI width of 0.
Data visualization and results analysis were performed using R version 4.0.5. Plots were produced using ggplot2, corrplot, ComplexUpset and PMXForest.
Evaluation
For the sake of simplicity, the notation of \({\beta }_{p,c}\) will be abbreviate to \(\beta\) in the following. Thus, covariate-parameter relationships for which an effect was simulated were indicated by \(\beta \ne 0\), and covariate-parameter relationships for which no effect was simulated were denoted by \(\beta =0\).
Model Selection Using SCM/SCM+
The final models selected by SCM and SCM+ for the 200 datasets simulated with the base and the covariate models were evaluated in order to check the correctness of the covariate model selection process.
Covariate Parameters Estimation Accuracy and Uncertainty Evaluation
The accuracy of covariate parameter estimates was assessed by calculating the estimation errors (EE) for the simulated covariates where \(\beta =0\), and the relative estimation errors (REE) for the simulated covariates where \(\beta \ne 0\):
where \(\widehat{\beta }\) and \(\beta\) are the estimated and simulated covariate parameters, respectively.
The uncertainty of covariate parameter estimates was evaluated by comparing the SE for the simulated covariates where \(\beta =0\), and RSE for the simulated covariates where \(\beta \ne 0\), with empirical SE and RSE, respectively. The RSE, empirical SE and empirical RSE computed as follows:
where \({\widehat{\beta }}_{s}\) is the estimated covariate parameter for the simulated dataset s and \(SE(\widehat{\beta })\), the estimated SE of the covariate parameter.
The estimated RSE/SE were considered as correctly estimated if the empirical RSE/SE were included between the 5th and 95th percentile of the estimated RSE/SE. If the empirical RSE/SE were below the 5th percentile or above the 95th percentile the estimated RSE/SE, the estimated RSE/SE were considered as overestimated or underestimated, respectively.
Covariate Ratios Estimation Accuracy, Precision and Uncertainty Evaluation
On the forest plots, the effect of covariate on the parameters of interest are represented as ratios relative to a typical individual, representative of the studied population.
For continuous covariates, the ratio between the covariate effect value computed at the 10th or 90th percentile of the observed covariate distribution (\(P10\) and \(P90\)) and the covariate effect value computed at the median (\(MED\)) of the same observed covariate distribution was considered. For instance, simulated and estimated P10 ratios were computed as follows:
The 90% CI of the estimated P10 ratio was then expressed as:
where \({z}_{0.95}\) is the 0.95 quantile of the standard normal distribution.
For categorical covariates, the ratio between the covariate effect value of one category and the parameter covariate effect value of the reference category (which was defined as the most frequent category in our study) was considered. The simulated and estimated ratios were then expressed as follows:
The 90% CI of the estimated ratio was then expressed as:
The accuracy of covariate ratio estimates was assessed by calculating the REE:
The relative root mean square error (RRMSE) were calculated to characterize the bias and the variance of covariate ratios over the 200 simulations as follows:
where \({\widehat{r}}_{s}\) is the estimated covariate ratio for the simulated dataset s.
Coverage rates were calculated to check the validity of the covariate ratios confidence interval. It mixed the evaluation of the bias and the uncertainty. There were computed as the proportion of estimated \({CI90}_{\widehat{r}}\) including the simulated covariate ratio \(r\), over the 200 simulations. Coverage rates were expected to be within the expected range of [0.850, 0.938], corresponding to the 95% prediction interval for a proportion following a binomial distribution with 200 trials and a probability of success of 0.90. By construction, coverage rates below the prediction interval (i.e. under 0.850) reflect bias ratio estimates (90% CI around the estimated ratio shifted relative to the simulated ratio value) or underestimated uncertainty (too small 90% CI around the estimated ratio). Conversely, coverage rates above the prediction interval (i.e. over 0.938) reflect overestimated uncertainty (too large 90% CI around the estimated ratio).
Covariate Clinical Relevance
Regarding the covariate clinical relevance decision, 5 different cases were assumed depending on the \({CI90}_{\widehat{r}}\) value in relation to the reference area of 0.8–1.2 and the reference line at 1, as illustrated on Fig. 1: relevant, non-relevant significant, non-relevant non-significant, insufficient information significant, insufficient information non-significant. Of note, the decisions insufficient information significant and insufficient information non-significant could have been grouped into one category insufficient information, as they are both inconclusive. An additional category, non-selected, was used to indicate when a covariate-parameter relationship was not selected by SCM or SCM+.
A covariate effect was considered relevant if the \({CI90}_{\widehat{r}}\) was completely outside the reference area of 0.8–1.2. It was considered as non-relevant if the entire \({CI90}_{\widehat{r}}\) was within the reference area. Finally, if the \({CI90}_{\widehat{r}}\) straddled the reference area, it was considered that there was insufficient information to identify the covariate parameter relevance.
