Abstract
We review our performance in the SAMPL5 challenge for predicting host–guest binding affinities using the movable type (MT) method. The challenge included three hosts, acyclic Cucurbit[2]uril and two octa-acids with and without methylation at the entrance to their binding cavities. Each host was associated with 6–10 guest molecules. The MT method extrapolates local energy landscapes around particular molecular states and estimates the free energy by Monte Carlo integration over these landscapes. Two blind submissions pairing MT with variants of the KECSA potential function yielded mean unsigned errors of 1.26 and 1.53 kcal/mol for the non-methylated octa-acid, 2.83 and 3.06 kcal/mol for the methylated octa-acid, and 2.77 and 3.36 kcal/mol for Cucurbit[2]uril host. While our results are in reasonable agreement with experiment, we focused on particular cases in which our estimates gave incorrect results, particularly with regard to association between the octa-acids and an adamantane derivative. Working on the hypothesis that differential solvation effects play a role in effecting computed binding affinities for the parent octa-acid and the methylated octa-acid and that the ligands bind inside the pockets (rather than on the surface) we devised a new solvent accessible surface area term to better quantify solvation energy contributions in MT based studies. To further explore this issue a, molecular dynamics potential of mean force (PMF) study indicates that, as found by our docking calculations, the stable binding mode for this ligand is inside (rather than surface bound) the octa-acid cavity whether the entrance is methylated or not. The PMF studies also obtained the correct order for the methylation-induced change in binding affinities and associated the difference, to a large extent to differential solvation effects. Overall, the SAMPL5 challenge yielded in improvements our solvation modeling and also demonstrated the need for thorough validation of input data integrity prior to any computational analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Blind challenges are unbiased exercises to test the accuracy of computational methods on problems of contemporary import. Both the Statistical Assessment of the Modeling of Proteins and Ligands (SAMPL) and the Drug Design Data Resource (D3R) grand challenges fielded validation benchmarks for the structure-based drug design community, thereby, encouraging improvements and innovations in the methods used in this field [1–5]. Our participations in the SAMPL challenge series focused on the introduction of new methods for estimation of thermodynamic quantities [6].
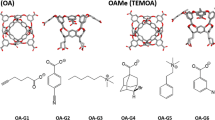
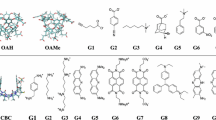
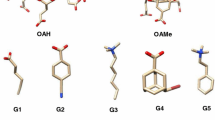
This year’s challenges focus on the calculations of binding thermodynamics of host–guest systems [7]. Host–guest systems have fewer atoms and degrees of freedom than protein–ligand systems. Importantly, they share the same fundamental statistical mechanical principles driving non-covalent binding as that of protein ligand systems, which makes them an excellent prototype for detailed study of ligand binding [4, 8–10]. The SAMPL5 host–guest binding challenge contains three different types of hosts: the acyclic Cucurbit[2]uril (CBClip), Octa-acids with hydroxyl groups at the cavity entrance (OAH) and methyl groups (OAMe). The acyclic CBClip, is a member of the cucurbituril-type synthetic host family that functions as a small molecular container [11] with two glycoluril units attached via methylene bridges and four sulfonic acids at the arms (refer Fig. 1).
The sulfonic acids impart water solubility to the receptor while the glycoluril moieties provide a cavity to partly surround hydrophobic ligands. Beyond its amphipathic qualities, CBClip possesses aromatic sidewalls to provide favorable π-stacking interactions between the receptor and certain guests, making it an appealing molecular container for hydrophobic drugs [12, 13]. As well as being excellent drug delivery agents, the members of cucurbituril family also play a role in the self-assembly of molecular machines, catalysis, and gene transfection [14–16]. The other two hosts, OAH and OAMe, differ from CBClips in that their basket-like shapes can completely envelop ligands in their binding pockets [17, 18]. OAH is a water-soluble molecular container also included in the SAMPL4 validation benchmarks [19]. The outer surface is hydrophilic due to the presence of eight ionizable carboxylates groups while the inside presents a deep hydrophobic cavity, around ~10 Å wide, with eight aromatic faces. The cavity is widest near its entrance, enabling OAH to hold guest molecules with a range of sizes [20, 21]. The third host provided in the current challenge, OAMe, is a methylated form of OAH with four methyl groups (as OMe) around the rim of the binding pocket. The presence of the four methyl groups at the entrance of the hydrophobic cavity of OAMe constricts the entrance to the binding pocket, which may alter the binding modes of certain guest molecules in OAMe relative to OAH [22]. The SAMPL5 challenge entails predicting the binding affinities of ten guests for CBClip and the affinities of six guests to each of OAH and OAMe, for a total of 22 predictions.
