Abstract
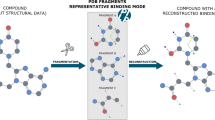
Here, we propose an in silico fragment-mapping method as a potential tool for fragment-based/structure-based drug discovery (FBDD/SBDD). For this method, we created a database named Canonical Subsite–Fragment DataBase (CSFDB) and developed a knowledge-based fragment-mapping program, Fsubsite. CSFDB consists of various pairs of subsite–fragments derived from X-ray crystal structures of known protein–ligand complexes. Using three-dimensional similarity-matching between subsites on one protein and another, Fsubsite compares the surface of a target protein with all subsites in CSFDB. When a local topography similar to the subsite is found on the surface, Fsubsite places a fragment combined with the subsite in CSFDB on the target protein. For validation purposes, we applied the method to the apo-structure of cyclin-dependent kinase 2 (CDK2) and identified four compounds containing three mapped fragments that existed in the list of known inhibitors of CDK2. Next, the utility of our fragment-mapping method for fragment-growing was examined on the complex structure of tRNA-guanine transglycosylase with a small ligand. Fsubsite mapped appropriate fragments on the same position as the binding ligand or in the vicinity of the ligand. Finally, a 3D-pharmacophore model was constructed from the fragments mapped on the apo-structure of heat shock protein 90-α (HSP90α). Then, 3D pharmacophore-based virtual screening was carried out using a commercially available compound database. The resultant hit compounds were very similar to a known ligand of HSP90α. As a result of these findings, this in silico fragment-mapping method seems to be a useful tool for computational FBDD and SBDD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fragment-based drug discovery (FBDD), which identifies low molecular weight ligands that bind to a target protein, is an increasingly popular method within the pharmaceutical industry [1,2,3,4]. The FBDD approach is based on biophysical screenings (NMR, X-ray, etc.) of a fragment library, which contains hundreds or thousands of small compounds (< 300 Da). The biophysical screening process has two aims; (i) detecting small ligands to a drug target and (ii) obtaining 3D information of the binders at the target site. In general, experimental fragment-based screening maps the position of each fragment to a chosen core as well as additional interactions on the protein surface. Subsequently, the various fragments can be linked or merged with each other (fragment-linking/fragment-merging). In addition, new functional groups can be added to the fragment (fragment-growing). Finally, the fragments will be integrated into a lead. Because fragment hits contain few functional groups for interactions with the target protein, the corresponding affinities tend to be very weak. Consequently, it is not easy to identify these fragments clearly using a standard biological assay [5]. The affinity of the lead compounds can then be determined experimentally.
In computational chemistry, early programs to map small molecules on a target protein were developed as the prototypes of docking programs before the appearance of FBDD [6]. These programs detected interaction points around a protein by probe molecules using grid searching methods (GRID) [7] and a random searching method (MCSS) [8]. Today, there are many fragment-mapping approaches based on docking [9,10,11,12,13,14]. These procedures evaluate likelihoods of the positions of fragments by force field potentials or empirical functions that are based on fittings of experimental affinities. Empirical functions, however, are less accurate because of small numbers of fragments available for fitting and less-diversity of their affinities. Force-field based approaches cannot distinguish between non-binders and weak-binders because the affinities of weak-binders (100 μM ~ 10 mM) are − 3 ~ − 6 kcal/mol of binding free energy, which is almost equal to the error (± 3 ~ 5 kcal/mol) for the computational methods employed to estimate the interaction [15, 16]. Therefore, it is difficult for docking programs to rank many poses of fragments correctly. This may be the reason why computer ligand-docking is unsuitable for identifying fragment-binding sites.
The another method is geometric pocket similarity search [17,18,19]. In a type of pocket similarity search, the positions of Cα (and Cβ) atoms of residues in the binding site of a target protein are compared to those of other proteins [18]. This procedure describes evolutional conservations [17] well but lacks for atomic level details. The another type represents only surface atoms of binding pockets as atom types or feature points and then compares the target pocket with the others using graph theory [19]. This pocket similarity search keeps atomic level details, but doesn’t make full use of residue information since only interfacial atoms are considered. The advantage of those pocket similarity search methods is that they use experimental data of X-ray crystallography well.
We have developed a new program, Fsubsite, to identify the similarity of local protein-surfaces between two proteins based on the molecular-superposing algorithm that was previously reported by ourselves [20]. In addition, we have also created a database consisting of subsite–fragment pairs, which is named Canonical Subsite–Fragment DataBase (CSFDB). On the surface of a target protein, Fsubsite searches for similar topographies to the subsites in CSFDB. All the subsites in CSFDB are examined by scanning the surface of the target protein using Fsubsite. To speed-up the process, Fsubsite treats each amino acid residue as a set of physicochemical feature points. Similarity between the matched site on the target protein and one of the subsites in CSFDB is calculated as the sum of pre-defined scores of matched feature points. Finally, fragment binding to the subsite in CSFDB is placed on the matched target site with high similarity (i.e. fragment-mapping).