The effect was found significant if the \({CI90}_{\widehat{r}}\) did not include the reference line at 1 which represents no change compared to the typical individual. On the other hand, it was non-significant if the \({CI90}_{\widehat{r}}\) included the reference line at 1.
For continuous covariates, the estimated P10 and P90 ratios could lead to 2 different conclusions about the clinical relevance of the covariate. This could also be the case for categorical covariates with more than 2 categories, i.e. for the RACE where a ratio was calculated for the BLK, ASN and OTH categories. In order to reach a single conclusion, a common decision based on more than one ratio was established see Table S1.3 in the Supplementary File 1.
Computational Efficiency
The computational efficiency of the 3 approaches was compared for the dataset n°1 simulated with the base and the covariate model using the total runtime, the number of NONMEM runs and the number of objective function evaluations. The number of NONMEM runs were also compared across the 200 simulated datasets under both simulated models.
Results
Results were presented for the reference model and the 3 evaluated approaches i.e. the full model, SCM and SCM+ applied to simulated datasets under the base and the covariate model. First, the models selected by SCM and SCM+ were evaluated. Then, the covariate parameter estimates and their associated uncertainty accuracy were evaluated. In addition, the covariate ratio estimates accuracy, precision, and uncertainty were studied. More importantly, the clinical relevance of the different covariates was assessed, which is the main objective of the article. Finally, the computational efficiency of the 3 approaches was compared.
In this section, covariate-parameter relationships for which an effect was simulated (i.e. \(\beta \ne 0\)) were referred to as “simulated covariate with an effect”. Similarly, covariate-parameter relationships for which no effect was simulated (i.e. \(\beta =0\)) were referred to as “simulated covariate with no effect”.
Model Selection Using SCM/SCM+
Figure 2 summarizes model combinations obtained with SCM and SCM+ on datasets simulated with the base and the covariate model. SCM and SCM+ mostly selected identical models (99.5% and 98.5% with the base and the covariate model, respectively). Therefore, only SCM results were discussed further.
The “true” model (i.e. the simulated model) was selected by SCM in 82% and 59% of the cases under the base model and the covariate model, respectively. Under both simulated models, BW was consistently included in the final model on CL/F and V/F. Regarding the other simulated covariates with an effect, they were retained in the final model in most instances, with percentages ranging from 85% (e.g. RACE on V/F) to 99% (e.g. ALB on CL/F). Simulated covariates with no effect were infrequently selected in only up to 4%, especially on KA.
Covariate Parameters
Estimation Accuracy
All covariate parameter estimates were unbiased with the full model and SCM, see Fig. S2.1 in the Supplementary File 2, where REE and EE boxplots were centered on 0. Under both simulated models, the REE of BW on CL/F and V/F ranged from − 5 to 6% with the full model and SCM, as for the reference model. Regarding the other simulated covariates with an effect, the REE boxplot sizes were similar among the reference model, the full model and SCM, ranging from − 19 to 18%. For simulated covariates with no effect, the EE boxplot sizes were flat for SCM compared to the full model.
Uncertainty Evaluation
Overall, the uncertainty of the covariate parameter estimates was acceptable with the full model and SCM approaches (RSE and the empirical RSE around 30%) and well estimated or slightly underestimated with SCM, see Fig. S2.2 in the Supplementary File 2. Under both simulated models, the RSE of BW on CL/F and V/F ranged from 2 to 8%, with the full model and SCM, aligning closely with the reference model. The reference and full models provided well estimated RSE while they were slightly underestimated with SCM under the covariate model. Regarding the other simulated covariates with an effect, RSE values varied between 16 and 36% with the reference model, the full model and SCM. As for the BW, RSE were correctly estimated with the reference and full models, but were slightly underestimated with SCM. For simulated covariates with no effect, the median SE values were higher and the SE boxplots sizes were larger for the full model compared to SCM. Nevertheless, SE were correctly estimated with the full model while slightly underestimated with SCM.
Covariate Ratios
Estimation Accuracy
Covariate ratio estimates were accurately estimated with the full model and SCM, see Fig. 3 where the REE boxplots were centered on 0. Under both simulated models, the REE of BW on CL/F and V/F ranged from − 5 to 4%, with the full model and SCM, as for the reference model. Regarding the other simulated covariates with an effect, the REE boxplot sizes were similar among the reference model, the full model and SCM, ranging from − 4 to 5%. For simulated covariates with no effect, the REE boxplot sizes were larger for the full model (ranging from − 11 to 7%) compared to SCM (ranging from − 5 to 4%).