Free energy calculations hinge on the ability to capture the favorable binding states and, no less important, a fair sampling of the chemical space within the appropriate ensemble. In the current exercise, the stiff binding sites of the hosts and some bulky guest structures all make it difficult to locate favorable states through searching the funnel-like energy landscapes enclosed by high energy barriers.
We apply our recently developed “Movable Type” (MT) method, which simulates the local partition functions utilizing Monte Carlo integration given the initial structures from a canonical ensemble as expressed in Eq. 1:
here the Helmholtz free energy A is estimated using the average of the sampled energy states (e −βEi) multiplied by the sampling volume (V M ). Given an N-particle physical space, a quantitative description of the ensemble volume is written as:
where, τ 1 to τ N are the coordinates of all the particles and D is the domain of definition for all of the particle coordinates. The distinctive feature of the MT method is that it numerically estimates the average of the local partition function given a defined sampling volume centered on an initial structure, instead of searching among actual physical structures within that defined volume. As such, the MT simulation method is collision-tolerant when more favorable contacts are within the sampling range, but can overestimate the partition function if unphysical states with low energies turn up in the sampling range. The SAMPL5 blind challenge provides an opportunity to review the MT performance in the binding free energy simulation in the relatively small Host–guest systems.
In our work, placement of the guest molecules in these three host molecules was carried out using docking and conformational search procedures to ensure that all guests are confined within the cavities of the hosts, even resulting in some “strained” binding modes. This provides an opportunity to inspect the behavior of the MT method in terms of its ability to solve collisions that a formal dynamics procedure might also overcome, as well as its propensity to include non-physical states with potentially lower energies.
Method
Structure preparation
All structures were taken from materials provided by the SAMPL5 organizers. The receptors OAH, OAMe and CBClip were processed using the protein preparation wizard utility accessible from Maestro interface of the Schrodinger suite 2015-4 [23]. Protonation states were assigned using PROPKA at pH 7.4 for CBClip and pH 11.5 for octa-acids based on the experimental conditions provided in the instruction manual [24, 25]. All four sulfonic acids were deprotonated at pH 7.4 for CBClip, imparting a net charge of −4, and carboxylates in the octa-acids were likewise deprotonated to impart a net charge of −8 at pH 11.5. The structures of each receptor structure were energy-minimized without constraints using the OPLS 2005 force field [26, 27].
Each of the 16 ligands was prepared using the Ligprep version 3.6 of the Schrödinger suite 2015-4 [28]. Epik was used to predict the protonation states and their associated energetic penalties [29]. The pH values used for assigning protonation states correspond to the experimental conditions. Glide version 6.9 from the same Schrödinger suite was used to perform docking of the ligands with the standard precision (SP) methodology [30–32]. The top 5 scoring docked poses were retained for further evaluation. Glide could not generate docked poses for some of the ligands: in the numbering given for the challenge these were ligands indexed 1, 2, 4 and 6 for the octa-acids and 6, 7, 8 and 10 for CBClip, even with its enhanced sampling feature. Conformational search was performed for these problematic ligands with the Embrace conformational search feature of Macromodel version 11.0 in the same Schrödinger suite [33, 34]. Ligand molecules were manually placed inside the binding pocket of the respective hosts in a reasonable orientation before beginning the conformational search of the complex. Host–guest complexes were minimized using the OPLS 2005 force field with implicit solvent [26, 27]. No explicit waters were added to the complex. Energy minimization of these structures was performed using Powell-Reeves conjugate gradient (PRCG) for 2500 steps with the convergence criterion of all gradient to be less than 0.05 kJ/Å mol [35]. Sampling was done using Mixed Torsion/Low MODe (MT/LMOD) sampling, a variant of low frequency normal mode sampling with random torsion moves added [36]. Receptors were held fixed during sampling and redundant conformers were eliminated using a RMSD criterion of 0.5 Å. This strategy was used to create up to 5 conformers for each ligand. Conformational sampling yielded reasonable binding modes to perform free energy calculations, except for ligand ID 8 of CBClip, which always clashed with its receptor.
