Abstract
We discuss the optimum experimental conditions to obtain assignment spectra for solid proteins at magic-angle spinning (MAS) frequencies around 100 kHz. We present a systematic examination of the MAS dependence of the amide proton T 2′ times and a site-specific comparison of T 2′ at 93 kHz versus 60 kHz MAS frequency. A quantitative analysis of transfer efficiencies of building blocks, as they are used for typical 3D experiments, was performed. To do this, we compared dipolar-coupling and J-coupling based transfer steps. The building blocks were then combined into 3D experiments for sequential resonance assignment, where we evaluated signal-to-noise ratio and information content of the different 3D spectra in order to identify the best assignment strategy. Based on this comparison, six experiments were selected to optimally assign the model protein ubiquitin, solely using spectra acquired at 93 kHz MAS. Within 3 days of instrument time, the required spectra were recorded from which the backbone resonances have been assigned to over 96 %.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sequential assignment of resonances in NMR spectra is a prerequisite to determine atomic-resolution structures and to gain insight into site-resolved dynamics of proteins. Successful assignments require spectra with high spectral resolution and sensitivity. To obtain both, magic-angle sample spinning (MAS) is mandatory for most solid materials (Andrew et al. 1958; Lowe and Norberg 1957). Most applications of solid-state NMR to proteins are presently based on the sequential assignment of the amide nitrogen and C′, Cα and Cβ resonances as well as the remaining sidechain 15N and 13C nuclei (Pauli et al. 2001; Schuetz et al. 2010; Huber et al. 2014; Luckgei et al. 2014). This provides a large set of resonance frequencies of nuclei which are subsequently used to measure inter-nuclear distance restraints (Castellani et al. 2002). In this scheme, protons are typically not used except for the initial cross-polarization (CP) step (Pines et al. 1973; Hartmann and Hahn 1962).
Fast MAS typically leads to an increase in coherence lifetimes (T 2′) (Lewandowski et al. 2011) in protein spectroscopy for all nuclei. This increase, however, is most pronounced for protons. Only above about 50 kHz MAS is the resolution in the proton spectral dimension of perdeuterated and 100 % back-exchanged proteins (Lewandowski et al. 2011; Knight et al. 2011; Barbet-Massin et al. 2014) sufficient enough to enable sensitive proton-detected experiments. Assignment experiments and strategies using proton detection differ from the ones developed for 13C detection (Schuetz et al. 2010), leading to a strategy more similar to the one used in solution-state NMR (Kay et al. 1990; Sattler et al. 1999; Cavanagh et al. 2007), as demonstrated at 60 kHz MAS (Knight et al. 2011; Barbet-Massin et al. 2014). The efficiency of the magnetization-transfer steps is of particular interest for the choice of the optimum experimental scheme. With solid-state NMR, one has the option to use either scalar or dipolar couplings as transfer mechanism for each step. The respective advantages will be investigated in detail in this paper. The MAS frequency strongly influences the coherence lifetimes (T 2′) but also the lifetime of spin-locked magnetization (T 1ρ). Therefore, it affects the relative efficiencies of the different polarization transfer steps. The longer T 2′ times associated with faster MAS make it more attractive to utilize magnetization transfers based on scalar couplings. These transfers have been successfully implemented in solid-state NMR before (Knight et al. 2011; Baldus and Meier 1996; Barbet-Massin et al. 2013; Lesage et al. 1999; Chen et al. 2007a, b) but are often limited in efficiency by T 2′ due to the long polarization-transfer periods which are necessary because of the relatively small size of the scalar coupling interaction.
Considering two extreme strategies, the first one would be to exclusively use dipolar-coupling based transfers, derived from traditionally 13C-detected schemes (Schuetz et al. 2010) and adapted to proton detection. The other one would be to only use scalar-coupling based transfers, as typically done in solution-state NMR (Sattler et al. 1999). In both cases, a resolved amide-proton-nitrogen plane greatly facilitates an unambiguous assignment. Previous investigations at 60 kHz spinning frequency have led to the conclusion that a mixture of scalar and dipolar transfers works best in the context of assignment spectra (Knight et al. 2011).
Here, we analyze—in a quantitative manner—a new regime at MAS frequencies around 100 kHz MAS. These spinning frequencies have recently been enabled by a new generation of MAS probes (Agarwal et al. 2013, 2014; ; Ye et al. 2014).
The goal of this contribution is to find the optimum experimental procedure at these spinning frequencies for a deuterated and 100 % back-exchanged model protein (ubiquitin). For this, we investigate, in detail, which sequential resonance assignment strategies are the most efficient under these conditions and using signal detection on protons exclusively. Our approach includes four steps: First we report on coherence life times under fast MAS and how they influence magnetization transfer; second we evaluate if dipolar or J-coupling based transfers are more efficient for the different steps; third we establish the best 3D experiments for assignments; and last we demonstrate sequential assignments of ubiquitin.
Materials and methods
Sample
100 %-HN-[2H,13C,15N] ubiquitin was prepared by over-expression of uniformly 2H,13C,15N-labeled ubiquitin in E. coli and crystallization in protonated MPD with H2O to re-protonate the exchangeable sites as previously described (Igumenova et al. 2004). The rotors were filled by ultracentrifugation with filling tools (Böckmann et al. 2009) with modifications for the 0.8 mm rotors.
NMR spectroscopy
All NMR experiments were carried out on a Bruker Biospin AVANCE III 850 MHz spectrometer. The experiments at 60 kHz MAS used a commercial 1.3 mm triple resonance MAS probe (Bruker Biospin), and the data at 93 kHz MAS were acquired with a 0.8 mm 700 nL/100 kHz proton detection optimized triple-resonance MAS probe with a 2H decoupling option, built in Tallinn, Estonia. The sample temperature of 10 °C was monitored by the frequency of the supernatant water resonance line (Böckmann et al. 2009). To achieve this sample temperature, the cooling unit (BCU II, Bruker) was running on nitrogen in strong mode with the gas flow set to 540 l/h. MPD was used as an internal chemical-shift standard. All proton-detected experiments listed below employ two dummy scans and a recycle delay of T 1(1H) × 1.28 = 1.0 s (see SI-Table 5 for table with relaxation times). All these experiments used MISSISSIPPI solvent suppression (Zhou and Rienstra 2008), WALTZ-64 (Zhou et al. 2007; Shaka et al. 1983) decoupling on 15N during acquisition, and frequency-swept low-power TPPM (Thakur et al. 2006) decoupling on 1H during J-coupling based transfers as well as during indirect evolution periods. Frequency-swept low-power TPPM was chosen as the 1H decoupling sequence after carefully optimizing different low-power pulse sequences for longest coherence decay times and picking the one with the best performance. This choice was later confirmed by yielding the highest transfer efficiency for J-coupling based transfers. The 13C-detected spectra were acquired with 1024 scans and a recycle delay of 1.0 and 13 s for CP and direct excitation, respectively.
All CP-based HN and NH transfers (60 and 93 kHz) were acquired with two adiabatic cross-polarization (Hediger et al. 1995) steps, which were matched to fulfill the double-quantum (DQ) (n = 1) Hartmann–Hahn condition using a 15 kHz RF-field on the 15N channel. Hard pulses were set to 100 and 50 kHz for the 1H and 15N nutation frequencies, respectively. In the INEPT-based spectrum, the J-coupling evolution delay was set to 2.2 and 2.5 ms for the H–N INEPT step.
For 3D spectra, the details are listed in SI-Table 1 to SI-Table 4. in the Supplementary Information. For selective 90° and 180° pulses on Cα and C′, Q5 and Q3 shapes (Emsley and Bodenhausen 1990) were used with 963 or 537 µs duration, respectively. For selective 90° and 180° pulses on the aliphatic region, Q5 and Gaussian shapes were used with 293 or 154 µs duration, respectively.
The 1D spectra used to calculate transfer efficiencies used the same parameters as the respective 3D experiments without the indirect dimensions, but were acquired with 512 scans each.
The spectra were processed in Topspin and analyzed in CCPNmr (Stevens et al. 2011) and MATLAB (MATLAB 2011R, The MathWorks Inc., Natick, MA, 2011).
Results
Coherence lifetimes and transfer efficiencies
Figure 1a shows that the proton coherence life times, T 2′, increase linearly with MAS frequencies in the range from 30 to 90 kHz, as measured on the amide proton signals (integral over amide region) from ubiquitin as function of the MAS frequency. In order to extract the bulk T 2′ times the experimental data was fitted to a mono-exponential decay; although the fit is not perfect, it gives an indication on the behavior of the T 2′ times with increasing spinning frequency. From the inset in Fig. 1a it seems that even at 90 kHz, the linewidth is still determined by the coherent second-order terms in the average-Hamiltonian expansion of the proton–proton interaction for the case of deuterated and 100 % re-protonated ubiquitin. True (incoherent) relaxation effects may only play a role for a minority of residues. In Fig. 1b the residue-specific T 2′ times of amide protons are compared at 60 and 93 kHz MAS. Noticeable differences are observed for different secondary structure elements with significantly shorter T 2′ times in α-helices and longer T 2′ for β-sheets. We ascribe these differences to the typical H–H dipolar interaction network in the two geometries: In α-helices, the closest two protons are within about 2.7 Å, compared to 3.5 Å in β-sheets. On average the coherence-decay times increase by a factor of 1.7. A comparison of the T 2′ relaxation times of amide 15N at 60 and 93 kHz MAS is shown in Fig. 1c, where an increase by a factor of 1.4 can be seen. The relevant T 2′ relaxation times at 93 kHz MAS are 16.3 ± 1.5 and 59 ± 1 ms for 1H and 15N, respectively, compared to 8.5 ± 1.0 and 40.6 ± 1.0 ms at 60 kHz MAS.