Boxplots of REE of estimated covariate ratios for simulated covariates were \(\beta \ne 0\) (left) and \(\beta =0\) (right) under the base (top) and covariate (bottom) model; for continuous covariates, only results for P10 are shown (results for P90 are given in Fig. S3.1); the 5th, 25th, 50th, 75th and 95th percentiles are displayed on the boxplot
Estimation Precision
Covariate ratio estimates were precisely estimated with the full model and SCM, as shown in Fig. 4 where the RRMSE did not exceed 20% and were mostly below 10%. Under both simulated models, the RRMSE of BW on CL/F and V/F ranged from 3 to 7%, with the full model and SCM, as for the reference model. Regarding the other simulated covariates with an effect, RRMSE values were comparable across all approaches with values ranging from 2 to 6% for the reference and full models and from 2 to 11% for SCM. For simulated covariates with no effect, RRMSE were higher for the full model (values up to 20%) compared to SCM (values up to 11%). Of note, compared to other covariate ratios, the AGE and INH on KA showed slightly higher RRMSE values with the full model and SCM.
Uncertainty Evaluation
Coverage rates are shown in Fig. 5. Under both simulated models, coverage rates of BW on CL/F and V/F were consistently around 0.90 and within the 95% prediction interval with the full model as for the reference model. With SCM, coverage rates were within the prediction interval under the base model and below or near the lower bound of the prediction interval under the covariate model, reflecting an underestimated uncertainty (see Fig. S2.2 in the Supplementary File 2).
Coverage rates of estimated covariate ratios for simulated covariates were \(\beta \ne 0\) (left) and \(\beta =0\) (right) under the base (top) and covariate (bottom) model; the target range of [0.850, 0.938] is shaded in gray and the value of 0.900 is indicated by a dashed line; for continuous covariates, only results for P10 are shown (results for P90 are given in Fig. S3.3)
Regarding the other simulated covariates with an effect, the two approaches showed some different results compared to the reference model. With the full model, the coverage rate of AGE on CL/F was slightly above the prediction interval, while it was within the prediction interval for the reference model, showing uncertainty likely to be overestimated (see Fig. S2.2 in the Supplementary File 2). With SCM, the coverage of BLK on V/F was above the prediction interval, while it was within the prediction interval for the reference model, highlighting an underestimated uncertainty (see Fig. S2.2 in the Supplementary File 2).
For simulated covariates with no effect, results differed between the full model and SCM. With the full model, most coverage rates were within the prediction interval (13 out of 16 under the base model and 10 out of 12 under the covariate model), while some were below the prediction interval, namely OTH on V/F and AGE on KA under both the base and covariate models in addition to ALB on CL/F under the base model. OTH on V/F and AGE on KA showed slightly biased ratios (see Fig. 3) and underestimated uncertainty (see Fig. S2.2 in the Supplementary File 2). For ALB on CL/F, only an underestimation of the uncertainty was observed (see Fig. S2.2 in the Supplementary File 2). With SCM, coverage rates of covariates with no effect were inflated as these covariates were not selected in between 96% and 100% of the time.
Of note, for continuous covariates, the results presented above in terms of accuracy, precision and uncertainty evaluation are focusing on the P10. P90 results are displayed in Supplementary File 3 and are similar to those obtained with the P10.
Covariate Clinical Relevance
Figure 6 shows the results of the covariates’ clinical relevance, with the different conclusions defined in Fig. 1. To provide a concrete illustration of the proposed analysis, Supplementary File 4 summarizes the results (in terms of parameter estimates and forest plots) obtained with the reference model, the full model and SCM for the dataset n°1 simulated with the base and the covariate model.
Covariate clinical relevance decisions for simulated covariates were \(\beta \ne 0\) (left) and \(\beta =0\) (right) under the base (top) and covariate (bottom) models; only percentages over or equal to 5% are shown; the 5 possible decisions about the covariate clinical relevance are the following: relevant (R), non-relevant significant (NRS), non-relevant non-significant (NRNS), insufficient information significant (IIS), insufficient information non-significant (IINS); with SCM/SCM+, covariates can also be non-selected (NSEL)
Under both simulated models, BW was found relevant with the full model and SCM in almost all cases, as with the reference model. In the minority of cases where the covariate was not found relevant, i.e. in between 0 and 1.5% of the cases depending on the approach and the simulated model considered, there was insufficient information to identify its clinical relevance. Of note, none of the methods identified that BW was not-relevant.
Regarding the other simulated covariates with an effect, SCM and full model reached similar conclusions as the reference model in the vast majority of the cases. However, SCM failed in up to 15% of cases simulated covariate with an effect (e.g. AGE on CL/F or RACE on V/F), while, by construction they were always included in the model for the full model.