The “Movable Type” method
The MT method was employed to rescore the affinities of binding modes generated by the docking described in the previous section. Briefly, the MT method uses an input conformation to portray an ensemble of states within the same vicinity. We assume that in the close neighborhood of a given conformation, all pairwise potentials regarding each atom α are independent. For a molecular system with N atoms, the local partition function regarding atom α is then expressed as:
where i indicates the index of arbitrary atom contacting with α. This approximation assumes orthogonal configurations τ αi of different atom pairs α-i within their respectively defined chemical space Τ αi , hence simulates a local partition function using Eq. 4:
Z′ can be easily generated when each atom pairwise Boltzmann factor term (f ij ) is independent in the MT calculation procedure, which means that instead of generating the molecular configurations of all the states, each sum of the Boltzmann factor on the right hand side of Eq. 4 can be calculated separately, within its defined sampling range τ 0 αi − Δτ ~ τ 0 αi + Δτ. τ 0 αi is the atom pairwise configuration from an initial reference structure in the MT calculation, and Δτ is the sampling amplitude for each atom pairwise configuration, which is typically set to 0.5 or 0.1 Å depending on the specific application. However, Z′ differs from the realistic partition function Z M in that it contains unphysical states which are not realistic in three-dimensional space due to the orthogonal treatment against the pairwise potentials. Z′ is further revised using a sampling volume (V M ) correction ahead of the average of Z′ through Monte-Carlo Integration on the right hand side of Eq. 1. A detailed explanation of the movable type method is present in the supporting information.
Expanding each pairwise interaction in the input conformation, the MT method creates a vector of discrete, uniformly distributed interactions as a function. The exponential average of each potential vector then contributes to the Helmholtz free energy as shown in Eq. 2. To estimate the host–guest binding free energy, local partition functions are summed through all the input configurations regarding the bound state and free state of the host–guest molecular system, as shown in Eq. 5 and 6.
KECSA [37] is a statistical potential developed in our group with a novel feature in the modeling of the “reference state” taken to calculate each atom pairwise probability distribution. The KECSA derivation always relies on structural database collection, but in separate versions different strategies have been applied to smooth the probability distribution function. In this exercise, MT1 refers to MT implementing a KECSA potential based on a Lennard–Jones potential formula to refit the atom pairwise probability distribution function. In contrast, MT2 refers to MT implementing a KECSA potential based on locally weighted regression smoothing (LOESS) [38, 39] of the probability distribution function.
Results and discussion
Blind challenge submissions
Due to the limited binding site volume of the host molecules and generally rigid guest molecular structures, small numbers of binding modes were generated for all host–guest complexes through the docking and conformational search procedures. We submitted two sets of binding affinity predictions for each of the host–guest complexes using the two scoring functions compiled with the MT algorithm, with a 0.5 Å sampling range for each host–guest binding modes obtained from the docking and conformational search studies. Binding free energies were calculated by collecting the Boltzmann factors from all the binding modes generated in this way.
For the OAH host–guest binding benchmark, MT1 calculations gave binding free energies with 1.26 kcal/mol mean unsigned error (MUE) and MT2 gave binding energies with 1.53 kcal/mol MUE. The statistical correlation between experimental and predicted binding affinities gave Pearson’s R values of 0.86 for MT1 and 0.81 for MT2. Both sets of submissions overestimated the binding free energy of the guests with sp3 nitrogen cation head groups (ligands G3 and G5) with a mean absolute error as 2.39 kcal/mol for MT1 and 1.95 kcal/mol for MT2. Binding energies for the other guests, all of which had carboxylate head groups, showed lower MUEs (0.69 kcal/mol for MT1 and 1.31 kcal/mol for MT2).
For the OAMe binding validation set, the predictions for G1, G2 and G3 are within 1 kcal/mol of the experimental binding affinity for MT1. G5 and G6 have slightly higher deviation from experimental binding affinities with the absolute errors of ~3 kcal/mol. The predictions for MT2 are similar to MT1 with the absolute difference of around ±1 kcal/mol except for G4. Due to the incorrect prediction of the OAMe-G4 binding free energy, the overall statistical correlation between the experimental and the predicted binding affinities for the OAMe validation dataset was quite poor with the Pearson’s R of −0.85 and −0.76 for MT1 and MT2. The errors were also higher for the binding affinity prediction when compared to the OAH-guest results with a MUE of 2.83 kcal/mol for MT1 and 3.06 kcal/mol for MT2.
