Abstract
Purpose
To evaluate whether the mitoscore of cleavage stage embryos might correlate with developmental kinetics and the ploidy status.
Materials
This retrospective single-center study involved all cycles between April 2016 and April 2018 in which preimplantation genetic testing for aneuploidy (PGT-A) on day 3 was performed. The mitochondrial DNA (mtDNA) content and embryo ploidy were determined on 375 single blastomere biopsies by next generation sequencing (NGS). After intracytoplasmic sperm injection, a time-lapse imaging system (embryoscope) was used to follow the development. The median mtDNA content of cleavage stage embryos (49.4) was used to stratify the embryos into two groups to compare embryo development and ploidy status: low mitoscore group (≤ 49.4) and high mitoscore group (> 49.4).
Results
The total number of euploid embryos was equal between both mitoscore groups (32.1% versus 33.5%; p = 0.854). However, embryos in the low mitoscore group had a significantly higher cell number on day 3 (8.13 ± 1.59 versus 7.62 ± 1.5; p = 0.0013) and showed a significantly faster development up until the 8-cell stage. Mitoscore was not different between euploid and aneuploid embryos, with the same blastomere number at the time of biopsy. Furthermore, absence of cavitation within 118 h after insemination was correlated with higher mitoscore values (60.22 ± 42.23 versus 50.97 ± 13.37; p = 0.006) and a lower chance of being euploid (17.1% versus 47.4%; p = 0.001).
Conclusion
mtDNA content of cleavage stage embryos correlates with time-lapse parameters. Early blastulation is correlated with a lower mtDNA content and a higher chance of euploidy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Embryologists are faced with the daily challenge of identifying the embryo with the highest implantation potential and chance for a live birth. Different invasive and non-invasive selection criteria have been used to give a direct or indirect indication of embryo implantation potential [1, 2]. Despite the selection of morphologically and chromosomally normal embryos, a significant percentage of these embryos will still fail to implant, and in most cases, the reason for failure is unknown [3].
Different theories have been proposed in an effort to explain failed implantation: asynchrony of the receptive endometrium and the potentially implanting blastocyst [4], elevated progesterone levels [5], and most recently, problems with the energy production of the developing embryo. As a result, the mitochondrion is now viewed as an exciting new viability paradigm in assisted reproductive techniques [6,7,8,9].
Mitochondria are membrane-bound, cytoplasmic organelles found in eukaryotic cells, and their size and number vary from cell to cell and depend on the cell’s volume and energy needs [7, 10]. The oocyte is the richest cell of the body in terms of the number of mitochondria and depends largely on their adenosine tri-phosphate (ATP) production to acquire competence for fertilization and early embryonic development [7, 11, 12]. The mitochondria contain their own circular double-stranded genome, the mitochondrial DNA (mtDNA), and is indicative for the number of mitochondria in the oocyte [13]. The number of mtDNA copies in the cells are prone to changes according to different conditions like (i) a decreased ovarian response [14], (ii) aging [8, 15], (iii) BMI [16, 17], (iv) smoking [16], (v) the type of stimulation [18], (vi) preimplantation embryonic stress [8], and even (vii) the method applied to determine the mtDNA content [19, 20]. Mature human oocytes contain around 100,000 mitochondria and 50,000 to 550,000 mtDNA copies, with a considerable degree of inter and intra-patient variability [10].
The mtDNA of oocytes and their cumulus cells have been examined, and lower levels are associated with fertilization failure or impaired embryo development [10, 16, 21, 22]. During cleavage divisions, the total number of mitochondria and mtDNA copy number are not subject to biogenesis and will therefore reduce per cell during preimplantation development [23,24,25]. mtDNA replication resumes around the time of blastocyst formation which coincides with a change in blastocyst energy demand, while mitochondrial replication will only be observed post-implantation [9, 26, 27]. However, substantial changes are noted in the mitochondrial morphology during preimplantation development to meet the energy demand of the embryo. From round oval mitochondria with a dense matrix and few arched cristae in the early stages of preimplantation development, the mitochondria elongate and develop an extensive array of cristae as they reach the blastocyst stage [13, 28, 29].