Under both simulated models, for simulated covariates with no effect, the full model and SCM led to different results, because of the way each method works. As no effects were simulated, the expectations would be to have covariates reported as non significant. In this manuscript, emphasis will however be made on the clinical relevance of the covariates. With the full model, regarding covariates on CL/F and V/F (except for the RACE), the approach found these covariates mostly non-relevant (from 78.5 to 98.5% of the cases), mainly non-significant (in between 74.5 and 90% of the cases). The RACE was also mainly found to be non-relevant non-significant but there was also insufficient information (from 28.5 to 33.5% of the time) to identify its clinical relevance. For the covariates on KA, it was mostly found that there was insufficient information (in between 79 and 90% of the cases) to identify their clinical relevance, and they were mainly non-significant (from 62 to 81% of the cases). With SCM, simulated covariates with no effect were mostly non-selected i.e. in between 96 and 100% of the cases. Both the full model and SCM approaches wrongly identify, in between 1 and 2% of the cases, that the AGE and INH on KA had a relevant effect. This could be explained by slightly biased estimates for these covariate ratios with the full model (see Fig. 3) and a slightly underestimated uncertainty for SCM (see Fig. S2.2 in the Supplementary File 2).
Computational Efficiency
The full model outperformed SCM/SCM+ in all aspects (total runtime, number of NONMEM runs and number of objective function evaluations), considering the simulated dataset no. 1, especially under the covariate model, see Table S5.1 in the Supplementary File 5. Under the base model, the full model was 4 times faster or took about the same time than SCM and SCM+, respectively. Furthermore, under the covariate model, the full model was 4 to 20 times faster than SCM/SCM+. The number of NONMEM runs was always equal to 6 with the full model due to the implementation settings (i.e. number of retries fixed to 5) while SCM/SCM+ required much due to the way the algorithms work. Regarding the number of objective function evaluations, the full model required 2 times fewer functions than SCM/SCM+ under the base model and 5 to 10 times fewer than SCM/SCM+ under the covariate model.
In addition, SCM+ showed better performances than SCM. Across the 200 datasets simulated with the base and the covariate models, SCM+ needed about 10 fewer runs than SCM to select a final model (see Table S5.2 in the Supplementary File 5), as about 2 and 3 parameter-covariate relationships were set aside from the set of investigated covariate-parameter relationships.
Discussion
The present work evaluated the ability of the full model, SCM and SCM+ approaches to correctly identify the covariate effects clinical relevance. For that purpose, a simulation study was performed based on a real case example led on hemophilia A patients treated with emicizumab. Thus, 2 models were considered for the simulation: a base model including 2 covariate-parameter relationships (BW on CL/F and V/F) and a covariate model including 6 covariate-parameter relationships (BW and AGE on CL/F and V/F, ALB on CL/F and BLK race on V/F) directly inspired from the base and the final population PK model reported in Retout et al. (2020) [30], respectively. It is important to note that these 2 models are simplified versions of the model published by Retout et al. (2020) [30] and shouldn’t be used as a reference to describe emicizumab PK. For the covariate model building, 7 covariates were considered with a total of 9 covariate-parameter relationships investigated on CL/F, 7 on V/F and 2 on KA (including all the covariate-parameter relationships for which an effect was simulated and others for which no effect was simulated).
Regarding covariates selection, SCM and SCM+ performed equally well, aligning with the work of Svensson et al. (2022) [7]. Both methods systematically select the “true” model (i.e. the simulated model) or a close one. However, it should be noted that in certain cases: (i) under both simulated models, up to 2 covariate-parameter relationships were selected by SCM/SCM+ while no effect was simulated, (ii) under the covariate model, up to 2 covariate-parameter relationships were not selected by SCM/SCM+ while an effect was simulated. This selection bias may be imputed to moderate correlation between some covariates (c.f. Fig. S1.2 of the Supplementary File 1). Indeed, it has been shown in the literature that competition between several moderately or highly correlated covariates leads to a loss of power to identify true covariates and a propensity to select false covariates with SCM [4].
As the covariate selection using SCM/SCM+ relies on a likelihood-ratio test, involving log-likelihood estimation, it might be valuable to compared these results with other estimation algorithms such as the importance sampling (IMP) or Stochastic Approximation Expectation Maximization (SAEM) that do not use linearization as FOCEi. In addition, SCM and SCM+ were run using a single set of values of forward, backward and cutoff p-values assumed to be the most commonly used [4, 5, 11, 14]. Additional investigations with alternative sets of p-values could be of interest, such as using 0.05 as equal forward and backward p-values, or using a cutoff p-value larger than the forward p-value, for example set at 0.1.
At last, only the correct form of covariate-parameter relationships was tested in this study i.e. power for the continuous covariates and additive for categorical covariates. With SCM and SCM+ it is possible to test automatically different types of covariate parameters relationships, whereas with the full model, all the different combinations will have to be made by hand. In all cases, an exploratory analysis of the data would enable the characterization of the covariate parameter relationships and thus choose the appropriate form to test.