The results for the OAH and OAMe systems are summarized in Table 1. Experimentally, the two octa-acids hosts do not distinguish between most of the guests in terms of binding free energy (with less than 1 kcal/mol binding ΔG difference), except that G3 has a 1.07 kcal/mol difference between OAH and OAMe while G4 has the most significant binding free energy difference (7.00 kcal/mol) between the two hosts. Both MT1 and MT2 capture the trend of the binding free energy changes for G2, G3 and G5 against the two hosts. The most serious error in binding free energy occurs in the OAMe-G4 complex: both MT1 and MT2 overestimated the binding affinity by roughly 9 kcal/mol.
The quality of binding affinity predictions was mixed for CBClip, as shown in Table 2. Predictions from MT1 are within 1.5 kcal/mol of the experimental values for G1, G2, G6, G7 and G9. Binding affinity prediction errors for G1 and G7 are slightly higher than 2 kcal/mol for MT2, but are consistent with the MT1 predictions for the rest of the guests. Neither MT1 nor MT2 was able to predict the affinities for G3, G4, G5 and G10. As stated in the Methods, we could not obtain a reasonable binding mode for G8 from either docking or conformational search, so no predictions were submitted for G8. The reason for the observed errors was ultimately traced to the structure preparation procedure, which is discussed further below. No obvious trend in the quality of predictions could be observed with respect to the ligand functional groups. The predicted binding affinity of the CBClip dataset did not show a clear correlation with the experimental binding affinities, with a Pearson’s R of −0.03 kcal/mol for the MT1 and −0.05 kcal/mol for the MT2 binding affinity predictions. The observed MUE for the MT1 predictions was 2.77 kcal/mol while for MT2 is was 3.36 kcal/mol.
Further analysis of the OAMe: G4 binding free energy
The difficulty of placing the bulky G4 guest structure in the cavity of the OAMe host molecule calls into question what the correct docked pose might be for this system in particular. In the original blind challenge, we only achieved the bound state conformations through a conformational search approach (docking failed) for some guests binding to the octa-acid hosts. Negative net charges on both the guest and host in these systems prevented our docking calculations from generating favorable binding poses. The disagreement between our calculated result and the experimental binding free energy may indicate misplacement of the G4 guest in the OAMe binding pocket, unphysical states in the MT calculation biasing the energy to be overly favorable, deficiencies in the KECSA potential, solvation model or any combination of these factors.
In contrast to docking, our conformational search approach buried the G4 guest in both of the OAH and OAMe cavities with the carboxylate group pointing out towards solution and the Bromine group surrounded by the hydrophobic inner surface of the hosts (see Fig. 2). The similarity of the calculated binding modes contrasts sharply with the disparity in experimental binding affinities and suggests at least two hypotheses. First, the methylated entrance of OAMe may be unable to accommodate the bulky brominated adamantyl guest, while there is enough room for it in the OAH host, implying that our docked poses are wrong. Alternatively, the MT free energy calculation does not capture the change in the binding affinity caused by the four-methyl groups—in particular solvation occlusion. Ultimately, we determined that the SASA calculation used in our solvent model did not account for the differential solvation effect and the details of how we were able to address this problem are given below. However, this did not address the issue of the preferred pose since some groups favored the G4 ligand binding to the surface of OAMe. To address this we carried out detailed simulations, which we present first prior to discussing the modifications of our solvation model. These simulations demonstrate that pocket binding is preferred over surface binding even for the OAMe-G4 complex.
To test these hypotheses, we turned to potentials of mean force (PMFs) generated through umbrella sampling and the Weighted Histogram Analysis Method (WHAM) [40]. The ligand and receptor were described by GAFF 1.8 atom types and bonded parameters [41], while partial charges for each system were taken from the materials distributed in the challenge. SPC/E waters were introduced as the solvent model [42]. A dummy particle with no charge or van-der Waals properties was positioned in the interior of the octa-acid cavity, bound to four tetrahedral carbon atoms at the base of the octa-acid receptor by harmonic distance restraints of 64 kcal/mol Å2 and to one of the ligand atoms by a harmonic distance restraint of 16 kcal/mol Å2. The dummy particle was given a mass of 10 Daltons to permit integration of the Newtonian equations of motion. The restraints and position of the dummy particle were chosen so as not to adversely affect the structure or dynamics of the octa-acids themselves while providing a means for pulling the ligand towards the interior of the cavity, not its side wall (see Fig. 3). We computed multiple PMFs pulling different ligand atoms towards the cavity interior. In each PMF, 96 windows of 2 ns dynamics were conducted, sequentially reducing the length of the umbrella sampling restraint from 24 Å to zero and discarding the first 200 ps of each window for equilibration. To check convergence, we replicated the same studies with 4 ns windows.