To verify the procedure, Fsubsite with CSFDB was performed for fragment-mappings onto well-known target proteins. The mapped fragments were compared with known ligands of CDK2. In addition, we attempted to use the mapped fragments for fragment-growing onto TGT and for building of a 3D-pharmacophore model on HSP90α.
Materials and methods
Extraction of subsite–fragment pairs from protein–ligand complexes
To obtain a subsite–fragment database, we used the database of 3D-structures of protein–ligand complexes (PDBbind v.2013) [21,22,23]. The core set of PDBbind is chosen from 38,918 complexes in PDB according to resolutions of X-ray crystal data (≤ 2.5 Å), clustering of proteins (sequence similarity cutoff ≥ 90%) and sampling of binding constants (the highest one, the lowest one, the one close to the mean value). The core set was composed of 195 protein–ligand complexes in 65 protein clusters. The diversity of the core set was considered to be suitable for this study. Ligands in the database were divided into fragments using Fragmenter of JChem 6.3.0 (ChemAxon Ltd., Budapest, Hungary, 2014). The RingChainRecap rule in Fragmenter was used for the fragmentation process (Fig. 1). This rule cuts non-ring single bonds starting from a ring atom, and thus splits a molecule into the ring systems and connecting chains. When cutting generates single heavy atoms, they are attached to the rings. A set of residues within 4 Å of each fragment was defined as the subsite using SYBYL-X 2.1.1 (Certara, L.P., Princeton, NJ, USA, 2013). As a result, 644 subsite–fragment pairs were obtained as the subsite–fragment set.
Fragment mapping
The Fsubsite program superposes each subsite of the fragment onto the mapping surface of a target protein using the SUPERPOSE algorithm [20]. The Fsubsite represents functional groups of amino acids as spheres of five types based on their physicochemical properties: hydrophobic (HP); aromatic (AR); hydrogen-bond donors (HD); hydrogen-bond acceptors (HA); and hydrogen-bond donors/acceptors (DA). Each type is represented as a sphere with a predefined radius (1.0 or 0.5 Å) and is assigned to functional groups in an amino acid residue (Fig. 2). HD, HA and DA are placed on nitrogen, oxygen, and sulfur atoms, respectively, which are possibly involved in forming hydrogen bonds. HP and AR are placed at the center of an aliphatic and aromatic ring, respectively. For an extended aliphatic side chain, HP is placed at the center of the aliphatic atoms. Usually, the radii of the property spheres are 1.0 Å. However, a radius of 0.5 Å is used for a donor and/or an acceptor atom (HD, HA, DA) in a ring that has a hydrophobic or an aromatic property sphere (HP or AR) at the center. In a carboxyl group and a guanidino group, the radii of HA and HD are 0.5 Å.
Property spheres on amino acids. The colors of the spheres indicate the following properties: hydrophobic (HP; white), aromatic (AR; yellow), hydrogen-bond donor (HD; cyan), hydrogen-bond acceptor (HA; red), and hydrogen-bond donor/acceptor (DA; violet). Large and small spheres indicate radii of 1 Å and 0.5 Å, respectively
The superposition is performed based on the scoring matrix (Table 1). Matched property spheres give a positive score, whereas mismatched property spheres give a negative score. When the planes of two aromatic rings match well, the score between two ARs is 4 points. Otherwise, the score of AR–AR is 3 point as is that of HP–HP. In order to avoid mapping the outer region of the mapping surface, the optional property of a hidden layer (HL) can be used. HLs are placed onto the residues of the outer region of a target protein and/or the binding fragments of subsites in the database. HLs are also used to avoid collision between mapped fragments and a lead compound in the binding site. The radii of HLs for the outer residues are set in the same manner as ordinary properties for the inner residues of the mapping surface. HLs for fragments or lead compounds are placed onto the center or the heavy atoms. If HL overlaps with other properties, a large negative score is assigned.
The procedures of Fsubsite are shown in Fig. 3. During superposing, each subsite of the fragment is moved on the mapping site of a target protein. Firstly, the rotational copies of the subsite are generated. The rotational angle is determined automatically as the farthest sphere from the center of the moving subsite could not jump over the diameters of the property spheres of the target site. The translation is carried out as each property sphere of the moving subsite jumps onto the property spheres of the target site, which are able to generate positive scores. Finally, the overlaps are optimized using a simplex algorithm according to the Matching_Score as the objective function (Eq. 1),
where i and j are matched property spheres of the subsite of I and the target site of J, respectively, rij is the distance, σi and σj are the radii of the property spheres, n+ and n− are numbers of positive and negative overlaps, cij is the element of the scoring matrix. The first term is the sum of the scores according to Table 1, and the second term is the position adjuster that ranges from 0 to 1. The position adjuster acts to make more positive overlaps and move away from negative overlaps. The Subsite_Similarity between the subsite and the target site is estimated as the value of the Matching_Score divided by the maximum value, which is obtained by self-matching of the subsite (Eq. 2). When similar subsites within the cutoff value of similarity are found, the fragments binding to the subsites in CSFDB can be mapped on the mapping surface of a target protein according to the translational and rotational vectors for the subsites.