Considering covariates for which an effect was simulated, results showed overall unbiased as well as precise estimates for covariate parameters and ratios using the 3 approaches. Covariate parameters uncertainty was well estimated with the full model while it tended to be underestimated with SCM/SCM+. Regarding the coverage rates, they were almost always within the 95% prediction interval around the target value of 0.9 with the full model, reflecting overall unbiased ratios and a valid associated uncertainty, and in fewer cases slightly below, reflecting slightly biased ratios or underestimated uncertainty. With SCM/SCM+, coverage rates were either near to the lower bound of the 95% prediction interval around the target value of 0.9 or below. Since ratio estimates were unbiased, lower coverage rates reflected an underestimated uncertainty. However, the coverage rate results must be interpreted cautiously because of the rather limited number of replicates leading to a rather large 95% prediction interval [0.850, 0.938].
Given that the 90% CI associated with the covariate ratio estimates was calculated using the parameter covariates’ SE, improper SE estimates may impact on covariates' clinical relevance evaluation. It would be interesting to compare the results obtained with different methods of SE calculation, for example by applying the Gallant correction [34] (which takes into account the number of estimated parameters relative to the available data to correct for possible underestimation), or using other approaches such as bootstrap [26] or SIR [27].
For covariates for which no effect was simulated, these covariates were not selected by SCM/SCM+ in a percentage ranging from 96 to 100% of the cases. By construction, for non-selected covariates, parameter estimates and their SE were set to 0 and ratio estimates were set to 1 with a CI width of 0. This implementation influenced all the results, leading to flat REE boxplot for covariate parameters and ratios, as well as over optimistic coverage rates (coverage rates automatically between 0.96 and 1 since the simulated and estimated ratio values were equal) despite the underestimated uncertainty observed for the few cases when the covariate is selected. For the full model, the results obtained for simulated covariates with no effect were similar to those obtained for simulated covariates with an effect, but with slightly more variable and less precise covariate parameters and ratios.
The main objective of this work was to study the appropriateness of different modeling approaches for making decisions on the clinical relevance of covariates. This included ensuring that clinically relevant covariates were not incorrectly reported as non-relevant and, conversely, that non-relevant covariates were not wrongly reported as clinically relevant.
The full model and SCM/SCM+ were able to well identify the clinical relevance of simulated covariates with an effect in the majority of the cases. However, SCM/SCM+ did not select some significant covariates (i.e. AGE on CL/F and V/F, RACE on V/F) in a percentage ranging from 7 to 15% of the cases. As mentioned above, this may be due to moderate correlation between some covariates (c.f. Fig. S1.2 of the Supplementary File 1) [4]. Simulated covariates with an effect were mostly found to be relevant, not-relevant significant or with insufficient information significant.
For simulated covariates with no effect, the full model and SCM/SCM+ gave very different results. As no effects were simulated, the expectations would be to have covariates reported as non-significant. The full model mainly found simulated covariate with no effect non-significant and mostly non-relevant or to a lesser extent that there was insufficient information to identify their clinical relevance. With SCM/SCM+, simulated covariates with no effect were mainly non-selected (i.e. between 96 and 100%). Since no effects are estimated for these covariates, it was chosen in this work to not represent these covariates on forest plots (cf. Figs. S4.1 and S4.2). Therefore, it is usually assumed that non-selected covariates have no relevant effect, without discussing that there might be cases where covariates are not selected because of insufficient information. This assumption is not needed with the full model. A covariate may not be selected by SCM because it has no effect or because there is not enough information to detect its effect; thus the two cases are not differentiated when the covariate is not selected, resulting in a loss of information. Whereas the full model will either identify that the covariate is not relevant or that there is not enough information to identify its relevance, which is in itself already informative. However, it is important to note that scientific plausibility must be taken into account when performing covariate analysis. Thus, if a covariate seems to plausibly have an effect on a parameter, but this relationship has not been selected by the method used, further investigations are usually performed. The full model and SCM/SCM+ wrongly identify (between 1 and 2% of the cases) that the AGE and STAT had a relevant effect on KA. Of note, KA was the PK parameter for which the sampling design was the least informative. The majority of the patients were sampled predose and 1 day post dose, only 37 patients were sampled 8 hours post dose while the KA of the emicizumab is about 13 h−1.
It is important to note that the results obtained on the covariate clinical relevance are closely linked to the study design (number of subjects, number of samples per subject and PK sampling times), the covariate distribution in the dataset but also to the magnitude of the covariate effect considered. Our results should therefore be put into perspective of our simulation study design i.e. a pool of phase I/II and III studies with either sparse and rich designs. However, the lack of classification (II or NR) for non-selected covariates is inherent to stepwise covariate modeling approaches (e.g. SCM or SCM+) and can therefore be generalized regardless of the design.