Potentials of mean force for binding of brominated adamantane (Guest ID 4) to OAH and to OAMe. A dummy particle (blue dot in the structures on the right), which had no non-bonded properties and so had no influence on the system except by the restraints tethering it to the four tetrahedral carbons at the base of the OA basket, was used to pull the ligand towards the center of the receptor cavity. Multiple PMFs were attempted for each case, using different window lengths and pulling from different atoms on the ligand, as labeled in the lower right panel. In five of the trials, the ligand did enter the cavity, and these successful binding events always had the orientations shown in the top two structures. If it failed to enter the cavity, the ligand bound to the outside of the basket in orientations like the one shown
The umbrella sampling studies confirm our hypothesis that the ligand does indeed bind inside the octa-acid basket receptor, even when the rim of the octa-acid is methylated. In both cases the binding pose, shown in Fig. 3, involves the ligand’s carboxylate group pointed out of the receptor’s mouth while the ligand’s bromine functional group points into one of the receptor’s four symmetry-related macrocyclic rings. Convergence, we found, was not to be determined by the length of individual windows but by having many runs in which the ligand either did or did not enter the cavity, after which its binding state was evaluated. While the ligand did not always enter the cavity, in five of twelve trajectories it did, and the PMFs obtained from these trajectories indicate substantially more favorable binding energies than those in which it did not. Having the ligand contact the OA receptor in the right orientation was not guaranteed, and our one-dimensional umbrella layout which allowed the ligand to move all around the receptor at large values of the distance restraint did nothing to direct the movement of the ligand into the cavity. A more thorough analysis using a two-dimensional WHAM and restraining the ligand to be certain distances from each of two particles positioned along the symmetry axis of the receptor would probably be able to guide the ligand into the pocket and yield a more converged estimate of the binding energy to OAH or OAMe. However, the PMFs computed when the ligand did enter the cavity yield binding energies of 14 kcal/mol for OAH (no error bar available, as only one trajectory showed binding inside the cavity) and 4.25 ± 0.5 kcal/mol for OAMe. Also, the PMFs indicate that methylation of the rim creates a 2–3 kcal/mol activation barrier to ligand binding which is not present in the hydrogenated case. Given that these PMFs were selected out of a wider assortment of possible outcomes, they can be expected to over-estimate the binding affinities by a small amount, but the results are in line with the known binding affinities for these host:guest systems.
The binding modes generated from the PMFs and the conformational search performed with Schrödinger’s software are quite similar, with sub-Ångstrom RMSDs between the results of each method for OAH-G4 and OAMe-G4. Moreover, both approaches place the G4 guest in nearly the same position inside the binding pocket of the hosts. The PMF calculation differentiates the binding modes for each octa-acid by ~10 kcal/mol, which further suggests that the MT calculation, KECSA potential or solvation model must be improved.
There is, in fact, a subtle difference between the binding poses generated by our PMFs and conformational searches: the conformation search pushes the ligand slightly further into the cavity than the molecular dynamics simulation, which in turn exposes less surface area of the G4 carboxylate group to the water solvent in the two OA cavities (see Figs. 2, 3). In the current study, we used an implicit solvent model called KMTISM to estimate the binding free energies [43], which was originally designed as a method for small molecule solvation free energy estimation. The solvation free energy is calculated as the integral of energy functions of the solvent accessible surface areas (SASA) regarding different atom types extended to a 6 Å cutoff distance, beyond which the solute–solvent contacts are omitted. However, since the original implementation was validated against small molecules the SASA model did not correctly account for solvent occlusion by the presence of other atoms in close proximity like the present case. This loss of sampling volume was not captured in our original KMTISM model and in our original SAMPL5 submission.
A modified solvation model was introduced to account for the solvation sampling volume loss as the ligand binds to the receptor (Fig. 4). Within the solvation sampling volume defined as the SASA multiplied by the 6 Å sampling cutoff distance with respect to each atom, any proximal atoms are considered with their volume of exclusion being defined as the volume of the sphere whose radius is the van der Waals radius of the proximal atom plus the water molecule radius. The solute-water contact energies are calculated at each discrete distance r (0.005 Å as the sampling step length) away from the solvent accessible surface of each solute atom, which is modeled as the solute atom-water contact energy (as a function of r) multiplied by the number of accessible water molecules at that distance. The number of water molecules at each discrete distance is simulated using the SASA excluding the cross section area of the volume of exclusion at that distance then divided by the water molecule cross section area.