T 2′ relaxation times of amide 1H and 15N in ubiquitin as measured in a spin-echo experiment (see Supporting Information, SI-Figure 1). For all experiments the sample temperature was kept at 10 °C. a The MAS-dependent T 2′ relaxation times (integral over all amide protons) for spinning frequencies from 30 kHz (magenta) to 90 kHz (red). The experimental data (dots) was fitted to the mono-exponential decay function (lines) to extract the T 2′ value. b Displays the site-specific T 2′ times of the amide protons, where each peak is individually fitted as in (a) for 60 kHz (green) and 93.3 kHz (brown). The error analysis is done using the bootstrap method. Structural elements are indicated at the top of the figure and highlighted in grey. c Depicts the comparison of the nitrogen T 2′ relaxation times (integral over all amide nitrogens) at 60 kHz (green) and 93.3 kHz (brown)
The increased T 2′ times at higher MAS frequencies are not only the basis for well-resolved proton spectra, they also influence the choice of magnetization-transfer schemes for assignment experiments. Different strategies are available to implement solid-state NMR experiments for the assignment of backbone resonances using fast MAS. Essentially, each transfer step in a multidimensional spectra can be performed using either recoupled dipolar interactions or using scalar couplings. Three effects are relevant: the maximum transfer efficiency, the influence of relaxation and the influence of the RF-field inhomogeneity.
-
1.
Efficiency: In the absence of relaxation, the efficiency of J-coupling or dipolar-coupling based experiments should be independent of the spinning frequency, as long as first-order recoupling sequences, e.g. cross polarization (CP) are used (Scholz et al. 2007). J-coupling based INEPT transfers lead to a 100 % efficient transfer in the case of an isolated spin pair. When using adiabatic CP, the theoretical transfer efficiency also approaches 100 %.
-
2.
Relaxation effects: For J-coupling based INEPT experiments, the relevant relaxation time is T 2′. In the presence of relaxation the optimum magnetization-transfer time (2τ) shortens from: \( \tau = \frac{1}{4J} \) (in the absence of relaxation) to \( \tau_{\text{opt}} = \frac{{\tan^{ - 1} \left( {\pi JT_{2}^{'} } \right)}}{2\pi J} \) assuming that both in-phase and anti-phase magnetization relax with the same time constant T 2′. The calculated optimum transfer efficiency for a simple INEPT step \( \varepsilon = \sin \left( {\pi \tau_{\text{opt}} 2J_{IS} } \right) \cdot \exp \left( {\frac{{ - 2\tau_{\text{opt}} }}{{T_{2}^{{\prime }} }}} \right) \) is illustrated in Fig. 2 as function of the T 2′ time, J-coupling strength, and optimum delay time \( \tau_{\text{opt}} \) calculated from both. A reasonably long T 2′ is the prerequisite for efficient transfer.
For a refocused INEPT step, the anti-phase magnetization—after being transferred to the S-spin—is allowed to evolve back into in-phase magnetization on the S-spin. This means the transfer efficiency ε of the refocused INEPT can be calculated as
The out-and-back INEPT transfer consists of two simple INEPT steps that go from the I-spin to the S-spin and back to the I-spin. The polarization is transverse only on the I-spin, therefore, only T 2′(I) matters, and the transfer efficiency is
The polarization can be partitioned between two destination spins. Both spin S can become polarized, when two coupling partners with strong enough J-coupling to mediate the transfer in the given delay time are present. This is the case, for example, for the transfer between Ni and Cαi and Cαi−1 (see below). Here, the transfer to the second destination nucleus (S2) has to be taken into account as well in order to calculate the transfer efficiency to one nucleus (S1):
Using the experimentally optimized delay time τ I , one can predict the transfer efficiency of INEPT steps based on the T 2′ times and J-coupling constants as summarized in Table 1. We will later use these calculated transfer efficiencies to compare them to the experimentally measured ones.
Table 1 indicates that the experimentally optimized delay times are in most cases very close to the calculated ones.
For cross-polarization (CP) experiments, the relevant relaxation time is T 1ρ. T 1ρ is also spinning-speed dependent and lengthens typically with faster spinning frequency (Lewandowski et al. 2011). Still, due to the finding that typically T 1ρ > T 2′ and d > J (where d denotes the dipolar-coupling constant) the relative performance of J-coupling based transfer increases with spinning frequency.
-
3.
A further point to be considered is RF-field inhomogeneity. Unlike the INEPT sequences, which only utilize short pulses, we have a strong influence of the RF-field distribution within measurement coil for the dipolar transfers. This is due to the Hartmann-Hahn matching condition getting narrower at faster spinning. A CP selects not only for the orientation of the N–H vector with respect to the external field, but also spatially along the rotor axis, even if the adiabatic sequences partially compensate for this effect. We will see that the first CP of a pulse sequence, therefore, has a lower transfer efficiency due to this crystallite selection and HH match section. The following CP steps operate on this already selected subset of spins and will introduce no further attenuation.
Individual transfer steps
At first we look at the individual transfer steps which will then be used in the different 3D assignment experiments described in the “3D Experiments for sequential assignments” section. We compare dipolar-coupling based transfers with scalar-coupling based transfers in the context of the 3D experiments where they will be used for as building blocks. This means, for example, that we will look at J-coupling based out-and-back transfers and at single CP steps at the same time. A scheme representing the basic assignment strategy is shown in Fig. 3. For a comparison to the X-nucleus based strategy see Fig. 4 of Schuetz et al. (2010).
Schematic overview of the connection possibilities in the proton-detected, triple-resonance correlation experiments used for the sequential backbone resonance assignment (including Cβ) in this study. The circled stops represent dimensions, and the solid and dashed lines indicate the connections for each experiment, starting at the H, N pair of residue(i) or (i ± 1), respectively. Dashed lines thus represent the same experiment as solid lines of the same color but starting at a different HN pair
For each transfer step we compare the signal intensity between the dipolar and scalar versions, as well as their theoretical predicted values according to Eqs. (1), (2) and (3). Each reported transfer efficiency in this paper was reproduced in three individual runs in different experimental sessions and with independent optimization with a standard deviation of σ ≤ 0.03.
N–H transfer steps
First we discuss the transfer from 1H to 15N and vice versa (Fig. 4), that is needed for a HNH/HSQC-type 2D experiment which delivers a characteristic fingerprint of a protein. Also, in combination with proton-detection, such an NH polarization-transfer block represents the final step for 3D experiments.
Illustration of H–N transfer steps as discussed in the text. Nuclei involved in the transfer are boxed, and dashed and solid lines indicate dipolar (through-space) transfers and J-coupling (scalar) transfers along the bond, respectively. a The HNH with CP steps (blue), b the HSQC with refocused INEPT steps (red)
In order to compare the efficiency of scalar-coupling based transfers with dipolar-coupling based transfers, we used the double INEPT experiment shown in Fig. 5a as a reference and inserted the polarization-transfer scheme to be characterized (Fig. 5b–d) after the first polarization-transfer step. For INEPT steps we concentrated on refocused INEPT (Bodenhausen and Ruben 1980) which, compared to simple INEPT steps, offers the advantage of being able to apply heteronuclear decoupling in the indirect dimension and to implement MISSISSIPPI for water suppression (Zhou and Rienstra 2008). MISSISIPPI was found perform better than water pre-saturation.
The complete HNH experiment is depicted in Fig. 5a. For one refocused H–N INEPT step, the expected transfer efficiency for ubiquitin, including decay due to transverse relaxation, is ε = 0.66 (see Table 1). The main loss comes from 1H relaxation, whereas the much longer 15N relaxation that is active during the refocusing period has a much lower impact. Therefore, no significant gain is to be expected from going to a single non-refocused INEPT step (ε = 0.70) instead of refocused INEPT step (ε = 0.66). Note that these expected transfer efficiency have an error or σ ≈ 0.03 due to the uncertainties in the T 2′ values.