It would be interesting to extend our work to other types of study design, in particular to sparse studies with low number of subjects and/or low number of informative PK samples per subject.
In addition, the performance of the full model and SCM/SCM+ on the covariate clinical relevance evaluation should be compared in more complex simulation settings. In our simulation, the number of covariates with a relevant effect was limited to BW, and the others simulated covariate had a relatively minor effect. Exploring a case study with a larger set of investigated covariates, including a wider range of effects would be of interest. Other covariate related characteristics could also be studied such as the impact of the covariate variability (small or large), covariate distribution shape, proportion of missing values and parameter-covariate relation type (linear, power, exponential …).
In our example, the RACE categorical covariate with more than 2 categories was included in order to provide an example in the comparison of the different covariate model building approaches. This covariate was implemented in such a way that an effect was estimated for each category in comparison with the reference (WHT). The decision-making rule proposed in this article was developed in order to take into account this kind of covariate implementation (see Table S1.3 in the Supplementary File 1). However, it’s important to note that it is possible to binarize the covariate by creating a number of dummy covariates equal to the number of categories—1. In that case, an effect would have been estimated for each covariate created, testing the effect of one category against all the others (e.g. effect of ASN comparatively to non ASN). By construction, the results would have been different as the estimated covariate effect is not the same (effect estimated comparatively to the REF or to all the others categories). This alternative parametrization has the advantage of reducing the degree of freedom to 1 for each covariates tested and avoids having to make a single decision on the covariate clinical relevance based on the conclusion of the different ratios estimated for each category.
In this work, the covariate “clinical relevance” was restricted to the evaluation of covariate effect on PK. However, the National Institutes of Health has defined clinical relevance as “the ability of a therapy to improve how the patient feels, functions, and/or survives” [35]. To accurately evaluate the clinical relevance of covariates, patient’s safety and efficacy should be taken into account.
In terms of computational efficiency, the full model performed better than SCM/SCM+ regarding the number of NONMEM runs and the number of objective function evaluations, resulting in faster total runtimes. Moreover, SCM+ outperformed SCM: (i) the number of runs was reduced by about 20% with SCM+ compared to SCM, considering the 200 datasets simulated with both models, while Svensson et al. (2022) [7] reported 44%, (ii) the number of objective function evaluations was reduced by around 15% and 50% with SCM+ compared to SCM, considering the dataset No. 1 simulated with the base model and the covariate model, respectively, while Svensson et al. (2022) [7] reported 70%. These differences can be explained by the higher number of covariates and covariate-parameter relationships tested in their study, with several covariates having a strong effect versus one for our work. Moreover, they simulated twice as many covariates having an effect, inducing in our case that the forward ends in fewer steps, which reduces the differences in the number of runs between SCM and SCM+.
No computational issues arose with the full model in our study. In fact, as our simulation study was inspired by a real case study, its design did not permit to challenge the full model on this point i.e. the simulated data were too informative (large number of patients with either rich or sparse PK sampling schema) for a middle dimensional problem (7 covariates and a total of 18 parameter-covariate relationships tested). Extending our comparative study of the performance of the 3 approaches to cases using sparse data (small number of subjects with sparse PK sampling) and/or a high-dimensional problem (large number of covariates and parameter-covariate relationships tested) would make it possible to assess whether computational problems could arise with the full model (given that a large number of parameters will have to be estimated with few data). In addition, it could be interesting to compare methods with and without prescreening, especially in the case of a high-dimensional problem. A pre-screening step on the empirical bayes estimates (EBE) could be added beforehand to reduce the size of the set of investigated covariate-parameter relationships or included in the full model.
To conclude, the full model, SCM and SCM+ provided satisfactory results, showing a good ability to identify the covariates' clinical relevance. However, the full model always makes it possible to distinguish cases where a covariate was non-relevant or if there was insufficient information to identify its clinical relevance, whereas this is not the case with SCM/SCM+ when the covariate was not selected. In addition, the full model showed better computational performances than SCM and SCM+. In the specific case of our simulation study design, the full model may therefore be preferred to SCM/SCM+ for assessing the clinical relevance of covariates. Our study was in a first instance a simulation case based on a real example comprising large amount of data relative to the number of investigated covariates and need to be extended to a more challenging design (i.e. high dimensional problem with a sparse dataset and/or a large number of covariates). Furthermore, our work needs to be applied to other methods such as LASSO [11] and FREM [20] for which an algorithm is implemented in PsN, or COSSAC [9] and SAMBA [12] available in Monolix and to go further with machine learning techniques [13,14,15] emerging in the pharmacometrics field for covariates search.