For each atom from the solute, the solvent accessible surface area (SASA) was generated in KMTISM. Previous calculations only used the multiplication of SASA and the sampling distance (6 Å) to account for the solvent sampling volume V solvent . Volumes occupied by other residues within each V solvent have been excluded in the updated solvent model applied in this study
We introduced this change into our solvation model and re-calculated the binding affinities. Table 3 compares the predictions of the OAMe-guests binding free energies using the old and the refined solvation model. The binding affinity prediction for the OAMe-G4 system reduced from −11.41 to −6.34 kcal/mol for MT1 and from −11.63 to −6.55 kcal/mol for MT2 by using the refined solvation model compared to −2.38 kcal/mol experimental. The simulations for the other OAMe-guests systems have corresponding changes when using the refined solvation model, with the overall MUE dropping from 2.83 to 1.42 kcal/mol for the MT1 procedure and from 3.06 to 1.54 kcal/mol for the MT2 procedure. Application of the refined solvation model was also introduced to the OAH-guests binding study. Results are listed in Table 4, including the binding affinity simulations using the MT1 and MT2 procedures with the old and refined solvation model. For both MT1 and MT2 procedures, the binding free energy simulation using the new solvation model showed a small change compared to the original submissions, with the MUE for the MT1 simulations compared to experiment being 1.34 kcal/mol using the refined solvation model, versus 1.26 kcal/mol for the original submission. The MUE for the MT2 simulations was 1.21 kcal/mol compared to 1.52 kcal/mol after use of the refined solvation model.
Generally, the refined solvation model lowered the OAMe-G4 binding affinity by ~5.1 kcal/mol compared to the original submission, hence, enabling the MT simulations to differentiate between the two OAMe-G4 binding process by ~2.3 kcal/mol, which illustrates the contribution of solvent occlusion to the two octa-acids-G4 binding affinities. Importantly, the binding affinity predictions for the other octa-acids-guest systems were not significantly affected by this new solvation model, resulting in an overall improvement in the prediction accuracy for the 12 host–guest systems, in terms of RMSE, from 3.26 to 1.83 kcal/mol for the MT1 procedure and from 3.53 kcal/mol to 1.48 kcal/mol for the MT2 procedure.
Structural issues for the CBClip guest set
After submitting our predictions for the SAMPL5 challenge, we discovered problems with the materials provided for CBClip guest structures. For CBClip, the bond orders were incorrect for ligands 4, 5, 8, 9 and 10. The atoms associated with these bonds are highlighted in Fig. 5.
We manually corrected the starting materials using the Build feature of Maestro [44]. After correcting the bond order issue and assigning proper formal charges, obvious improvements in the MT calculations were seen using the same procedure applied in the original submission. Plots of the predicted versus experimental binding affinities are shown for the original and corrected submissions in Fig. 6. The original submissions had no discernable correlation to the experimental binding affinities, but after correcting the structures the correlations rebounded to (Pearson’s R) 0.68 for MT1 and 0.53 for MT2. At the same time, there is a huge drop in MUEs, going from 2.77 in the original submission to 1.42 after corrections for MT1, and from 3.36 to 2.15 for MT2. The fact that our MT rescoring gave poor performance when the structures were wrong and incorrectly charged, but then gave improved signal when the charges and atom types were corrected, indicates an ability to discriminate against decoy atom typing.
It shows the correlation plots of predicted binding affinities with the experimental binding affinities (kcal/mol) for CBClip and its 10 guests obtained using MT1 (top panel) and MT2 (bottom panel). The correlation plots for predictions in original submission are in left and after correcting structures are in right
Figure 7 shows the comparison of the experimental binding affinities of individual CBClip host–guest complexes, the predicted binding affinities (kcal/mol) from the original submission, the predictions obtained after structure correction and the binding affinity predictions after correcting the structure and including the new solvation model using both MT1 and MT2 energy functions. The performance of MT1 gets worse after incorporating the new solvation model in the energy function. The Pearson’s R for the corrected structures with experiment (kcal/mol) reduces from 0.68 to 0.57 after inclusion of the new solvation model and the RMSE errors increase from 1.75 to 2.1 kcal/mol. On the other hand, the MT2 energy function performs better with the incorporation of the new solvation model. The Pearson’s R correlation coefficient increases from 0.53 to 0.68. The errors were roughly the same with the MUE around 2.15 kcal/mol. The binding affinity predictions after correcting the structures and incorporating the new solvation model for all the CBClip-guest complexes are listed in Table 5.