The ratio of the integrals, I, of the reference experiment of Fig. 5a without, and with two additional refocused INEPT transfers (Fig. 5b), allows us to extract the efficiency of two refocused H–N/N–H INEPT steps without the need for 15N direct detection: \( \varepsilon = \frac{I\left( B \right)}{I\left( A \right)} = 0.33 \). Here we assume that the forward and backward INEPT transfers have the same efficiency. Therefore, the experimental efficiency for a single refocused H–N INEPT step is: \( \varepsilon \left( {{\text{HN}}^{\text{INEPT}} } \right) = \sqrt {\frac{I\left( B \right)}{I\left( A \right)}} = 0.57 \). Even when considering the error on the calculated value of ε = 0.66 ± 0.03, the experimental efficiency is lower. The reason for this could be the influence of the RF-field inhomogeneity which leads to incomplete excitation by the \( \frac{\pi }{2} \)-pulse and deteriorates the refocusing of the π-pulses.
The efficiency of an HNH experiment with two adiabatic CP transfers (back and forth) was also determined by comparing the intensity of the corresponding 1D spectra. Unlike in the INEPT, we assume here a difference in efficiency between the first CP step and the second CP step. The transfer efficiencies of the first and subsequent CP steps were determined by a series of 1D experiments referenced to each other: (1) HNH (Fig. 5a) with insert (d) which is two CP steps inside a decoupled HSQC, (2) with insert (c) which is four CP steps inside the decoupled HSQC, and (3) only (a) which is a normal decoupled HSQC. Referencing the experiment with insert (c) to the one with insert (d) yields the polarization-transfer efficiency of two subsequent CP steps after filtering through the first CP \( \left( {\varepsilon = \frac{I\left( C \right)}{I\left( D \right)} = 0.81} \right) \). Therefore, the efficiency of a single subsequent CP is \( \varepsilon = 0.90 \). Referencing the experiment with insert (c) to (a) of Fig. 5 gives the combined efficiency of the first and a subsequent CP step \( \left( {\varepsilon = \frac{I\left( D \right)}{I\left( C \right)} = 0.52} \right) \). With the known efficiency for a subsequent CP, the efficiency of the first CP step is \( \varepsilon \left( {{\text{HN}}^{{ 1 {\text{st CP}}}} } \right) = 0.58 \).
Therefore, the total efficiency for the HNH experiment using dipolar-coupling based transfers amounts to \( \varepsilon \left( {{\text{HNH}}^{\text{CP}} } \right) = 0.58 \, \times \, 0.90 = 0.52 \) which is 1.7 times higher than the total efficiency of \( \varepsilon \left( {{\text{HNH}}^{\text{INEPT}} } \right) = 0.57 \, \times \, 0.57 = 0.33 \) for the HNH experiment with two refocused INEPT steps. This is also evidenced in Fig. 6a where the integral of the signal obtained in the CP-based experiment is 1.9 times larger than the INEPT-based one. We note that these numbers depend to some degree on the protein investigated (due to the relaxation parameters) and on the probehead used (due to the RF-field inhomogeneity).
a Fourier transform of the first FID of the 1H,15N 2D acquired with dipolar-coupling based transfers (CP) or scalar-coupling based transfers (refocused INEPT), showing the ratio of both integrals to be 1.9. b Residue-specific SNR analysis of the HNH and HSQC spectra after 16 scans. The SNR was calculated by dividing the height of each peak by the RMS of a selected noise region. Grey shadows indicate regions with α helices or β sheets. c Schematic drawing of ubiquitin (Vijay-Kumar et al. 1987) with missing resonances in purple and the 10 peaks with lowest SNR in gray, all from the decoupled HSQC experiment
A site-specific SNR analysis was performed for ubiquitin (Fig. 6b) in order to evaluate whether signals from all amino acids can be observed in the scalar-coupling transfer-based experiment. It results that, with one exception (25Asn), all peaks that are found in the spectrum with CP transfers were also detected in the spectrum with refocused INEPT steps. It can, however, be observed that the intensities in the dipolar-coupling based experiment are on average higher than in the scalar-coupling based one. The ten peaks with the lowest SNR are all found in the scalar-coupling based experiment. A total of 15 resonances were missing in both spectra. They are marked in red in the schematic drawing of Fig. 6c and include 1Met, and the C-terminal 8 residues, 9The, 24Glu and 53Gly which are known to exhibit dynamics (Lienin et al. 1998; Schanda et al. 2010). The ten peaks with the lowest SNR are marked in blue and are predominantly located in the α-helical and loop regions.
The different intensities observed in the J-coupling mediated HNH (decoupled HSQC) are directly correlated to the site-specific proton T 2′ relaxation times (Fig. 1b). If looking at the first transfer step, however, i.e. a single H–N transfer, the efficiencies of dipolar-coupling or J-coupling-mediated polarization transfer are very similar. The relaxation loss in the first transfer step of the INEPT experiment matches approximately the loss observed in the CP due to crystallite and RF inhomogeneity in our setup. Despite this fact, it is still more advantageous to use the CP, as the following CP steps profit from the selection and will have higher efficiency than the first one. If we extrapolate the linear increase of T 2′ times of the amide protons with increasing spinning frequencies (Fig. 1a), the break-even point for the HNH experiment using two refocused INEPT steps with the one using CP steps for the HN and NH transfer would be around 180 kHz MAS (with an estimated T 2′(1H) = 28 ms) and under the assumption that T1ρ does not lead to changes in the CP efficiency. Already around 100 kHz MAS, however, the INEPT might be more efficient than CP in certain cases, as in β-sheet rich proteins (see e.g. 14Thr, 15Leu, 16Glu), or at a lower extent of back-protonation, or in the presence of fast motion. In the case of ubiquitin though, CP presents the better option.
CA–N transfer
Two experiments that both transfer magnetization between N and Cα will be compared in the following. Both CA–N steps will be embedded in a pulse sequence that starts on the N–H proton and also ends there (see the dashed lines in Fig. 7. The long 15N T 2′ times of about 60 ms at 93 kHz MAS allow us to implement scalar-coupling based out-and-back transfer from N to CA as in the typical solution-state HNCA experiment (Kay et al. 1990; Grzesiek and Bax 1992). Unlike for the HN transfer, the INEPT steps are implemented without refocusing the coherences. The out-and-back style transfer allows the in-phase magnetization to evolve into anti-phase and back into in-phase with the transverse magnetization on 15N (longer T2′ times than the CA) which leads to less loss by relaxation. The dipolar alternative is a CA–N transfer step with CP which will be combined with a direct H–CA transfer and N–H CP step.
J-coupling based out-and-back N–CA–N versus CA–N CP. Schematic drawings of transfer pathways between H, N, and CA as they will be used for 3D experiments: a HNCANH using J-coupled out-and-back N–CA–N transfer (purple), and b HCANH using a CA–N CP (red). The nuclei involved in the transfer are boxed and the two transfer steps that are compared here are emphasized with dark colors while faded colors indicate the rest of the experiment. Dipolar-coupling based transfers (through space) are indicated with dashed lines, J-coupling based transfers (scalar, via bonds) are shown as solid line. Numbers point out the order of the transfers. In this section, the spectra are acquired as 1D experiments to compare integrals of the amide region in the 1H dimension
An important difference needs to be taken into account when comparing these two transfers. For J-coupling or dipolar-coupling based methods, the splitting of polarization between Cαi or Cαi−1 is different. As the N–Cαi 1J-coupling and the N–Cαi−1 2J-coupling constants are comparable, with 11 and 7 Hz respectively, polarization is transferred to both Cα when applying the scalar-based transfer (Fig. 7a, transfer step 2 and 3). On contrast, when using dipolar couplings to transfer polarization, the 1/r3-distance dependence of the dipolar interaction leads to a much weaker transfer to the Cαi−1. Compared to the Cαi (d N,C = 1.46 Å), the Cαi−1 (d N,C = 2.41 Å) is 1.7 times as far away which will result in a reduction of transfer efficiency by roughly a factor of 4.5 and makes transfer to the Cαi−1 weak (Fig. 7b, transfer step 2). This effect will be further amplified by dipolar truncation effects which favor the transfer across shorter distances (Bayro et al. 2009). This difference will result in different information content when using one or the other building block for HNCA 3D experiments.
The determination of the N–CA–N out-and-back transfer efficiency is accomplished by implementing the out-and-back transfer as an H–N–CA–N–H, where the H–N and N–H transfer is CP-based. This is then acquired as a 1D experiment and referenced to an H–N–H experiment without N–CA–N transfer: \( { \varepsilon }\left( {\text{NCAN}} \right) = \frac{{I\left( {\text{HNCANH}} \right)}}{{I\left( {\text{HNH}} \right)}} \). Experimentally one obtains \( { \varepsilon }\left( {\text{NCAN}} \right) = 0.22 \). The expected value for ubiquitin for the transfer from N–Cαi and N–Cαi−1 and back can be calculated according to Eq. ( 3) and is \( \varepsilon = 0.16 \) for Cαi and \( \varepsilon = 0.09 \) for Cαi−1. This results in a total of \( \varepsilon = 0.25 \), which is close to the experimental value of \( \varepsilon = 0.22 \).