In the Fsubsite software package, core programs are written in Fortran 90/95 and utilities in Perl or Python. The supported operating systems (OSs) are Linux, and Apple Mac OS X. Molecular graphic software capable of reading/writing the Tripos Mol2 file format (such as SYBYL-X) is required for input and visualization. The Fsubsite program can run on a distributed processing system as a Linux cluster using Open Grid Scheduler/Grid Engine 2011.11 (http://gridscheduler.sourceforge.net, 2011).
Generating a Canonical Subsite–Fragment DataBase
In order to reduce computational costs of fragment-mapping, similar subsites of the subsite–fragment set were grouped into one representative subsite using sieve theory.
-
i.
The largest subsite in the subsite–fragment list was chosen on the basis of the scores by self-matching of subsites.
-
ii.
Similarities between the largest subsite and the remaining subsites in the list were evaluated.
-
iii.
The largest subsite was adopted as the representative subsite, and then the subsites within the cutoff value of similarity were defined as similar subsites.
-
iv.
Bound fragments were aligned into the representative subsite by superposing similar subsites.
-
v.
The bound fragment of the representative subsite was chosen as the representative fragment and then the bound fragments of similar subsites were registered as members (Fig. 4).
-
vi.
The largest and similar subsites were eliminated in the subsite–fragment list.
-
vii.
These processes were repeated until the list was emptied.
Finally, we obtained 482 representative subsite–fragment pairs to be admitted to the Canonical Subsite–Fragment DataBase (CSFDB).
For comparison between subsite- and fragment-similarities, chemical similarities of the fragments were calculated as Tanimoto similarities between 64-bit fingerprints of 3D distances of atom pairs using Canvas 2.4 in Small-molecule Drug Discovery Suite 2015-2 (Schrödinger, LLC., New York, NY, USA, 2015) [24, 25]. All atoms of fragments were represented as Carhart atom types: C, O, N, S, F, Cl, Br, I, P, Si, B, Se, As, and Y (any other) [26]. All atom pairs in each fragment were considered for calculations of distances. The distances were not 2D topological distances (bond paths) but 3D distances in the binding conformation of each fragment in the X-ray structure of the original protein–ligand complex. Pearson correlation coefficient between subsite- and fragment-similarities were calculated. R 3.1.3 was used as a statistical software (The R Foundation for Statistical Computing, https://www.r-project.org/, 2015).
Protein preparation for fragment mapping
The fragment-mapping method was tested using several proteins; CDK2, TGT and HSP90α. The ligand-unbound (apo) structures of CDK2 (PDB ID: 4ek3) and HSP90α (PDB ID: 1yer) were used in order to validate the influence of fluctuations in target proteins. For fragment-growing, the complex structure of TGT with a small ligand was used (PDB ID: 1s38).
For CDK2 and HSP90α, the residues within 4 Å from the ligand bound to the holo structure was defined as the mapping surface for the apo structure. The ligands of the holo structures of PDB IDs (CDK2: 2vtq, HSP90α: 2xjx) were used. The mapping surface of TGT was defined using our HBOP program [27, 28] for binding site detection. HBOP detected additional binding space around the bound small ligand on the basis of hydrophobicity. The mapping surface for TGT was defined as residues within 4 Å from the grid points of the detected hydrophobic site.
In order to avoid mapping on the outer region, the property spheres of HL were put on residues from 4 to 8 Å of the grid points or the ligand. For TGT, additional HLs were placed onto the heavy atoms of the lead compound in order to avoid collision between mapped fragments and the lead compound.
Examination of mapped fragments
To verify the effectivity of our mapping procedure, we examined whether a 3D-database search using information of mapped fragments obtained from our method was able to retrieve known active compounds for the target protein. Fsubsite was first carried out on CDK2, which is a popular target for FBDD. In this case study, we assumed CDK2 to be an unknown kinase. All fragments in the CSFDB except for the fragments derived from CDK2-ligand complexes were mapped onto the apo structure of CDK2 to obtain a set of mapped fragments. All combinations of the three in the mapped fragments without obstructing each other were generated using the FAS program (produced by Dr. H. Yamasaki and Dr. Y. Nishibata). Then, the CDK2 ligands in the general-PL set of PDBbind v.2013 were three-dimensionally compared with the obtained triplet fragments using UNITY in SYBYL-X 2.1.1. The UNITY flex search was used for treatment of ligand flexibility and then Lipinski’s rule of 5 was turned off. The spatial point constraint was put on the center of each fragment with a tolerance of 1.0 Å. As the representation of the inside of the target protein, the heavy atoms of the mapping and exclusive residues for Fsubsite used as the excluded volume constraint with a van der Waals radii scale factor of 0.2 was applied in order to treat induced fits.