It displays the experimental binding affinity (black), predicted binding affinities in original submission (red), predicted binding affinities after correcting the structures (yellow) and predicted binding affinities after incorporating new solvent model and correcting the structures for CBClip and its 10 guests. The top graph shows the binding affinity predictions (kcal/mol) obtained from MT1 and the bottom graph from MT2
Collision forgiveness in MT simulation
When atom pairwise collisions are introduced from the docking or conformational search procedures, the MT method lowers the contact energy if a more favorable pairwise distance region is close by. A crucial question is whether this collision tolerance can help to improve the binding affinity prediction or merely introduces errors into the calculation. In this exercise, we purposefully designed a validation set with mild collisions between the host–guest contacts. Conformational search was done with the additional condition that the host structures were kept rigid through-out the whole process. Unsurprisingly, most of the complexes generated in this manner contained at least a few strained contacts. A validation set, with mild collisions, was created via inclusion of complex conformations having less than three atom pairwise distances within the collision region according to the KECSA potential function. A shift towards high-energy values were observed for single point calculations (no MT sampling) using the KECSA scoring function against this test set. MT simulation was then introduced with 0.1 and 0.5 Å sampling ranges against the same set of conformations. The results and comparisons are shown in Table 6, with an overall improvement in terms of RMSE via introduction of sampling using MT simulation. With such a small validation set, it is far from a detailed analysis of the collision forgiveness afforded by the MT method, but it gives a sense that MT simulation does improve a poor initial structure. However, a few overestimations are observed in the calculations for some systems, i.e. the simulation against the OAME-guest 6 binding free energies, which points to the need to further refine MT.
Conclusions
The SAMPL blind challenge provides an excellent validation platform to assist in the rapid development of tools aimed at addressing the phenomenon of molecular recognition. Our participation utilized the “Movable Type” method, an in-house free energy simulation algorithm, applied to the conformations generated from the docking and conformational search procedures that form the Schrödinger small-molecule drug discovery suite. Our blind submissions estimated the binding free energies for the three host–guest validation sets with MUE’s distributed from 1.26 to 3.36 kcal/mol. We performed best on the OAH-guest set with MUEs of 1.26 kcal/mol for MT1 and 1.53 kcal/mol for MT2, and Pearson R values of 0.86 and 0.81, respectively. On the other hand, both the OAMe-guest and CBClip-guest sets proved to be more challenging for our computational approach.
In the OAMe-guest set, the OAMe-G4 guest gave a ~9 kcal/mol error for both of our submissions. In particular, the computed binding affinity for both OAMe-G4 and OAH-G4 were nearly identical with the former occluding the carboxylate of G4 from solvent via the presence of the larger—OMe groups around the cavity opening. This lead us to explore the nature of our solvent model, which we discovered did not accurately account for solvent occlusion by the—OMe groups. Using a revised solvation model that accounts for solvent occlusion appropriately lead to a significant improvement (reducing the binding affinity by ~5 kcal/mol) in our binding free energy prediction. In discussions there were questions about whether or not G4 bound to the surface of the entrance in OAMe or bound akin to the OAH guest where G4 was buried into the pocket. We favored the latter rather than the former and to address this we decided to carry out extensive MD PMF studies.
In the follow-up study, PMF calculations using umbrella sampling were performed against both the OAMe-G4 and the OAH-G4 complexes, which differ only around their entrances (four methyl groups vs. four hydrogens, respectively), to dig deeper into the binding mode of these systems. The umbrella sampling results suggested similar binding modes for both systems with the G4 ligand placed in almost identical locations for both the OAH and OAMe cavities. The PMF calculation differentiated the two binding procedures by ~10 kcal/mol in terms of free energy, with the OAH-G4 binding affinity being too negative by ~5 kcal/mol relative to experiment. From these results we conclude that G4 binds nearly identically to OAH and OAMe and that differential solvation effects are largely responsible for the observed differential binding affinities.
Bond order issues and incorrect formal charges were present in the provided material for the CBClip-guest validation set, which we did not detect in our first submission. Significant improvement was observed after correcting these problems. Collision forgiveness of the MT method was also studied by introducing a validation set with mild collisions in the host–guest binding modes. For 9 out of 12 test cases in this validation set, a 0.5 Å sampling range improved the MT binding affinity simulation with less than 2 kcal/mol absolute errors for each of the cases. However, the MT simulation for the three OAMe-G6 binding modes resulted in a more than 3 kcal/mol overestimation hinting at a deleterious impact arising from the generation of non-physical states in MT simulations.