The corresponding dipolar experiment, an adiabatic CP step between CA and N, involves a transfer over a 1 kHz dipolar coupling (1.46 Å distance). The experimental value for this transfer step can be determined by comparing the integral (I) of the 1D signal of the H–CA–N–H to the H–N–H integral of the 1D signal in the experiment without CA–N transfer, and then multiplying it by the efficiency of the first H–N transfer over the efficiency of the H–CA transfer: \( \varepsilon \left( {\text{CAN}} \right) = \frac{{I\left( {\text{HCANH}} \right)}}{{I\left( {\text{HNH}} \right)}} \cdot \frac{{ \varepsilon \left( {\text{HN}} \right)}}{{ \varepsilon \left( {\text{HCA}} \right)}} \). The transfer efficiency of the H–CA CP was determined to \( \varepsilon \left( {\text{HCA}} \right) = 0.31 \) (see Supporting Information). Therefore, \( \varepsilon \left( {\text{CAN}} \right) = 0.39 \). In a later section we will examine whether it is of advantage to only have one correlation using a higher efficiency transfer (dipolar), or whether it makes sense to obtain two correlations and only acquire one experiment, but with more scans (out-and-back transfer).
CO–N transfer
The comparison of the two transfer strategies between Ni and C′i−1 follows the same principle ideas as the comparison for Ni and Cαi steps in the previous section. Contrary to the N–CA–N step, the N–CO–N out-and-back transfer mainly correlates Ni with C′i−1 (Fig. 8a, transfer 2, 3). The reason for this lies in the strong N–C′i−1 coupling of J = 15 Hz, and the negligible coupling of N–Ci′ with J = −0.2 Hz. Analogously to the previous section, the expected transfer efficiency is calculated to ε = 0.42 according to Eq. ( 2) using the values from Table 1. The experimental transfer efficiency is measured by comparing an H–N–CO–N–H transfer with an H–N–H transfer, in analogy to the HNCANH. The transfer efficiency is determined to be \( \varepsilon \left( {\text{NCON}} \right) = \frac{{I\left( {\text{HNCONH}} \right)}}{{I\left( {\text{HNH}} \right)}} = 0.41 \) which is in good agreement with the theoretical value.
J-coupling based out-and-back N–CO–N transfer versus CO–N CP. Schematic drawings of transfer pathways between H, N, and CO as they will be used for 3D experiments: a HNCONH using J-coupled out-and-back N–CO–N transfer (brown), and b HCONH using a CO–N CP (tan). The nuclei involved in the transfer are boxed and the two transfer steps that are compared here are emphasized with dark colors while faded colors indicate the rest of the experiment. Dipolar-coupling based transfers (through space) are indicated with dashed lines, J-coupling based transfers (scalar, via bonds) are shown as solid line. Numbers point out the order of the transfers. In this section, the spectra are acquired as 1D experiments to compare integrals of the amide region in the 1H dimension
In the corresponding dipolar experiment, an adiabatic CP step between C′i−1 and Ni over a distance of 1.33 Å (corresponding to a 1.3 kHz coupling) (Fig. 8b, transfer 2), the experimental transfer efficiency was determined by comparing the H–CO–N–H to an H–N–H taking into account the efficiency ratio of the preparation step which is H–N CP in one case and H–CO in the other one: \( \varepsilon \left( {\text{CON}} \right) = \frac{{I\left( {\text{HCONH}} \right)}}{{I\left( {\text{HNH}} \right)}} \cdot \frac{{ \varepsilon \left( {\text{HN}} \right)}}{{ \varepsilon \left( {\text{HCO}} \right)}} \). The H–CO CP transfer efficiency was determined to be 0.27 (see Supporting Information). Therefore, the CO–N CP transfer efficiency is \( \varepsilon \left( {\text{CON}} \right) = 0.57 \).
CA–CO transfer
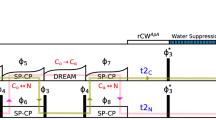
It has been shown that, at 60 kHz MAS, the proton-detected version of an N(CO)CA (Pauli et al. 2001; Knight et al. 2011) can be further improved by changing the single CA–CO J-coupling based INEPT step to an J-coupled out-and-back CO–CA–CO block (Barbet-Massin et al. 2013; Yamazaki et al. 1994). Another possibility would be to use a DREAM transfer (Verel et al. 1998, 2001).
The J-coupled out-and-back CO–CA transfer (Fig. 9a, step 2, 3) is expected to yield a transfer efficiency of ε = 0.68 [using Eq. (2), see Table 1 for values used]. Experimentally, the extracted transfer efficiency is given by the intensity ratio of HCOCACONH over HCONH: \( \varepsilon \left( {\text{COCACO}} \right) = \frac{{I\left( {\text{HCOCACONH}} \right)}}{{I\left( {\text{HCONH}} \right)}} = 0.51 \).
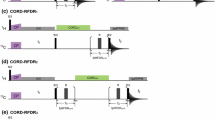
J-coupling based out-and-back CO–CA–CO versus CA–CO DREAM. Schematic drawings of transfer pathways to correlate Hi, Ni, and CAi − 1 as they will be used for 3D experiments: a HCOCACONH using J-coupled out-and-back CO–CA–CO transfer (dark blue), and b HCACONH using a CA–CO DREAM (light blue). The nuclei involved in the transfer are boxed and the two transfer steps that are compared here are emphasized with dark colors while faded colors indicate the rest of the experiment. Dipolar-coupling based transfers (through space) are indicated with dashed lines, J-coupling based transfers (scalar, via bonds) are shown as solid line. Numbers point out the order of the transfers. In this section, the spectra are acquired as 1D experiments to compare integrals of the amide region in the 1H dimension
The extraction of the transfer efficiency for the DREAM experiment (Fig. 9a, step 2) includes several steps, as it cannot be directly compared to either an (H)CONH or an (H)CANH. When taking the ratio between 1D spectra of the HCACONH and the HCONH one also needs to consider the different initial CP step and multiply it by the efficiency of the H–CO CP over the H–CA CP: \( \varepsilon \left( {\text{CACO}} \right) = \frac{{I\left( {\text{HCACONH}} \right)}}{{I\left( {\text{HCONH}} \right)}} \cdot \frac{{ \varepsilon \left( {\text{HCO}} \right)}}{{ \varepsilon \left( {\text{HCA}} \right)}} \). Although the C′ is spatially closer to the amide N (2.02 Å) than the Cα (2.13 Å), the H–CA CP is slightly more efficient than the H–CO step with transfer efficiencies of ε = 0.31 and ε = 0.27, respectively. With the ratio between the H–CO and H–CA steps, this results in a transfer efficiency of \( \varepsilon \left( {\text{CACO}} \right) = 0.41 \) for the CA–CO DREAM. This is slightly lower than the experimentally measured transfer efficiency of a refocused CA–CO INEPT step of \( \varepsilon \left( {\text{CACO}} \right) = 0.45 \).
CO–CA transfer
The opposite is true for the comparison between a refocused CO–CA INEPT (Fig. 10a) step and a CO–CA DREAM (Fig. 10b). Both transfers occur with the same pathway, for example, in an HCACBCACONH. Therefore, to determine the experimental transfer efficiency, the ratio between the HCACBCACONH and HCACBCANH steps were taken, multiplied by the ratio of the CA–N over the CO–N step: \( \varepsilon \left( {\text{COCA}} \right) = \frac{{I\left( {\text{HCACBCONH}} \right)}}{{I\left( {\text{HCACBCANH}} \right)}} \cdot \frac{{ \varepsilon \left( {\text{CAN}} \right)}}{{ \varepsilon \left( {\text{CON}} \right)}} \). The experiment using INEPT for the CO–CA step yielded a \( \varepsilon \left( {\text{COCA}} \right) = 0.37 \) transfer efficiency while the one using DREAM was \( \varepsilon \left( {\text{COCA}} \right) = 0.43 \).
CO–CA refocused INEPT versus CO–CA DREAM. Schematic drawings of transfer pathways to connect Hi, Ni, COi as they will be used for 3D experiments: a HCOCANH using INEPT transfer (magenta), and b HCOCANH using a CO–CA DREAM (purple). The nuclei involved in the transfer are boxed and the two transfer steps that are compared here are emphasized with dark colors while faded colors indicate the rest of the experiment. Dipolar-coupling based transfers (through space) are indicated with dashed lines, J-coupling based transfers (scalar, via bonds) are shown as solid line. Numbers point out the order of the transfers. In this section, the spectra are acquired as 1D experiments to compare integrals of the amide region in the 1H dimension
CA–CB transfer
In the deuterated and back-exchanged sample the polarization to and from CB is best transferred via the CA spin. This leads to two choices for out-and-back type CA–CB–CA transfer. One possibility is J-coupling based transfer as for the CO–CA–CO or N–CO/CA–N transfer (Fig. 11b). The other possibility is two DREAM transfers (Fig. 11a).