Additional fragments for fragment-growing
TGT is known to be a good target for computational de novo design [29] and virtual screening [30], and so we chose the complex structure of TGT with a small ligand and mapped additional fragments around the small ligand using Fsubsite (Fig. 5a) [31, 32]. The reduced type of CSFDB eliminating the subsite–fragment pairs derived from TGT-ligand complexes was used for the mapping. The mapped fragments were grouped according to their chemical structures and then their center positions were clustered using a clustering radius of 3 Å. In each cluster, the fragment with the most neighbors was chosen as the representative (Fig. 5b).
The position of the methyl group of the lead compound was chosen as the starting point for fragment-growing (Fig. 5c). In the vicinity of the starting point, the nearest and most clustered fragment was selected for the fragment-growing process. To bridge between the starting point and the fragment, a de novo design program, RACHEL (SYBYL-X 1.3, Certara, L.P., 2011), was used with CHARLIE mode (Fig. 5d). This procedure generated 50 compounds. In CHARLIE mode, the enrichment of generated ligand diversity (SEARCH_DIVERSITY) = 0.25 and the maximum splice atom overlap error (SEARCH_SP_ERROR) = 2.5 Å were used for generating more compounds and looser links, respectively. The used chemical building blocks were derived from the approved drugs found in DrugBank 5.0.5 [33,34,35,36].
In order to validate the adaptability of the obtained compounds, these compounds were docked into the protein. The compounds and the protein were prepared by LigPrep 4.1 and Protein Preparation Wizard, respectively, in Small-molecule Drug Discovery Suite 2017-1 (Schrödinger, LLC., 2017). The docking calculations were performed using Glide 7.4 in SP mode [37, 38]. When the fragment part of the docked compound held on the position of the mapped fragment, we adopted the compound as a candidate ligand (Fig. 5d).
Using mapped fragments for virtual screening
We demonstrated a virtual screening using the obtained fragment map by Fsubsite. Because we assumed HSP90α to be an unknown protein, the fragments derived from HSP90α-ligand complexes were excluded from the CSFDB and only the apo structure of HSP90α was used for mapping. 3D pharmacophores were generated from all mapped fragments using DISCOtech in SYBYL-X 2.1.1. DISCOtech, DONOR_ATOM, ACCEPTOR_ATOM, HYDROPHOBIC, AROMATIC, POSITIVE_N, and NEGATIVE_CENTER were used as pharmacophore points with tolerances for matching of 1.0 Å and for constraint of 1.0 Å. We modified the dialog menu of DISCOtech to set ‘% of Molecules Allowed to Miss’ at 0, i.e. generating all pharmacophore points on the largest fragment and then clustering. The virtual screening was performed using UNITY flex search without Lipinski’s rule of 5. The excluded volume constraints were put on the heavy atoms of the mapping and exclusive residues for Fsubsite with a van der Waals radii scale factor of 1.0. The HSP90α ligands in the general-PL set of PDBbind v.2013 were used for validation. The Enamine Kinase Hinge Region directed Library (18,020 compounds, Enamine Ltd, Ukraine, 2016) was used as the compound database for the virtual screening because kinases utilize the same substrate as ATP for HSP90α.
Computational hardware
The making of the subsite–fragment set was performed on a Mac Pro (Apple Inc., Cupertino, CA, USA; Intel Xeon Processor 5160; 2 Cores; 3 GHz; 2 central processing units [CPUs]). The UNITY flex searches and the RACHEL calculation were carried on an hp Z600 workstation (HP inc., Palo Alto, CA, USA; Intel Xeon Processor X5570; 4 Cores; 2.93 GHz; 2 CPUs). The reduction of the subsite–fragment set into the canonical database, the fragment-mapping calculations and the Glide re-docking were performed using two Linux clusters, 13 nodes of an HPC5000-XH2UTwin-D24 (HPC SYSTEMS Inc., Tokyo, Japan; Intel Xeon Processor E5-2690 v3; 12 Cores; 2.6 GHz; 26 CPUs in total) and 30 nodes of a Dell PowerEdge 1950 III (Dell Inc., Round Rock, TX, USA; Intel Xeon Processor X5460; 4 Cores; 3.16 GHz; 60 CPUs in total). The computation time for fragment-mapping was about 1 day on our Linux clusters.
Results
Canonical Subsite–Fragment DataBase
Firstly, we performed the calculations for redaction of 644 subsite–fragment pairs using 75% of the cutoff of Subsite_Similarity. However, in some cases the adjacent subsite–fragment pairs in the same protein–ligand complexes were grouped into the same clusters. Therefore, 90% of the similarity cutoff was used in cases of self-scores below 40 for the representative subsites. Using this procedure we obtained 482 representatives of the Canonical Subsite–Fragment DataBase (Table S1).
The molecular weights and numbers of heavy atoms of fragments in the CSFDB are shown in Fig. 6. The molecular weights ranged from 26 to 712 and the average was 106.6. Nineteen fragments had a molecular weight of more than 300, i.e. upper limit for lead-like molecules [39]. The range and average of numbers of heavy atoms were 2–50 and 7.4, respectively. Twenty-four fragments comprised 20 or more heavy atoms. In these cases, any increase in fragment size was not expected to enhance potency [40]. These larger fragments consisted of peptides, macrocyclic or polycyclic compounds, and a long-chain fatty acid.