References
European Medicines Agency (2022) ICH guideline E11A on pediatric extrapolation - Scientific guideline. https://www.ema.europa.eu/en/ich-guideline-e11a-pediatric-extrapolation-scientific-guideline. Accessed 4 Oct 2023
Dartois C, Brendel K, Comets E et al (2007) Overview of model-building strategies in population PK/PD analyses: 2002–2004 literature survey. Br J Clin Pharmacol 64:603–612. https://doi.org/10.1111/j.1365-2125.2007.02975.x
Hutmacher MM, Kowalski KG (2015) Covariate selection in pharmacometric analyses: a review of methods. Br J Clin Pharmacol 79:132–147. https://doi.org/10.1111/bcp.12451
Ahamadi M, Largajolli A, Diderichsen PM et al (2019) Operating characteristics of stepwise covariate selection in pharmacometric modeling. J Pharmacokinet Pharmacodyn 46:273–285. https://doi.org/10.1007/s10928-019-09635-6
Jonsson EN, Karlsson MO (1998) Automated covariate model building within NONMEM. Pharm Res 15:1463–1468. https://doi.org/10.1023/a:1011970125687
Lindbom L, Pihlgren P, Jonsson N (2005) PsN-Toolkit—a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Programs Biomed 79:241–257. https://doi.org/10.1016/j.cmpb.2005.04.005
Svensson RJ, Jonsson EN (2022) Efficient and relevant stepwise covariate model building for pharmacometrics. CPT Pharmacomet Syst Pharmacol 11:1210–1222. https://doi.org/10.1002/psp4.12838
Lindbom L, Ribbing J, Jonsson EN (2004) Perl-speaks-NONMEM (PsN)—a Perl module for NONMEM related programming. Comput Methods Programs Biomed 75:85–94. https://doi.org/10.1016/j.cmpb.2003.11.003
Ayral G, Si Abdallah J-F, Magnard C, Chauvin J (2021) A novel method based on unbiased correlations tests for covariate selection in nonlinear mixed effects models: the COSSAC approach. CPT Pharmacomet Syst Pharmacol 10:318–329. https://doi.org/10.1002/psp4.12612
Mandema JW, Verotta D, Sheiner LB (1992) Building population pharmacokineticpharmacodynamic models. I. Models for covariate effects. J Pharmacokinet Biopharm 20:511–528. https://doi.org/10.1007/BF01061469
Ribbing J, Nyberg J, Caster O, Jonsson EN (2007) The lasso—a novel method for predictive covariate model building in nonlinear mixed effects models. J Pharmacokinet Pharmacodyn 34:485–517. https://doi.org/10.1007/s10928-007-9057-1
Prague M, Lavielle M (2022) SAMBA: A novel method for fast automatic model building in nonlinear mixed-effects models. CPT Pharmacomet Syst Pharmacol 11:161–172. https://doi.org/10.1002/psp4.12742
Terranova N, Venkatakrishnan K, Benincosa LJ (2021) Application of machine learning in translational medicine: current status and future opportunities. AAPS J 23:74. https://doi.org/10.1208/s12248-021-00593-x
Sibieude E, Khandelwal A, Hesthaven JS et al (2021) Fast screening of covariates in population models empowered by machine learning. J Pharmacokinet Pharmacodyn 48:597–609. https://doi.org/10.1007/s10928-021-09757-w
Ronchi D, Tosca EM, Bartolucci R, Magni P (2023) Go beyond the limits of genetic algorithm in daily covariate selection practice. J Pharmacokinet Pharmacodyn. https://doi.org/10.1007/s10928-023-09875-7
Xu XS, Yuan M, Zhu H et al (2018) Full covariate modelling approach in population pharmacokinetics: understanding the underlying hypothesis tests and implications of multiplicity. Br J Clin Pharmacol 84:1525–1534. https://doi.org/10.1111/bcp.13577
Gastonguay MR (2011) Full covariate models as an alternative to methods relying on statistical significance for inferences about covariate effects: a review of methodology and 42 case studies. Twentieth Meeting, Population Approach Group in Europe, Athens, Grece. https://www.page-meeting.org/pdf_assets/1694-GastonguayPAGE2011.pdf. Accessed 4 Oct 2023
Yngman G, Nordgren R, Freiberga S, Karlsson MO Linearization of full random effects modeling (FREM) for time-efficient automatic covariate assessment
U.S. Food & Drug Administration (2022) Population pharmacokinetics, guidance for industry. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/population-pharmacokinetics. Accessed 4 Oct 2023
Yngman G, Bjugård Nyberg H, Nyberg J et al (2022) An introduction to the full random effects model. CPT Pharmacomet Syst Pharmacol 11:149–160. https://doi.org/10.1002/psp4.12741