Recently, studies assessed the normalized mtDNA content, the so-called “mitochondrial score” or “mitoscore,” as a parameter for embryo viability and as a predictor for implantation [8, 30]. Both studies suggested that an increased number of mtDNA copies at the blastocyst stage is associated with a decreased chance of implantation. However, both studies reported conflicting results on the value of this parameter for cleavage stage embryos and only a limited number of cleavage stage embryos were analyzed [8, 30, 31]. There are discordant results related with the mtDNA copy number and cleavage stage embryo quality, and so far, no correlation has been described regarding the ploidy status and mtDNA copy number of cleavage stage embryos [30, 32].
As embryo vitrification is not allowed in the United Arab Emirates, it is important to explore the validity of the cleavage stage mitoscore as a potential tool in selecting embryos with the highest viability as currently; there is still a gap in the data linking the mitochondrial function to embryo viability [33]. Therefore, the aim of this retrospective analysis was to evaluate whether the mitoscore of cleavage stage embryos might correlate with developmental kinetics up to the blastocyst stage and the ploidy status.
Material and methods
Study design
In this single-center retrospective study, the data retrieved between April 2016 and April 2018, the blastomere mtDNA copy number was obtained from day 3 cleavage stage embryos, which underwent biopsy for preimplantation genetic testing for aneuploidies (PGT-A). As the fertilization law of the United Arab Emirates does not allow embryo cryopreservation, only day 3 cleavage stage biopsy was applicable in order to allow a fresh euploid day 5 transfer. All embryos were divided into two groups based on the median mitoscore value [34]. The obtained mitoscore was correlated with the morphokinetic parameters recorded by time-lapse imaging and with ploidy status. Sub-analysis was performed for ploidy outcome and ability to cavitate based on the number of blastomeres at the time of biopsy. This retrospective data analysis was approved by the Ethics committee of IVI Middle East Fertility Clinic (REC008).
Ovarian stimulation, oocyte retrieval, and ICSI
Patients underwent ovarian stimulation using standardized protocols (GnRH-agonist protocol, either short or long protocol, GnRH-antagonist protocol). Stimulation medication was either recFSH (recombinant follicle-stimulating hormone) or HMG (human menopausal gonadotropin); the dosage was chosen according to the ovarian reserve parameters [35]. When ≥ 3 follicles ≥ 17 mm were identified on ultrasound scan, final oocyte maturation was achieved by administration of either 5.000 IU/10.000 IU of hCG (Pregnyl®, NV Organon) or use of “dual trigger” (5.000 IU hCG plus 0.3 mg decapeptyl) or in an effort to avoid ovarian hyperstimulation syndrome 0.3 mg GnRH-agonist was administered [36]. Oocyte retrieval was carried out 36 hours after administration of trigger for final oocyte maturation under ultrasound guidance. ICSI procedure was performed according to previously described techniques [37].
Time-lapse analysis and embryo development
Time-lapse images were recorded every 20 min. Embryos were placed in the embryoscope (Vitrolife, Göteborg, Sweden) immediately after ICSI and the following quantitative parameters were assessed: pronuclear fading (tPNf), two-cell division (t2) to nine-cell division (t3-9+), time of formation of morula (tM), and start of blastulation (tSB). Additionally, relevant kinetic parameters were calculated: s2 is the time of synchrony of the second cell cycle (t4-t3), cc2 represents the timing of the second cell cycle (t3-t2), s3 is the time of synchrony of the third cell cycle (t8-t5), and cc3 represents the timing of the third cell cycle (t5-t2). Analysis was based on the digital images acquired by the time-lapse monitoring (TLM) system and the evaluation on an external computer with the software for analysis (EmbryoViewer workstation, Vitrolife). A single embryologist analyzed the data and calculated relevant kinetic parameters.
Blastocyst formation was verified 118 h post-injection by checking the cavitation capacity [38, 39].
Embryo biopsy and culture conditions
Embryos were cultured in SAGE Quinn’s Advantage Sequential medium (SAGE, Målov, Denmark) using time-lapse system embryoscope™ (6%CO2, 5%O2).