The Pearson correlation coefficient between subsite- and fragment-similarities was very good when the Matching_Score between subsites was 30 and over, i.e. between larger subsites (n = 2736, r = 0.902). Using Matching_Score ≥ 20, the Pearson correlation coefficient was moderate (n = 38,883, r = 0.603).
Mapped fragment on CDK2
In this case study, we assumed CDK2 to be an unknown kinase. When Matching_Score ≥ 24 and Subsite_Similarity ≥ 75% were used as cutoff for mapping, we obtained a total of 11 mapped fragments using the reduced type of CSFDB eliminating fragments derived from CDK2-ligand complexes. In order to identify CDK2 ligands that comprise mapped fragments, the obtained fragments were used as 3D-queries in a UNITY flex search (Table 2; Fig. 7). The FAS program generated 74 combinations of the three fragments that did not interfere with each other. Due to each other’s steric hindrance it was not possible to select combinations of four or more fragments from the mapped fragment. For each combination, the chemical structures of the fragments and their centers were used as 3D-queries with the excluded volume constraint of the protein surface. The three CDK2 ligands in PDBbind (PDB IDs: 2vtq, 2vtt, and 2vu3) were composed of a triplet of fragments (Fig. 8a). Moreover, one ligand of the other kinase (PDB ID: 2xch) was matched with another triplet (Fig. 8b).
Fragment-growing on TGT
In this case study, similar subsites of other proteins were found and their binding fragments mapped on the complex of TGT with the lead compound. Using cutoff values for smaller subsites (Matching_Score ≥ 10 and Subsite_Similarity ≥ 80%), 46 fragments were obtained as the result. 46 fragments were mapped to 1242 locations at different frequencies. For example, benzene was mapped to 206 locations (Fig. 5a), and naphthalene was 1 location (Fig. 14a). The adjacent fragments were grouped into representatives by clustering (Fig. 5b). Because benzene was the most mapped fragment and one of the benzene clusters was close to the starting point of the lead compound, this moiety was chosen as the additional fragment (Fig. 5c).
The de novo design program of RACHEL built bridges between the benzene fragment and the lead compound (Fig. 5d), and then 50 compounds were generated. The obtained compounds were docked into TGT using the docking program of Glide (Fig. 9). The fragment parts of the docked compounds were compared with the position of the mapped fragment. Based on this analysis, we adopted 42 of the 50 compounds as candidate ligands because the fragment parts were located in the vicinity of the mapped fragment. Three of the 42 candidates were very similar to known ligands of TGT (Fig. 10) [31, 32, 41, 42].
Comparison of the docked candidate with the known ligand of TGT: a the benzene fragment in the vicinity of the lead compound (PDB ID: 1s38), b docking pose of candidate No. 707, and c X-ray pose of a similar known ligand of TGT (PDB ID: 1q66). The colors indicate the following: lead compound (brown), target fragment (green), candidate (white), and known ligand (purple)
Virtual screening for HSP90α
On the assumption that HSP90α is an unknown target, we obtained a total of 6 mapped fragments using the reduced type of CSFDB eliminating fragments derived from HSP90α-ligand complexes (Table 3; Fig. 11a). Matching_Score ≥ 20 and Subsite_Similarity ≥ 75% were used as cutoff for mapping. Because the smaller fragments overlapped into the larger fragments, the fragment-combination approach was not used. Therefore, a total of 15 points as the 3D-pharmacophore were generated from all mapped fragments using DISCOtech (Fig. 11b). In the validation, the two compounds of HSP90α ligands in PDBbind (PDB IDs: 2yi0 and 2yi7) satisfied 8 points, which is more than half of the 3D-pharmacophore (Figs. 11c, 12a). Only four hits were obtained in the virtual screening of the commercially available compound database using the same conditions (Figs. 11d, 12b).
3D-pharmacophore model from mapped fragments on apo HSP90α. a Mapped fragments, b 3D-pharmacophore model, c aligned pose of the known ligand of 2yi7, and d aligned pose of the virtual hit Z990826384. The colors of the pharmacophore points indicate the following properties: DONOR_ATOM (violet), ACCEPTOR_ATOM (green), HYDROPHOBIC (cyan), AROMATIC (yellow), POSITIVE_N (red), and NEGATIVE_CENTER (blue)
Discussion
Bioisosteres in subsite
In CSFDB, representative fragments as well as fragment members belong to a representative subsite (Fig. 4, Table S1). This arrangement means that these fragments are able to be replaced by one another [19, 43]. As shown in Fig. 4, the hydrophobic subsite of the S1 cleft in the trypsin family of proteins may be able to accept various aromatic rings (Canonical No. 106 in Table S1). Using the Fsubsite program, medicinal and computational chemists are readily able to obtain poses of the fragment members using the translational and rotational vector for the representative fragments mapped onto the target protein.