CA–CB–CA double DREAM to a J-coupling based out-and-back CA–CB–CA. Schematic drawings of transfer pathways between H, N, and CB as will be used for 3D experiments: a HCACBNH using a CA–CB–CA DREAM transfer (light green), and b HCACBCANH using an out-and-back CA–CB–CA J-coupling based transfer (dark green). The nuclei involved in the transfer are boxed and the two transfer steps that are compared here are emphasized with dark colors while faded colors indicate the rest of the experiment. Dipolar-coupling based transfers (through space) are indicated with dashed lines, J-coupling based transfers (scalar, via bonds) are shown as solid lines. Numbers point out the order of the transfers. In this section, the spectra are acquired as 1D experiments to compare integrals of the amide region in the 1H dimension
Using the values in Table 1, the theoretical transfer efficiency calculates according to Eq. (2) to ε = 0.36.
The experimentally measured transfer efficiency depends slightly on the transfer pathway used around the CA–CB–CA transfer. When the transfer is surrounded by another J-coupled out-and-back transfer as in the HNCACBCANH, the efficiency is slightly higher compared to just having an H–CA CP preceding. This is due to the T 2′ filtering induced by another J-coupling based transfer where residues with longer T 2′ times are favored (see Fig. 1b). However, both values \( \varepsilon \left( {\text{CACBCA}} \right) = \frac{{I\left( {\text{HCACBCANH}} \right)}}{{I\left( {\text{HCANH}} \right)}} = 0.42 \) (for the HCACBCANH) and \( \varepsilon \left( {\text{CACBCA}} \right) = \frac{{I\left( {\text{HNCACBCANH}} \right)}}{{I\left( {\text{HNCANH}} \right)}} = 0.45 \) (for the HNCACBCANH) are significantly higher than predicted.
The DREAM experiment was carefully optimized for both directions with regards to matching condition, carrier frequency, shape of the adiabatic amplitude modulation, and duration (Westfeld et al. 2012). Technically the transfer efficiency cannot be determined in this case, as the polarization that remains on Cα cannot be phase-cycled out and adds with the same phase to the signal. When normalizing the double DREAM to the reference experiment by taking the ratio of HCACBCANH over HCANH, one can get a rough estimate of \( \varepsilon \left( {\text{CACBCA}} \right) = \frac{{I\left( {\text{HCACBCANH}} \right)}}{{I\left( {\text{HCANH}} \right)}} = 0.23 \) for the combination of two DREAM transfers (\( \varepsilon = 0.48 \) per step).
A summary of the transfer efficiencies of the elementary steps for resonance-assignment experiments for deuterated and 100 % back-exchanged proteins, as measured on the model system ubiquitin, is given in Table 2.
3D Experiments for sequential assignments
In this section, we evaluate the experiments necessary for a sequential walk involving the HN, N, C′, Cα and Cβ spins. Sequential NH pairs can indeed be connected via the C′, the Cα or the Cβ spins. The three spins show different spectral resolution and their chemical shifts have different information content. Furthermore, the transfer efficiencies of the corresponding experiments are different. As in strategies using heteronuclear detection (Schuetz et al. 2010), experiments which connect the amide nitrogen to the surrounding carbon spins are the most straightforward and efficient ones—both in implementation and resulting signal. The forward and backward walks (inside the same or to the previous amino acid) have different efficiencies for the Cα and C′ spins due to the different number of transfers involved. Experiments connecting the more remote Cβ spins use multiple relayed transfers which result in a lower overall transfer efficiency, and therefore, a lower SNR. However, as in classical approaches, they contain precious additional information about spin systems and show a better dispersion which increases from C′ to Cα to Cβ. These different walks present complementary procedures, and a combination of them will likely result in the most complete assignment, as will be shown in the “Sequential assignment of ubiquitin” using the example of ubiquitin. The three procedures will be detailed in the following, and a complete list of different experiments with their individual transfer pathways is given in the Supporting Information, SI-Table 1.
Backbone walk using the Cα resonances
In order to perform a walk along the Cα resonances, one needs to connect the Hi, Ni pair with the Cαi (intra-residue) and Cαi−1 (inter-residue) resonances. This can be achieved in different ways as illustrated in Fig. 12. As discussed before, the (HN)CANH with N–CA–N out-and-back transfer based on scalar couplings (Fig. 12: purple pathway) splits the magnetization from Ni to both Cαi and Cαi−1. This will lead to correlations of Hi, Ni, Cαi,i−1 in one spectrum. This corresponds to the common HNCA scheme in solution-state NMR, except that the initial HN INEPTs are exchanged to CPs here. In principle, this one spectrum would suffice for the Cα backbone walk, as illustrated in Fig. 13a. The difference in peak intensity of the stronger Cαi and weaker Cαi−1 resonance signal in the (HN)CANH is due to the different coupling strengths of the J-couplings (7–11 Hz for N–Cαi and 4–8 Hz for N–Cαi−1) and allows one, to some degree, to differentiate between them.
Schematic drawing of connections to correlate the Cα for sequential assignment. There are two different possibilities: (1) The purple line connecting Cαi and Cαi−1 with Hi, Ni, which would correspond to the (HN)CANH, or (2) a combination of the blue and red line, where the red line connects HiNiCαi as in the (H)CANH and the blue line connects HiNiCαi−1 which could be either an (H)CA(CO)NH with CA–CO DREAM transfer, or a (HCO)CA(CO)NH with out-and-back J-coupling based CO–CA–CO transfer
Another way is to combine one spectrum for the Cαi correlation and one spectrum for the Cαi−1 (Fig. 13b). In this section, we first compare the transfer to the Cαi resonances in the scalar (HN)CANH experiment (Fig. 14a) to the transfer to the Cαi resonance in an dipolar (H)CANH experiment (Fig. 14b) Next, we compare two different pulse sequences to obtain the Cαi−1 correlations, the dipolar (H)CA(CO)NH (Fig. 14c) and the scalar (HCO)CA(CO)NH (Fig. 14d). In the following, we compare these 3D experiments based on mean SNR and information content by looking at the residue-specific SNR. It should be noted that the mean SNR was calculated from assigned peaks only. It is meant to give an impression of the performance of an experiment concerning sensitivity and efficiency, while the information content is evaluated separately investigating the number of missing peaks in the spectrum. In the end, we evaluate which strategy works better: Having one spectrum for both correlations or combining two separate experiments showing one correlation each.
Pulse sequences of the triple-resonance experiments used for the Cα backbone walks. a (HN)CANH using NCAN J-coupling based transfer as in the solution-state experiment (Grzesiek and Bax 1992) and applied to solid-state NMR (Linser et al. 2008) but substituted with HN/NH CP steps. b (H)CANH (Knight et al. 2011) based on (H)CAN (Pauli et al. 2001), c (HCO)CA(CO)NH as in (Barbet-Massin et al. 2013), d (H)CA(CO)NH with DREAM based on NCOCA (Schuetz et al. 2010). The schematic transfer pathway figures show the ordering of the transfer steps and circle the nuclei of the dimensions in the 3D experiment. Solid and dashed lines stand for J-coupling and dipolar-coupling based transfers, respectively. Orange boxes and shapes indicate adiabatic CP transfer steps, black narrow and broad rectangles stand for 90° and 180° pulses, respectively. Narrow and broad shaped pulses correspond to for 90° and 180° selective pulses, respectively. DREAM transfers are indicated by the dark blue shape. J-coupling based transfers are underlayed in green (homonuclear transfer) and purple (heteronuclear transfer), z-filters are marked with brown, and MISSISSIPPI water suppression with blue. During all J-coupling evolution times and indirect dimensions 1H decoupling (fslp TPPM (Thakur et al. 2006)) was applied and during acquisition 15N WALTZ64
HiNiCαi correlation
An (H)CANH with all dipolar-coupling based transfers (Fig. 12: red line, pulse sequence: Fig. 14b) yields the correlation of Cαi to Hi, Ni only. The total efficiency amounts to:
This is within the error of the total efficiency of the (HN)CANH experiment with:
The theoretical ratio is thus 1.1. However, in one experiment the polarization is split while in the other it only ends up on one peak. Despite this, the out-and-back J-coupling based (HN)CANH shows a higher SNR of the individual peaks as shown in Fig. 15a. On average the Cαi have a SNR of 75, whereas in the dipolar-coupling based (H)CANH the average SNR is 67, where the ratio of 1.1 confirms the theoretical value. However, as seen before in the HNH with refocused INEPT steps, the J-coupling based experiment shows low-intensity to no peaks for residues in certain mobile regions of the protein (8Leu–10Gly, 23Ile–25Asn, 52Asp–53Gly). This can also be seen for the (HN)CANH where six peaks are missing when compared to the dipolar experiment. Using the INEPT (HN)CANH transfer for the Cαi assignment thus leads to a higher SNR, but fewer assigned resonances.