Fertilization was assessed 17–20 h post-ICSI and confirmed by the presence of two pronuclei and two polar bodies.
Embryo biopsy was performed on day 3. Only embryos with five or more nucleated blastomeres and less than 25% fragmentation were biopsied. Day 3 embryo biopsy can be summarized as follows: embryos were placed in a droplet containing Ca2+/Mg2+-free medium (G-PGD, Vitrolife, Göteborg, Sweden), the zona pellucida was perforated by pulses of laser (OCTAX, Navi lase Herborn, Germany), and one blastomere was withdrawn from each embryo. Individual blastomeres were placed in 0.2-ml PCR tubes containing 2 μL PBS. For blastomere washing and handling, 1% polyvinylpyrrolidone (PVP) was used [40]. After biopsy, the embryos were further cultured in blastocyst medium (SAGE, Målov, Denmark). From day 3 onwards, the number of evaluated embryos was declining due to either arrest of the embryo(s) or embryo transfer on day 4.
Next generation sequencing
To analyze biopsy samples, a next generation sequencing (NGS) platform was used (Resproseq, Life-Thermofisher, USA). Whole genome amplification (WGA) protocol was performed on all individual samples. After WGA, library preparation consisted of the incorporation of individual barcodes for the amplified DNA of each embryo. Sequencing was performed in a 316 or 318 chip using the personal genome sequencing machine (Life-Thermofisher, USA) after isothermal amplification and enrichment. For sequencing analysis and data interpretation, the ion reporter software was employed.
Determination of mitochondrial DNA copy number
mtDNA copy number was detected from the same sample in Igenomix, Dubai, UAE. NGS yields one value for mtDNA and one value for nuclear DNA (nDNA) quantity, and the ratio of mtDNA to nDNA is the principal mode to assess mtDNA quantity per cell which is defined as the mitoscore [41]. Crucially, using nDNA values for normalization, it is assumed that the composition of nDNA is equal across samples.
Statistical analysis
The median mtDNA content was used to stratify the embryos into two groups to compare embryo development and ploidy status. Student’s t test was used to compare developmental kinetics and to compare embryo cellularity and mitoscore, and χ2 test was used to compare the ploidy status between low and high mitoscore groups. Cellularity was checked based on the number of mitotic divisions at the time of biopsy: embryos that underwent 3 mitotic divisions had 5–8 blastomeres, while embryos with 4 mitotic divisions had 9–16 blastomeres at the time of biopsy. All p values were based on two-tailed tests, with statistical significance indicated by p < 0.05. Data analysis was performed using the statistical software R (version 3.5.0).
Results
The mitoscore was calculated in 375 day 3 blastomeres obtained from 66 patients undergoing PGT-A. The number of tested embryos per patient ranged from 1 to 13 with a mean of 5.5 embryos biopsied. The mean female age was 34.9 years (range, 18–44 years). The indications for PGT-A are presented in Table 1. The results of the genetic testing showed that of the 375 analyzed embryos, 123 (32.8%) were euploid and 252 (67.2%) aneuploid.
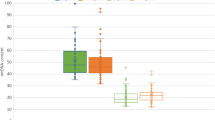
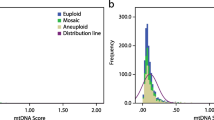
Mitoscore values from day 3 embryo biopsies ranged from 24.0 to 353.3 with a mean of 53.21 ± 23.34 (Fig. 1). Mitoscore values were categorized using the median mitoscore value of 49.4; therefore, the low mitoscore group comprised the embryos with a mitoscore value of ≤ 49.4 (187 embryos) and the high mitoscore group included embryos with mitoscore values > 49.4 (188 embryos). When stratifying the ploidy status between the low and high mitoscore group, no difference was found in the ploidy status: 32.1% versus 33.5%; p = 0.854. As the mitoscore may be affected by the number of mitotic divisions, the mitoscore was compared based on the number of blastomeres at the time of biopsy and was significantly higher for embryos with less mitotic divisions (≤ 8 blastomeres, 55.0 versus 47.5; p = 0.0093). There was no difference in mitoscore values between euploid and aneuploid embryos with the same blastomere number (Table 2).