Matching_Score and Subsite_Similarity
When Matching_Score ≥ 30, Subsite_Similarity strongly correlated with similarity between fragments. These results implied that similar fragments bind to similar subsites if the subsites are relatively large. However, most cases where Matching_Score ≥ 30 involved the same proteins or homologous proteins (Table S1). We considered that situations where the Matching_Score ≥ 20 would be suitable for choosing fragments for drug design or virtual screening because correlation between the Subsite_Similarity and the fragment similarity was moderate.
Validity of mapped fragments on CDK2
As seen in Fig. 8 and Table 2, four of the 11 mapped fragments were used as part of known CDK2 ligands (fragments of Nos. 1, 4, 5, and 6). These three ligands came from the CDK2-ligand complexes (PDB IDs: 2vtq, 2vtt, and 2vu3) [44] (Fig. 8a). Furthermore, the ligand of 3-phosphoinostitide-dependent protein kinase-1 (PDB ID: 2xch) can also inhibit CDK2 [45] (Fig. 8b).
As seen in Fig. S3, the fragments of Nos. 1 and 6 were in good agreement with the known ligand, but the position of the fragment of No. 5 was different due to induced fit. Since the X-ray structure of the complex of 2xch’s ligand with CDK2 is not available, the agreement with the fragment of No. 4 is unknown.
Ability for fragment-growing on TGT
The benzene fragment in the vicinity of the starting point of the lead compound was located in the hydrophobic pocket of Val45, Leu68, Leu100 and Val282, which was targeted by Meyer et al. (Fig. 9a) [42]. The benzene fragments of the docked compounds were retained in the hydrophobic pocket (Fig. 9b). The docking poses of the three candidates resembled the X-ray poses of similar known ligands (Fig. 9c).
Grädler group identified interaction sites of hydrogen-bond donors around Asp102 and Asp280 using the de novo design program, LUDI [29, 46]. Our Fsubsite mapped the ethylamine fragment with an amino group in the vicinity of Asp102 and Asp280 (Fig. 13a). Indeed, the docking pose of a generated ligand by RACHEL and Glide was in good agreement with the X-ray structure of a known similar ligand with an amino group (Fig. 13b, c) [32, 41].
Comparison of the generated ligand with the known ligand of TGT targeting Asp102 and Asp280: a the ethylamine fragment in the vicinity of the lead compound (PDB ID: 1s38), b docking pose of a generated ligand, and c X-ray pose of a similar known ligand of TGT (PDB ID: 1q65). The colors indicate the following: lead compound (brown), target fragment (green), candidate (white), and known ligand (purple)
Interestingly, the naphthalene fragment was mapped to a position somewhat distant from the lead compound (Fig. 14a). This naphthalene fragment was hydrophobic interacting with Val282. In the X-ray pose of a known ligand with a naphthalene group, the naphthalene group of the ligand interacts with Val282, the same as the mapped fragment does (Fig. 14b, c) [47].
Comparison of the mapped fragment with the known ligand of TGT targeting Val282: a the naphthalene fragment in the vicinity of the lead compound (PDB ID: 1s38), and b X-ray pose of a known ligand of TGT (PDB ID: 2zqr) with a naphthalene group. The colors indicate the following: lead compound (brown), target fragment (green), and known ligand (purple)
3D-pharmacophore model for HSP90α
We generated a 3D-pharmacophore model using all the mapped fragments on the apo surface of HSP90α. Virtual screening successfully identified the two known ligands of HSP90α (i.e. PDB IDs of 2yi0 and 2yi7; see Figs. 11c, 12a) [48]. Moreover, these aligned poses on the 3D-pharmacophore were consistent with the binding poses of the X-ray crystal structures (Fig. 15). The root-mean-square distances between the aligned and X-ray poses were 1.13 Å for 2yi0 and 1.12 Å for 2yi7.
As shown in Figs. 11d and 12b, four compounds were identified by virtual screening that satisfied 8 points of the 3D-pharmacophore as described above for the known ligands. Instead of carrying out an assay, a similarity search was performed using SciFinder (American Chemical Society, 2017). A similar compound HDI4-04 (CAS ID: 1031565-15-7) was identified displaying 80% structural similarity that exhibited inhibitory activity against HSP90α with an IC50 of 0.35 μM [49]. HDI4-04 satisfied 6 points of our 3D-pharmacophore (Fig. 16). Therefore, we concluded that the mapped fragments were successfully converted to the appropriate 3D-pharmacore model for the given target protein.
Induced fit
The overall differences between the apo and holo backbones were rmsd = 0.50, 1.49 Å for CDK2 and HSP90α, respectively. Comparison of the ligands suggests the mapped fragments on apo proteins are adequate (Supporting Information). Thus, in some cases Fsubsite is able to identify credible fragments on the apo surface of a given target protein. However, if the target protein undergoes a significant induced fit, additional protein structures obtained from X-ray crystal studies or by molecular dynamics need to be analyzed.