Comparison of the residue-specific SNR for a Cαi resonances and b Cαi−1 resonances. For the Cαi resonances the scalar-coupling based (HN)CANH (purple) is compared to the completely dipolar-coupling based (H)CANH (red). For the Cαi−1 the out-and-back J-coupling (HCO)CA(CO)NH (dark blue) is compared to an (H)CA(CO)NH using DREAM (light blue) and the Cαi−1 of the (HN)CANH (purple). α-helices and β-sheets are underlayed with grey. In (a) yellow highlights illustrate differences in information content of both spectra, in (b) the yellow bars indicate peaks that only appear in the (H)CA(CO)NH with DREAM. The mean SNR is calculated only from existing peaks
HiNiCα i−1 correlation
We assess two different experiments that yield Hi, Ni, Cαi−1 correlations: the (H)CA(CO)NH with all dipolar-coupling based transfers and with DREAM for the CA–CO transfer (Fig. 14c), and the (HCO)CA(CO)NH with CO–CA–CO out-and-back J-coupling based transfers and the remaining transfers with CP steps (Fig. 14d).
The (H)CA(CO)NH benefits from the slightly larger efficiency of the H-CA CP compared to the H–CO CP, but suffers from the less efficient DREAM transfer from CA to CO. The total efficiency is:
This is slightly lower than the total transfer efficiency of the (HCO)CA(CO)NH:
The theoretical ratio is thus 1.1. In Fig. 15 the difference in the mean SNR is distinctly larger than one would expect, as the ratio amounts to 61/42 = 1.5, which clearly favors the scalar version with respect to signal/noise.
A combination of two dipolar-coupling based experiments provides more complete assignments than one J-coupling based experiment showing both Cαi and Cαi−1. In the case of the Cαi resonances it is clear that the scalar-coupling based (HN)CANH experiment is more efficient. For the Cαi−1 resonances the situation is more difficult. If only one experiment was used, the number of scans could be doubled to have the same total experimental time. For the Ni–Cαi−1 correlation in the (HN)CANH experiment with four scans the SNR is 36 as shown in Fig. 15. With eight scans the SNR would be 51, higher than the DREAM-based (H)CA(CO)NH, but still lower than the out-and-back J-coupling based (HCO)CA(CO)NH. So, based on SNR, the (HCO)CA(CO)NH is the best choice. However, both the (HCO)CA(CO)NH and the (HN)CANH have missing peaks that do appear in the DREAM-based experiment. Therefore, despite the higher overall information content of the (HN)CANH, there are more peaks missing, compared to the combination of the two experiments. In conclusion, the (HN)CANH enables a complete backbone walk along the Cα, but is more susceptible to having weaker resonances, possibly stemming from mobile residues, or strongly coupled spins, disappear into the noise. Another disadvantage of using one spectrum for Cαi and Cαi−1 correlations is spectral crowding of resonances, especially when investigating larger proteins. For ubiquitin there already were eight peaks that were either assigned to more than one resonance or displayed significant overlap.
If one’s goal is the most complete assignment, the best combination for the sequential walk along the Cα resonances are the two experiments which use all dipolar-couplings, that is, the (H)CANH for Cαi and the (H)CA(CO)NH with DREAM for Cαi−1.
Backbone walk along the C′ resonances
For a backbone walk connecting the NiHi to the two neighboring C′, we have to use two spectra: An intra-residue one correlating Hi, Ni, Ci′, and an inter-residue one correlating Hi, Ni, C′i−1 (Fig. 16). For the Hi, Ni, Ci′ correlation (pink lines), we compare two (H)CO(CA)NH, where one has a CO–CA DREAM transfer (Fig. 17a), and the other one uses INEPT (Fig. 17b). All other transfer steps are accomplished by CP. For the Hi, Ni, C′i−1 correlation (brown lines), we look at an (HN)CONH with N–CO–N out-and-back scalar-coupling based transfer and dipolar transfers for all other steps (Fig. 17c), and an (H)CONH with all dipolar-coupling based transfers (Fig. 17d).
Schematic drawing of connections that correlate HiNiCi′ (pink) and HiNiC′i−1 (brown) for sequential assignment. The brown line could be achieved by an (H)CONH or an (HN)CONH. For the dotted pink line one could use an (H)CO(CA)NH with CO–CA DREAM or CO–CA INEPT. Both experiments utilize transfer steps via the Cα which is depicted as contact to the Cα stop without highlighting the stop itself. The combination of one of each allows for a sequential walk along the C′ resonances via the HN pair
Pulse sequences of the triple-resonance experiments used for the C′ backbone walks. a (H)CO(CA)NH with DREAM, the proton-detected version of the NCACO (Schuetz et al. 2010), b (H)CO(CA)NH with INEPT as shown in (Barbet-Massin et al. 2014), c (HN)CONH using NCON J-coupling based transfer as in the solution-state experiment (Kay et al. 1990) and applied to solid-state NMR (Linser et al. 2008) but substituted with HN/NH CP steps, d (H)CONH (Knight et al. 2011) based on (H)CON (Pauli et al. 2001). The schematic transfer pathway figures show the ordering of the transfer steps and circle the nuclei of the dimensions in the 3D experiment. Solid and dashed lines stand for J-coupling and dipolar-coupling based transfers, respectively. Orange boxes and shapes indicate adiabatic CP transfer steps, black narrow and broad rectangles stand for 90° and 180° pulses respectively. Narrow and broad shaped pulses correspond to for 90° and 180° selective pulses, respectively. DREAM transfers are indicated by the dark blue shape. J-coupling based transfers are underlayed in green (homonuclear transfer) and purple (heteronuclear transfer), z-filters are marked with brown, and MISSISSIPPI water suppression with blue. During all J-coupling evolution times and indirect dimensions 1H decoupling (fslp TPPM(Thakur et al. 2006)) was applied and during acquisition 15N WALTZ64 (Zhou et al. 2007)
HiNiCi′ correlation
The comparison of the (H)CO(CA)NH (Fig. 18a, b) is relatively straightforward since the transfer pathway is the same and the only difference is the CO–CA transfer. From the previous section we know that the DREAM with ε = 0.43 is more efficient than the INEPT-step transfer which is ε = 0.37. This results in total efficiencies of
using DREAM, and
using INEPT.
Comparison of experiments needed to connect a Ci′ and b C′i−1. For each case two experiments are compared, for Ci′ an (H)CO(CA)NH with CO–CA DREAM (purple) with one using CO–CA INEPT (pink), and for the C′i−1 an (H)CONH with all dipolar transfers (tan) and an (HN)CONH with J-coupling based N–CO–N transfer (dark brown). Yellow highlights point to the differences in information content. The displayed mean SNR is based on existing peaks only
The ratio between the two efficiencies is 1.2. When comparing the residue-specific SNR in Fig. 18a the (H)CO(CA)NH with DREAM (purple) has a higher SNR as expected with 23 compared to 19 for the (H)CO(CA)NH with INEPT (pink). The ratio between the two corresponds with 1.2 to the calculated ratio. Furthermore there are six resonances (7Thr, 8Leu, 10Gly, 22Thr, 51Glu, 52Asp, 71Leu) that can only be found in the version with DREAM. The 3D experiment for Ci′ correlations are thus best done with CO–CA DREAM.
HiNiC′i−1 correlation
For the connection of the HiNiC′i−1 spins, the comparison is less straightforward, as the transfer pathways for J-based and dipolar-based experiments are different. Despite the N–CO–N transfer being less efficient with ε = 0.41 than the simple CO–N CP step with ε = 0.57, the sequence using the N–CO–N transfer benefits from the initial H–N CP which is higher than the H–CO CP. Therefore, in total, for the (HN)CONH experiment Fig. 17c one expects an efficiency of
The total transfer efficiency for the (H)CONH Fig. 17d on the other hand is:
The calculated ration is 1.5. This ratio is close to that reflected in the SNR of the individual resonances in Fig. 18b. The (HN)CONH (brown) has a SNR of 110 while the (H)CONH (tan) has 64, resulting in a slightly higher ratio of 1.7. Here, too, the J-coupling based experiment shows less assigned peaks than the dipolar-coupling based one. However, the (HN)CONH is only missing three peaks (9Thr, 23Ile, 25Asn) where most shifts could even be complemented with information from one of the (H)CO(CA)NH experiment using DREAM. We thus suggest that the 3D experiment for C′i−1 correlations is best done with N–CO–N using INEPT.
In conclusion, the best combination for the C′-backbone is to walk forward using the (H)CO(CA)NH where the CO–CA step is done with a dipolar DREAM and backward using an (HN)CONH with a scalar-coupling based out-and-back transfer.
Backbone walk using the Cβ resonances
The assignment of Cβ resonances (Fig. 19) not only supplements the backbone assignments but also facilitates the identification of spin systems and their assignment to residue types, and with that the sequential backbone walk for overlapped resonances. The choice for the most efficient CA–CB–CA transfer has already been discussed. We use the out-and-back J-coupling based transfer with complete evolution to CB for all following experiments as it is more efficient (ε = 0.42) than a double DREAM step (ε = 0.23). Nevertheless, there are multiple pulse sequences that can be used to assign Cβi and Cβi−1 which will be discussed in the following.