As embryos with less mitotic divisions show significantly higher mitoscore values, comparison of all time-lapse parameters between both mitoscore groups enables us to find at which developmental stage this difference occurs (Table 3). The average cell number at which the biopsy was performed was significantly higher in the low mitoscore group (p = 0.001). Embryos from the low mitoscore group performed all cleavages up to the 8-cell stage significantly earlier compared to the high mitoscore group. However, cc2, cc3, s2, and s3 are not different between both mitoscore groups.
As embryos with lower mitoscore values develop faster from the time of pronuclear fading, it was questioned if these embryos have a higher potential to develop into a blastocyst that is also euploid. Therefore, blastocyst development was analyzed on the subset of embryos (n = 274) that were cultured until day 5 by verifying the cavitation capacity 118 h post-injection. Mitoscore values on day 3 were significantly higher in embryos that did not cavitate before 118 h: 60.22 ± 42.23 versus 50.97 ± 13.37 (p = 0.006). Based on the blastomere number at the time of biopsy, only embryos with 5 blastomeres showed a significantly lower mitoscore if they were able to cavitate (59.4 ± 24.8 vs. 88.8 ± 25.5; p = 0.016; Table 4). If cavitation capacity was verified between euploid and aneuploidy embryos, this difference was still visible for the embryos with 5 blastomeres (59.4 ± 28.4 vs. 97.8 ± 26.6; p = 0.014; Table 4). Of the 76 embryos without cavitation, only 13 (17.1%) were euploid, while 94 (47.4%) of the 198 cavitating embryos were euploid (p = 0.001).
Discussion
In this retrospective study, developmental kinetics and ploidy status of embryos with a low or high mitoscore on day 3 were compared. When comparing developmental patterns, the low mitoscore group showed a significantly faster development up to the 8-cell stage. Mitoscore values were significantly higher in embryos with less mitotic divisions. The ratio of euploid embryos was not significantly different between both mitoscore groups and was not affected by the number of blastomeres at the time of biopsy. Interestingly, in a subgroup of patients followed to day 5, the day 3 mitoscore values were significantly higher in embryos that failed to cavitate before 118 h post-injection and cavitating embryos had a significantly higher chance of being euploid.
Only limited data is available analyzing day 3 mitoscore values [8, 30, 31]. In one such study, Fragouli and colleagues [8] concluded that cleavage stage embryos from reproductively younger women contained significantly higher mtDNA levels than from older women. In contrast, our study demonstrated no significant difference in age (34.7 vs. 33.9 years) and euploidy status (32.1% vs. 33.5%) in the low and high mitoscore group. Analysis based on the blastomere number at the time of biopsy was not able to show a difference in mitoscore between euploid and aneuploid embryos. A recent study analyzing 50 frozen-thawed zygotes was also unable to demonstrate any significant difference between the mitoscore on day 3 and the ploidy status [31], indicating that embryonic mosaicism is an important barrier of day 3 biopsy outcome [42, 43]. As the fertilization law of the United Arab Emirates does not allow embryo cryopreservation, only day 3 biopsy was applicable in order to allow fresh euploid day 5 transfer. Due to mosaicism implicated with cleavage stage biopsy, the ploidy rate could be affected, hence being a limitation to our findings. However, when stratifying according to cavitation capacity, a lower mtDNA content and higher euploidy status were observed in embryos able to cavitate 118 h post-injection. Based on the blastomere number at the time of biopsy, only embryos with 5 blastomeres showed a significantly lower mitoscore for embryos able to cavitate on day 5 (p = 0.016) as well as for aneuploid embryos (p = 0.014). It is unclear why only embryos with 5 blastomeres show a significant difference between cavitation capacity and ploidy; though embryos with only 3 mitotic divisions have higher mitoscore values, unequal division of blastomeres, and hence cytoplasm, could be responsible for the observed difference [32]. Diez-Juan and colleagues [30] concluded that the implantation potential of 205 day 3 embryos is decreased if increased mtDNA levels are present. As this study only took into account euploid embryos, it is unclear if aneuploid embryos present equal mtDNA levels or not.