Conclusion
We have developed a new knowledge-based fragment-mapping method that utilizes a subsite-similarity search. Our method was validated using the apo surface of CDK2 and the analysis identified appropriate fragments that were mapped onto the target protein. For fragment-growing, we mapped fragments around the lead compound in the complex of TGT. The mapped fragment was linked to the lead compound and the generated products were very similar to known ligands of TGT. We then tested the ability of the mapped fragments to generate a 3D-pharmacophore model on the apo HSP90α. Using the obtained 3D-pharmacophore model, virtual screening of a commercially available compound database was performed, which generated hits that closely resembled a known ligand of HSP90α. Thus, the in silico fragment-mapping method developed here will be a useful tool for computational FBDD and SBDD.
Fsubsite is freely available for academic use. For more information, please contact us by e-mail.
References
Oster L, Tapani S, Xue YF, Kack H (2015) Successful generation of structural information for fragment-based drug discovery. Drug Discov Today 20(9):1104
Wang T, Wu MB, Chen ZJ, Chen H, Lin JP, Yang LR (2015) Fragment-based drug discovery and molecular docking in drug design. Curr Pharm Biotechnol 16(1):11
Murray CW, Verdonk ML, Rees DC (2012) Experiences in fragment-based drug discovery. Trends Pharmacol Sci 33(5):224
Erlanson DA (2012) Introduction to fragment-based drug discovery. In: Davies GT, Hyvönen M (eds) Fragment-based drug discovery and x-ray crystallography. Springer, Berlin, p 1
Wyss DF, Wang YS, Eaton HL, Strickland C, Voigt JH, Zhu ZN, Stamford AW (2012) Combining NMR and X-ray crystallography in fragment-based drug discovery: discovery of highly potent and selective BACE-1 inhibitors. In: Davies TG, Hyvonen M (eds) Fragment-based drug discovery and X-ray crystallography, 317. Springer, Berlin, p 83
Rognan D (2012) Fragment-based approaches and computer-aided drug discovery. In: Davies TG, Hyvonen M (eds) Fragment-based drug discovery and X-ray crystallography, 317. Springer, Berlin, p 201
Goodford PJ (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem 28(7):849
Miranker A, Karplus M (1991) Functionality maps of binding-sites—a multiple copy simultaneous search method. Proteins Struct Funct Genet 11(1):29
Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo LQ, Xia B, Beglov D, Vajda S (2015) The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc 10(5):733
Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S (2009) Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics 25(5):621
Sandor M, Kiss R, Keseru GM (2010) Virtual fragment docking by glide: a validation study on 190 protein-fragment complexes. J Chem Inf Model 50(6):1165
Kawatkar S, Wang HM, Czerminski R, Joseph-McCarthy D (2009) Virtual fragment screening: an exploration of various docking and scoring protocols for fragments using glide. J Comput Aided Mol Des 23(8):527
Nikolic D, Blinov N, Wishart D, Kovalenko A (2012) 3D-RISM-Docc: a new fragment-based drug design protocol. J Chem Theory Comput 8(9):3356
Imai T, Oda K, Kovalenko A, Hirata F, Kidera A (2009) Ligand mapping on protein surfaces by the 3D-RISM theory: toward computational fragment-based drug design. J Am Chem Soc 131(34):12430
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10(5):449
Weis A, Katebzadeh K, Soderhjelm P, Nilsson I, Ryde U (2006) Ligand affinities predicted with the MM/PBSA method: dependence on the simulation method and the force field. J Med Chem 49(22):6596
Skolnick J, Gao M, Roy A, Srinivasan B, Zhou HY (2015) Implications of the small number of distinct ligand binding pockets in proteins for drug discovery, evolution and biochemical function. Bioorg Med Chem Lett 25(6):1163
Gao M, Skolnick J (2013) APoc: large-scale identification of similar protein pockets. Bioinformatics 29(5):597
Jalencas X, Mestres J (2013) Chemoisosterism in the proteome. J Chem Inf Model 53(2):279
Iwase K, Hirono S (1999) Estimation of active conformations of drugs by a new molecular superposing procedure. J Comput Aided Mol Des 13(5):499
Cheng TJ, Li X, Li Y, Liu ZH, Wang RX (2009) Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model 49(4):1079
Wang RX, Fang XL, Lu YP, Yang CY, Wang SM (2005) The PDBbind database: methodologies and updates. J Med Chem 48(12):4111
Wang RX, Fang XL, Lu YP, Wang SM (2004) The PDBbind database: collection of binding affinities for protein–ligand complexes with known three-dimensional structures. J Med Chem 47(12):2977
Duan J, Dixon SL, Lowrie JF, Sherman W (2010) Analysis and comparison of 2D fingerprints: Insights into database screening performance using eight fingerprint methods. J Mol Graph Model 29(2):157
Sastry M, Lowrie JF, Dixon SL, Sherman W (2010) Large-scale systematic analysis of 2D fingerprint methods and parameters to improve virtual screening enrichments. J Chem Inf Model 50(5):771