Schematic drawing of connections to correlate the Cβ for sequential assignment. There are two different possibilities: (1) The orange line connecting CBi and CBi−1 with Hi, Ni, which would correspond to the (HNCA)CB(CA)NH. (2) A combination of the green and yellow line, where the green line connects Hi, Ni Cβi as in the (HCA)CB(CA)NH and the yellow line connects HiNiCBi−1 which could be either an (HCA)CB(CACO)NH or an (HCOCA)CB(CACO)NH
HiNiCβi correlation
As already mentioned, not only the CA–CB–CA transfer step has to be considered, but the preceding or following steps also need to be taken into account. The easiest way might be to go directly from H to Cα and then to Cβ in a (HCA)CB(CA)NH, which will correlate Hi, Ni and Cβi (Fig. 20a). Another possibility would be to use the out-and-back J-coupling based N–CA–N building block around the CA–CB–CA transfer in an (HNCA)CB(CA)NH (Fig. 20b). This corresponds to the building block in the common solution-state HNCACB (Wittekind and Mueller 1993). Analogous to the (HN)CANH this will lead to correlations of both the Cβi and Cβi−1 with the Hi, Ni spin pair. Figure 21a shows that the Cβi resonances of the (HNCA)CB(CA)NH have almost the same SNR (57) as the Cβi in an (HCA)CB(CA)NH where the polarization is not split (56). For the sake of completeness, the residue-specific SNR of the DREAM-based (HCA)CB(CA)NH (sequence not shown) is also shown in Fig. 21a, as we have previously seen that dipolar-based experiments have fewer peaks disappear into the noise. Here, however, it is clear that with a SNR of 13 it offers no advantage compared to the (HCA)CB(CA)NH with out-and-back J-coupling based transfer. Using an (HNCA)CB(CA)NH (INEPT N–CA–N transfer around the CA–CB–CA transfer) yields the same SNR as an (HCA)CB(CA)NH for Cβi. However, the (HCA)CB(CA)NH contains peaks for four more resonances and is therefore chosen to assign the Cβi resonances because of the higher information content.
Pulse sequences of the triple-resonance correlation experiments for walks along Cβ. a (HCA)CB(CA)NH as in (Barbet-Massin et al. 2013), b (HNCA)CB(CA)NH as the solution-state experiment (Wittekind and Mueller 1993), again with HN/NH CP steps, and aiming for a complete transfer from CA to CB to only get CB correlations, c (HCA)CB(CACO)NH as in (Barbet-Massin et al. 2014), d (HCOCA)CB(CACO)NH with COCACO out-and-back J-coupling based transfers (Yamazaki et al. 1994). The schematic transfer pathway figures show the ordering of the transfer steps and circle the nuclei of the dimensions in the 3D experiment. Solid and dashed lines stand for J-coupling and dipolar-coupling based transfers, respectively. Orange boxes and shapes indicate adiabatic CP transfer steps, black narrow and broad rectangles stand for 90° and 180° pulses respectively. Narrow and broad shaped pulses correspond to for 90° and 180° selective pulses, respectively. J-coupling based transfers are underlayed in green (homonuclear transfer) and purple (heteronuclear transfer), z-filters are marked with brown, and MISSISSIPPI water suppression with blue. During all J-coupling evolution times and indirect dimensions 1H decoupling [fslp TPPM (Thakur et al. 2006)] was applied and during acquisition 15N WALTZ64
Comparison of the residue-specific SNR for a Cβi resonances and b Cβi−1 resonances. For the Cβi resonances the scalar-coupling based (HCA)CB(CA)NH (dark green) is compared an (HNCA)CB(CA)NH with an additional NCAN out-and-back transfer around the CACBCA transfer (orange), and an (HCA)CB(CA)NH with DREAM (light green). The yellow highlights point to residues where the Cβi can be found in the (HCA)CB(CA)NH but not in the (HNCA)CB(CA)NH. For the CBi−1 the INEPT-based (HCA)CB(CACO)NH (yellow) is compared to an COCACO out-and-back (HCOCA)CB(CACO)NH (gray) and the CABi−1 of the (HNCA)CB(CA)NH (orange). Here the yellow highlights indicate Cβi−1 resonances that are found in the (HNCA)CB(CA)NH, but not in the (HCA)CB(CACO)NH. α-helices and β-sheets are underlayed with gray. The mean SNR is calculated only from existing peaks
HiNiCβi−1 correlation
We investigated two different alternatives to complement the correlations to the Cβi resonances with the Cβi−1 resonances. The first experiment, an (HCA)CB(CACO)NH (Fig. 20c), uses a CA–CO INEPT step, which is slightly more efficient than the CA–CO DREAM (ε = 0.45 vs. ε = 0.41). The second experiment is an (HCOCA)CB(CACO)NH where the CA–CB–CA building block is embedded in a CO–CA–CO out-and-back J-coupling based building block (Fig. 20d). The first experiment has the advantage of a slightly more efficient H–CA CP step, while the second one benefits from the out-and-back CO–CA–CO compared to the single CA–CO INEPT step. The total transfer efficiencies amount to the same value when calculated:
and
The theoretical values compare with a ratio of 1. From Fig. 21b it is clear, however, that experimentally this is not the case, since a ratio of 1.75 is observed: the (HCOCA)CB(CACO)NH has a distinctly lower SNR with 24 than the (HCA)CB(CACO)NH experiment with 42. The decision for one experiment is complicated, however, by the information content. The (HCOCA)CB(CACO)NH allows one to assign three more residues (23Ile, 59Thr, 64Glu), while the (HCA)CB(CACO)NH has only one residue (71Leu) that is missing in the (HCOCA)CB(CACONH). In order to find the best strategy, we therefore have to look at the possible combinations.
The most complete assignment of Cβ is achieved by combining the (HCA)CB(CA)NH with the (HNCA)CB(CA)NH: For the Cβi resonances our best choice was the (HCA)CB(CA)NH based on the same SNR compared to the (HNCA)CB(CA)NH, but with a higher information content. However, the (HNCA)CB(CA)NH not only provides Cβi, but also Cβi−1 peaks. Compared to both other choices for the Cβi−1 assignment, the (HNCA)CB(CA)NH has the highest information content. Three resonances show exclusively peaks in this spectrum, and four more are missing in either the (HCA)CB(CACO)NH or (HCOCA)CB(CACO)NH. Therefore, the best combination for the walk along the Cβ resonances consists of the (HCA)CB(CA)NH for the forward walk, and the (HNCA)CB(CA)NH for the backbone walk using the Cβi−1 resonances of the latter. Overall it is also here the case that the single experiment will lead to more missing residues in the assignment.
The results for all spectra are summarized in the Supporting Information SI-Table 6. The pulse sequences that were chosen based on evaluation in the last section are marked in red.
Sequential assignment of ubiquitin
Based on the results of the previous sections, we chose a combination of six experiments for the sequential assignments of ubiquitin: (H)CANH and (H)CA(CO)NH for the assignment along Cα, (HN)CONH and (H)CO(CA)NH with DREAM for the path connecting C′, and (HNCA)CB(CA)NH and (HCA)CB(CA)NH for the walk along Cβ (underlined pulses sequences in Figs. 14, 17 and 20, marked in red in Supporting Information, SI-Table 6). The connections of the 3D correlation experiments are illustrated in Fig. 22c. The individual experimental times are listed in the table in Fig. 22a and yield a total acquisition time of about 35 h. This means that about 3 days of measurement time are required for six experiments to assign 98 % of the C′, Cα and Cβ resonances in ubiquitin using 500 μg (59 nmol) of protein. The assignment is deposited in the BMRB under accession number 26604. Using only this set of experiments, the C′, Cα and Cβ resonances could be assigned to 96, 99, and 97 % for residues 1–71 (omitting glycines for CB), and the amide nitrogen und protons to 99 % (residues 2–71, omitting prolines). For the Cα and Cβ, only residue 37Pro was missing, while for the C′, in addition, 24Gly and 71Leu were left unassigned. A representative walk along residues 43–49 is shown in Fig. 22b to give an impression of the quality of the spectra.
a Table with experimental times for chosen assignment spectra. b Representative strip plot with peaks of the six 3D spectra assigned to their respective backbone resonance signal. Asterisks indicated aliased peaks. c Connectivity plot showing the correlations of the six experiments used for the assignment of ubiquitin
Conclusions
We have observed a linear increase of T 2′ coherence decay times with increasing MAS frequency up to the measured limits of 93 kHz MAS with deuterated and 100 % back-exchanged ubiquitin at a static field strength corresponding to a proton frequency of 850 MHz. Despite the increase in T 2′ coherence decay times at 93 kHz MAS, compared to lower MAS frequencies, in most cases the dipolar-based transfers outperform INEPT-based transfers as building blocks for assignment experiments. INEPT-based transfers in some cases show a higher efficiency, especially in regions of the proteins with β-sheets, but they are also more susceptible to having resonances disappear into the noise where dynamics or a higher local proton density lead to shorter T 2′ times. When using dipolar-based transfers as building blocks, the average SNR might be lower, however, it is still sufficient for finding and picking peaks with the advantage of being able to assign more resonances. Generally, the SNR measured here is quite high considering the sample amount of <500 µg. The dipolar-coupling based transfers are further expected to yield even higher efficiencies using a coil with a lower RF-field inhomogeneity.