Amann LF, Wicha SG (2023) Operational characteristics of full random effects modelling (‘frem’) compared to stepwise covariate modelling (‘scm’). J Pharmacokinet Pharmacodyn. https://doi.org/10.1007/s10928-023-09856-w
European Medicines Agency (2018) Investigation of subgroups in confirmatory clinical trials - Scientific guideline. https://www.ema.europa.eu/en/investigation-subgroups-confirmatory-clinical-trials-scientific-guideline. Accessed 26 Oct 2023
Menon-Andersen D, Yu B, Madabushi R et al (2011) Essential pharmacokinetic information for drug dosage decisions: a concise visual presentation in the drug label. Clin Pharmacol Ther 90:471–474. https://doi.org/10.1038/clpt.2011.149
Marier J-F, Teuscher N, Mouksassi M-S (2022) Evaluation of covariate effects using forest plots and introduction to the coveffectsplot R package. CPT Pharmacomet Syst Pharmacol 11:1283–1293. https://doi.org/10.1002/psp4.12829
European Medicines Agency (2018) Investigation of bioequivalence - Scientific guideline. https://www.ema.europa.eu/en/investigation-bioequivalence-scientific-guideline. Accessed 4 Oct 2023
Thai H-T, Mentré F, Holford NHG et al (2014) Evaluation of bootstrap methods for estimating uncertainty of parameters in nonlinear mixed-effects models: a simulation study in population pharmacokinetics. J Pharmacokinet Pharmacodyn 41:15–33. https://doi.org/10.1007/s10928-013-9343-z
Dosne A-G, Bergstrand M, Harling K, Karlsson MO (2016) Improving the estimation of parameter uncertainty distributions in nonlinear mixed effects models using sampling importance resampling. J Pharmacokinet Pharmacodyn 43:583–596. https://doi.org/10.1007/s10928-016-9487-8
Cuzick J (2005) Forest plots and the interpretation of subgroups. Lancet 365:1308. https://doi.org/10.1016/S0140-6736(05)61026-4
Hahn AW, Dizman N, Msaouel P (2022) Missing the trees for the forest: most subgroup analyses using forest plots at the ASCO annual meeting are inconclusive. Ther Adv Med Oncol 14:17588359221103200. https://doi.org/10.1177/17588359221103199
Retout S, Schmitt C, Petry C et al (2020) Population pharmacokinetic analysis and exploratory exposure-bleeding rate relationship of emicizumab in adult and pediatric persons with hemophilia A. Clin Pharmacokinet 59:1611–1625. https://doi.org/10.1007/s40262-020-00904-z
Oldenburg J, Mahlangu JN, Kim B et al (2017) Emicizumab prophylaxis in hemophilia A with inhibitors. N Engl J Med 377:809–818. https://doi.org/10.1056/NEJMoa1703068
Mahlangu J, Oldenburg J, Paz-Priel I et al (2018) Emicizumab prophylaxis in patients who have hemophilia A without inhibitors. N Engl J Med 379:811–822. https://doi.org/10.1056/NEJMoa1803550
Yoneyama K, Schmitt C, Portron A et al (2023) Clinical pharmacology of emicizumab for the treatment of hemophilia A. Expert Rev Clin Pharmacol 16:775–790. https://doi.org/10.1080/17512433.2023.2243213
Gallant AR (1975) Seemingly unrelated nonlinear regressions. J Econ 3:35–50. https://doi.org/10.1016/0304-4076(75)90064-0
National Center for Advancing Translational Sciences Toolkit. Clinical relevance - Glossary. https://toolkit.ncats.nih.gov/glossary/clinical-relevance. Accessed 17 Nov 2023
Acknowledgements
This work was financed by a CIFRE agreement (Conventions Industrielles de Formation par la Recherche) and was conducted under the supervision of the ANRT (Association Nationale de la Recherche et de la Technologie). The CIFRE agreement is a partnership between a public laboratory and a company, here the UMR (Unité Mixte de Recherche) 1137 and INSTITUT ROCHE, respectively. The authors are grateful to Kamill Jaworski for his technical support in the simulation implementation.
Author information
Authors and Affiliations
Contributions
M.P., S.M., S.R. and F.M. designed the simulation study. M.P. implemented and performed the simulations. M.P. produced the results. M.P., S.M., S.R. and F.M. analyzed the results. M.P. wrote the manuscript. S.M., S.R. and F.M. reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Philipp, M., Buatois, S., Retout, S. et al. Impact of covariate model building methods on their clinical relevance evaluation in population pharmacokinetic analyses: comparison of the full model, stepwise covariate model (SCM) and SCM+ approaches. J Pharmacokinet Pharmacodyn (2024). https://doi.org/10.1007/s10928-024-09911-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10928-024-09911-0