Carhart RE, Smith DH, Venkataraghavan R (1985) Atom pairs as molecular features in structure–activity studies: definition and applications. J Chem Inf Comput Sci 25(2):64
Oda A, Yamaotsu N, Hirono S (2009) Evaluation of the searching abilities of HBOP and HBSITE for binding pocket detection. J Comput Chem 30(16):2728
Yamaotsu N, Oda A, Hirono S (2008) Determination of ligand-binding sites on proteins using long-range hydrophobic potential. Biol Pharm Bull 31(8):1552
Grädler U, Gerber H-D, Goodenough-Lashua DM, Garcia GA, Ficner R, Reuter K, Stubbs MT, Klebe G (2001) A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase. J Mol Biol 306(3):455
Brenk R, Naerum L, Grädler U, Gerber H-D, Garcia GA, Reuter K, Stubbs MT, Klebe G (2003) Virtual Screening for submicromolar leads of tRNA-guanine transglycosylase based on a new unexpected binding mode detected by crystal structure analysis. J Med Chem 46(7):1133
Stengl B, Meyer EA, Heine A, Brenk R, Diederich F, Klebe G (2007) Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding. J Mol Biol 370(3):492
Meyer EA, Furler M, Diederich F, Brenk R, Klebe G (2004) Synthesis and in vitro evaluation of 2-aminoquinazolin-4(3H)-one-based inhibitors for tRNA-guanine transglycosylase (TGT). Helv Chim Acta 87(6):1333
Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A, Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y, Wishart DS (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42(D1):D1091
Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS (2011) DrugBank 3.0: a comprehensive resource for ‘Omics’ research on drugs. Nucleic Acids Res 39(suppl_1):D1035
Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M (2008) DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36(suppl_1):D901
Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34(suppl_1):D668
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47(7):1750
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47(7):1739
Congreve M, Carr R, Murray C, Jhoti H (2003) A rule of three for fragment-based lead discovery? Drug Discov Today 8(19):876
Reynolds CH, Tounge BA, Bembenek SD (2008) Ligand binding efficiency: trends, physical basis, and implications. J Med Chem 51(8):2432
Brenk R, Meyer E, Reuter K, Stubbs MT, Garcia GA, Diederich F, Klebe G (2004) Crystallographic study of inhibitors of tRNA-guanine transglycosylase suggests a new structure-based pharmacophore for virtual screening. J Mol Biol 338(1):55
Meyer EA, Brenk R, Castellano RK, Furler M, Klebe G, Diederich F (2002) De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity. ChemBioChem 3(2–3):250
Desaphy J, Rognan D (2014) sc-PDB-Frag: a database of protein–ligand interaction patterns for bioisosteric replacements. J Chem Inf Model 54(7):1908
Wyatt PG, Woodhead AJ, Berdini V, Boulstridge JA, Carr MG, Cross DM, Davis DJ, Devine LA, Early TR, Feltell RE, Lewis EJ, McMenamin RL, Navarro EF, O’Brien MA, O’Reilly M, Reule M, Saxty G, Seavers LCA, Smith DM, Squires MS, Trewartha G, Walker MT, Woolford AJA (2008) Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a novel cyclin dependent kinase inhibitor using fragment-based X-ray crystallography and structure based drug design. J Med Chem 51(16):4986
Angiolini M, Banfi P, Casale E, Casuscelli F, Fiorelli C, Saccardo MB, Silvagni M, Zuccotto F (2010) Structure-based optimization of potent PDK1 inhibitors. Bioorg Med Chem Lett 20(14):4095
Böhm H-J (1992) The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J Comput Aided Mol Des 6(1):61
Hörtner SR, Ritschel T, Stengl B, Kramer C, Schweizer WB, Wagner B, Kansy M, Klebe G, Diederich F (2007) Potent inhibitors of tRNA-guanine transglycosylase, an enzyme linked to the pathogenicity of the shigella bacterium: charge-assisted hydrogen bonding. Angew Chem Int Ed 46(43):8266
Sharp SY, Roe SM, Kazlauskas E, Čikotienė I, Workman P, Matulis D (2012) Co-crystalization and in vitro biological characterization of 5-aryl-4-(5-substituted-2-4-dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. PLoS ONE 7(9):e44642
Xu X-L, Sun H-P, Liu F, Jia J-M, Guo X-K, Pan Y, Huang H-Z, Zhang X-J, You Q-D (2014) Discovery and bioevaluation of novel pyrazolopyrimidine analogs as competitive Hsp90 inhibitors through shape-based similarity screening. Mol Inf 33(4):293
Acknowledgements
This work was supported in part by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number JP15K07899. We would like to thank Dr. Hiroyuki Yamasaki and Dr. Yoshihiko Nishibata (Pharmaceutical Informatics, School of Pharmacy, Kitasato University) for providing the FAS program.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yamaotsu, N., Hirono, S. In silico fragment-mapping method: a new tool for fragment-based/structure-based drug discovery. J Comput Aided Mol Des 32, 1229–1245 (2018). https://doi.org/10.1007/s10822-018-0160-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0160-8