After having evaluated the single transfer steps, as well as their application in triple-resonance, 3D assignment experiments, we selected the six most efficient experiments to assign the backbone and Cβ of ubiquitin. Using only these spectra, 96 % of the C′, Cα, and Cβ resonances could be assigned with spectra acquired in <3 days. These results depend of course on the parameters of the probe (especially in the case of the transfer efficiencies of single steps), and also on the sample used. However, the different structural and dynamical elements in ubiquitin allow one to infer suitable choices for other proteins as well.
Still, there generally is considerable potential to improve the SNR of the assignment experiments. We find an overall transfer efficiency of the 3D experiments described between 0.023 and 0.214 (Table 6). Even the best of our experiments loses 78 % of the intensity, leading to an increase of the experimental time, compared to a perfect experiment, by a factor of roughly 22.
Due to the higher gyromagnetic ratio of 1H, proton-detected experiments open an avenue for bio-molecular NMR using considerably smaller sample amounts than presently required. It has already been shown that proton-detected distance-restraint experiments from ubiquitin at 100 kHz MAS allow one to obtain a de-novo atomic resolution structure from 500 µg of protein (Agarwal et al. 2014) and further improvements are imminent, including optimized pulse sequences, improved probe circuits, and even higher MAS frequencies.
References
Agarwal V et al (2013) Amplitude-modulated low-power decoupling sequences for fast magic-angle spinning NMR. Chem Phys Lett 583:1–7
Agarwal V et al (2014) De Novo 3D structure determination from sub-milligram protein samples by solid-state 100 kHz MAS NMR spectroscopy. Angew Chem Int Ed 53(45):12253–12256
Andrew ER, Bradbury A, Eades RG (1958) Nuclear magnetic resonance spectra from a crystal rotated at high speed. Nature 182(4650):1659
Baldus M, Meier BH (1996) Total correlation spectroscopy in the solid state. The use of scalar couplings to determine the through-bond connectivity. J Magn Reson Ser A 121(1):65–69
Barbet-Massin E et al (2013) Out-and-back 13C-13C scalar transfers in protein resonance assignment by proton-detected solid-state NMR under ultra-fast MAS. J Biomol NMR 56(4):379–386
Barbet-Massin E et al (2014) Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J Am Chem Soc 136(35):12489–12497
Bayro MJ et al (2009) Dipolar truncation in magic-angle spinning NMR recoupling experiments. J Chem Phys 130(11):114506
Böckmann A et al (2009) Characterization of different water pools in solid-state NMR protein samples. J Biomol NMR 45(3):319–327
Bodenhausen G, Ruben DJ (1980) Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett 69(1):185–189
Castellani F et al (2002) Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420(6911):98–102
Cavanagh J et al (2007) Protein NMR spectroscopy—principle and practice. In: Cavanagh J et al (eds) Protein NMR spectroscopy, 2nd edn. Academic Press, Burlington, pp 781–817
Chen L et al (2007a) Backbone assignments in solid-state proteins using J-based 3D heteronuclear correlation spectroscopy. J Am Chem Soc 129(35):10650–10651
Chen L et al (2007b) J-based 2D homonuclear and heteronuclear correlation in solid-state proteins. Magn Reson Chem 45(Suppl 1):S84–S92
Emsley L, Bodenhausen G (1990) Gaussian pulse cascades: new analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem Phys Lett 165(6):469–476
Grzesiek S, Bax A (1992) Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson (1969) 96(2):432–440
Hartmann SR, Hahn EL (1962) Nuclear double resonance in the rotating frame. Phys Rev 128(5):2042–2053
Hediger S, Meier BH, Ernst RR (1995) Adiabatic passage Hartmann-Hahn cross polarization in NMR under magic angle sample spinning. Chem Phys Lett 240(5–6):449–456
Huber M et al (2014) Solid-state NMR sequential assignment of Osaka-mutant amyloid-beta (Aβ1–40 E22Δ) fibrils. Biomol NMR Assign 9:7–14
Igumenova TI et al (2004) Assignments of carbon NMR resonances for microcrystalline ubiquitin. J Am Chem Soc 126(21):6720–6727
Kay LE et al (1990) Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson (1969) 89(3):496–514
Knight MJ et al (2011) Fast resonance assignment and fold determination of human superoxide dismutase by high-resolution proton-detected solid-state MAS NMR spectroscopy. Angew Chem Int Ed Engl 50(49):11697–11701
Lesage A, Bardet M, Emsley L (1999) Through-bond carbon–carbon connectivities in disordered solids by NMR. J Am Chem Soc 121(47):10987–10993
Lewandowski JZR et al (2011a) Enhanced resolution and coherence lifetimes in the solid-state NMR spectroscopy of perdeuterated proteins under ultrafast magic-angle spinning. J Phys Chem Lett 2(17):2205–2211
Lewandowski JR et al (2011b) Site-specific measurement of slow motions in proteins. J Am Chem Soc 133(42):16762–16765
Lienin SF et al (1998) Anisotropic intramolecular backbone dynamics of ubiquitin characterized by NMR relaxation and MD computer simulation. J Am Chem Soc 120(38):9870–9879
Linser R, Fink U, Reif B (2008) Proton-detected scalar coupling based assignment strategies in MAS solid-state NMR spectroscopy applied to perdeuterated proteins. J Magn Reson 193(1):89–93
Lowe IJ, Norberg RE (1957) Free-induction decays in solids. Phys Rev 107(1):46–61
Luckgei N et al (2014) Solid-state NMR sequential assignments of the C-terminal oligomerization domain of human C4b-binding protein. Biomol NMR Assign 8(1):1–6
Pauli J et al (2001) Backbone and side-chain 13C and 15N signal assignments of the α-Spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 Tesla. ChemBioChem 2(4):272–281
Pines A, Gibby MG, Waugh JS (1973) Proton-enhanced NMR of dilute spins in solids. J Chem Phys 59(2):569–590
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc 34(2):93–158
Schanda P, Meier BH, Ernst M (2010) Quantitative analysis of protein backbone dynamics in microcrystalline ubiquitin by solid-state NMR spectroscopy. J Am Chem Soc 132(45):15957–15967
Scholz I, Meier BH, Ernst M (2007) Operator-based triple-mode Floquet theory in solid-state NMR. J Chem Phys 127(20):204504
Schuetz A et al (2010) Protocols for the sequential solid-state NMR spectroscopic assignment of a uniformly labeled 25 kDa protein: HET-s(1-227). ChemBioChem 11(11):1543–1551
Shaka AJ et al (1983) An improved sequence for broadband decoupling: WALTZ-16. J Magn Reson (1969) 52(2):335–338
Stevens T et al (2011) A software framework for analysing solid-state MAS NMR data. J Biomol NMR 51(4):437–447
Thakur RS, Kurur ND, Madhu PK (2006) Swept-frequency two-pulse phase modulation for heteronuclear dipolar decoupling in solid-state NMR. Chem Phys Lett 426(4–6):459–463
Verel R et al (1998) A homonuclear spin-pair filter for solid-state NMR based on adiabatic-passage techniques. Chem Phys Lett 287(3–4):421–428
Verel R, Ernst M, Meier BH (2001) Adiabatic dipolar recoupling in solid-state NMR: the DREAM scheme. J Magn Reson 150(1):81–99
Vijay-Kumar S, Bugg CE, Cook WJ (1987) Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol 194(3):531–544
Westfeld T et al (2012) Properties of the DREAM scheme and its optimization for application to proteins. J Biomol NMR 53(2):103–112
Wittekind M, Mueller L (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J Magn Reson Ser B 101(2):201–205
Yamazaki T et al (1994) A Suite of triple resonance NMR experiments for the backbone assignment of 15 N, 13C, 2H labeled proteins with high sensitivity. J Am Chem Soc 116(26):11655–11666
Ye YQ et al (2014) Rapid measurement of multidimensional 1H solid-state NMR spectra at ultra-fast MAS frequencies. J Magn Reson 239:75–80
Zhou DH, Rienstra CM (2008) High-performance solvent suppression for proton detected solid-state NMR. J Magn Reson 192(1):167–172
Zhou Z et al (2007) A new decoupling method for accurate quantification of polyethylene copolymer composition and triad sequence distribution with 13C NMR. J Magn Reson 187(2):225–233
Acknowledgments
We thank Riccardo Cadalbert for expressing the protein and Emilie Testori and Alons Lends for help with crystallization and rotor filling. This work was supported by the Agence Nationale de la Recherche (ANR-11-BSV8-021-01, ANR-12-BS08-0013-01), the ETH Zurich, the Swiss National Science Foundation (Grant 200020_124611), the Centre National de la Recherche Scientifique (CNRS), PUT126 and ETF9229 from Estonian Ministry of Science and Education.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Penzel, S., Smith, A.A., Agarwal, V. et al. Protein resonance assignment at MAS frequencies approaching 100 kHz: a quantitative comparison of J-coupling and dipolar-coupling-based transfer methods. J Biomol NMR 63, 165–186 (2015). https://doi.org/10.1007/s10858-015-9975-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-9975-y