References
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) J Comput Aided Mol Des 24(4):259
Guthrie JP (2009) J Phys Chem B 113(14):4501
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) J Comput Aided Mol Des 28(4):305
Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK (2012) J Comput Aided Mol Des 26(5):475
Skillman AG (2012) J Comput Aided Mol Des 26(5):473
Benson ML, Faver JC, Ucisik MN, Dashti DS, Zheng Z, Merz KM (2012) J Comput Aided Mol Des 26(5):647
Yin J, Henriksen NM, Slochower DR, Shirts MR, Chiu MW, Mobley DL, Gilson MK (2016) J Comput Aided Mol Des. doi:10.1007/s10822-016-9974-4
Chang CE, Gilson MK (2004) J Am Chem Soc 126(40):13156
Chen W, Chang CE, Gilson MK (2004) Biophys J 87(5):3035
Houk KN, Leach AG, Kim SP, Zhang XY (2003) Angew Chem Int Ed 42(40):4872
Liu SM, Ruspic C, Mukhopadhyay P, Chakrabarti S, Zavalij PY, Isaacs L (2005) J Am Chem Soc 127(45):15959
Gilberg L, Zhang B, Zavalij PY, Sindelar V, Isaacs L (2015) Org Biomol Chem 13(13):4041
Zhang B, Isaacs L (2014) J Med Chem 57(22):9554
Hettiarachchi G, Nguyen D, Wu J, Lucas D, Ma D, Isaacs L, Briken V (2010) PLoS One 5(5):e10514. doi:10.1371/journal.pone.0010514
Lagona J, Mukhopadhyay P, Chakrabarti S, Isaacs L (2005) Angew Chem Int Ed 44(31):4844
Rogers KE, Ortiz-Sanchez JM, Baron R, Fajer M, de Oliveira CAF, McCammon JA (2013) J Chem Theory Comput 9(1):46
Choudhury R, Gupta S, Da Silva JP, Ramamurthy V (2013) J Org Chem 78(5):1824
Porel M, Jayaraj N, Kaanumalle LS, Maddipatla MVSN, Parthasarathy A, Ramamurthy V (2009) Langmuir 25(6):3473
Gibb CLD, Gibb BC (2014) J Comput Aided Mol Des 28(4):319
Gibb CLD, Gibb BC (2004) J Am Chem Soc 126(37):11408
Liu SM, Whisenhunt-Ioup SE, Gibb CLD, Gibb BC (2011) Supramol Chem 23(6):480
Gan HY, Benjamin CJ, Gibb BC (2011) J Am Chem Soc 133(13):4770
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) J Comput Aided Mol Des 27(3):221
Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH (2011) J Chem Theory Comput 7(2):525
Rostkowski M, Olsson MHM, Sondergaard CR, Jensen JH (2011) BMC Struct Biol. doi:10.1186/1472-6807-11-6
Jorgensen WL, Tiradorives J (1988) J Am Chem Soc 110(6):1657
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) J Phys Chem B 105(28):6474
LigPrep (2015) Version 3.6. Schrödinger, LLC, New York
Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M (2007) J Comput Aided Mol Des 21(12):681
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) J Med Chem 47(7):1739
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) J Med Chem 49(21):6177
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) J Med Chem 47(7):1750
Macromodel (2015) Schrödinger, LLC, New York
Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC (1990) J Comput Chem 11(4):440
Polak E, Ribiere G (1969) Rev Fr Inf Rech Oper 3(16):35
Kolossvary I, Guida WC (1996) J Am Chem Soc 118(21):5011
Zheng Z, Merz KM (2013) J Chem Inf Model 53(5):1073
Cleveland WS (1979) J Am Stat Assoc 74(368):829
Cleveland WS (1981) Am Stat 35(1):54
Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM (1992) J Comput Chem 13(8):1011
Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25(9):1157
Berendsen HJC, Grigera JR, Straatsma TP (1987) J Phys Chem 91(24):6269
Zheng Z, Wang T, Li PF, Merz KM (2015) J Chem Theory Comput 11(2):667
Maestro (2015) Schrödinger, LLC, New York
Acknowledgments
We would like to acknowledge the SAMPL5 organizers for providing the data and platform for the blind challenge and global communication. NB would like to acknowledge Mr. Dario Gioia for numerous discussions related to docking of host–guest systems.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nupur Bansal and Zheng Zheng have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bansal, N., Zheng, Z., Cerutti, D.S. et al. On the fly estimation of host–guest binding free energies using the movable type method: participation in the SAMPL5 blind challenge. J Comput Aided Mol Des 31, 47–60 (2017). https://doi.org/10.1007/s10822-016-9980-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9980-6