In contrast to the limited data available on day 3 mitoscore values, this value has been more extensively studied at the blastocyst stage with conflicting results. While increased mtDNA levels have been linked to aneuploidy [8, 17], two studies were unable to find this difference between euploid and aneuploid blastocysts [19, 31]. When analyzing the implantation potential of euploid blastocysts, increased mtDNA was mostly associated with an increased chance of implantation failure [8, 30, 44, 45]. On the other hand, some studies could not find a difference in implantation potential based on the mtDNA content [15, 19, 46]. When comparing mitoscore values of cleavage stage embryos and blastocysts, the blastocyst mitoscore appears to be a stronger predictor of pregnancy as compared to the cleavage stage mitoscore value [30, 44, 45]. Furthermore, blastocyst morphology remains an essential tool for embryo transfer selection. Some studies demonstrated that blastocysts which experienced more mitotic divisions—higher trophectoderm grading—were found to contain lower quantities of mtDNA [15, 45, 46].
While it is thought that mtDNA will evenly segregate between the different blastomeres upon each cell division, a lower blastomere number on day 3 is expected in the high mitoscore group. Indeed, day 3 embryos had on average 8.13 and 7.62 blastomeres in the low or high mitoscore groups, respectively (p = 0.001), indicating a dilution effect with every cell division. However, when looking at the blastomere number at the time of biopsy, no difference was noted between euploid and aneuploidy embryos. Coinciding with the significantly higher blastomere number we observed in the low mitoscore group, these embryos showed a significantly faster pronuclear fading and performed their first mitotic divisions 1–4 h earlier compared to embryos with a high mitoscore. While these morphokinetic differences were seen from t2 until t8, the timings between different mitotic divisions (cc2, cc3, s2, and s3) were not different. This indicates that the difference is set at the moment of pronuclear fading and follows the same pattern up until the 8-cell stage. Unfortunately, we were unable to perform the morphokinetic analysis based on the number of blastomeres at the time of biopsy, as the number of embryos is some groups were too low. Ho and colleagues [31] also looked at a possible correlation between the morphological cleavage stage grading, time-lapse parameters, and cleavage events but could not correlate this to the day 3 mitoscore values. However, this study was conducted on a very small group (n = 50) of frozen-thawed zygotes and cryopreservation, particularly slow-freezing and has a dramatic impact on mitochondrial survival [47]. Embryo quality on day 3 in this study was also poor which could be a possible cause of not finding any difference in grading and mitoscore [31]. When taking into account only the blastomere number on day 3, as in the current study, it clearly demonstrates that the total amount of mtDNA was diluted over cleavage divisions, resulting in a lower mitoscore if embryos underwent more mitotic divisions. However, this hypothesis has been questioned by previous studies as analysis of all 8 blastomeres from a single embryo showed major differences in mtDNA content between all 8 blastomeres [32], and blastomeres also inherit different proportions of mtDNA mainly according to their cytoplasmic volume [48]. As we do not know the original mtDNA content of the oocyte and we do not know the exact volume of the cell at the time of biopsy, it is hard to draw conclusions based on a single parameter measured at one specific time during development. Also, only on a subgroup of embryos—that was able to be fertilized and develop into a good quality embryo on day 3—were included in this analysis, disabling us to draw conclusions on embryos with failed fertilization or with impaired embryo development or developmental arrest [32, 48]. Day 3 biopsy in combination with mitoscore analysis seems an attractive tool to allow fresh transfer on day 5 of a euploid blastocyst with the highest viability. However, the high rate of mosaicism observed in day 3 biopsy samples makes it difficult to rely on a single mononucleated blastomere to predict ploidy status [42].
The differences in development pattern between euploid and aneuploid embryos are not new. In 2012, Chavez and colleagues [49] showed that euploid day 2 embryos have very strict and tightly clustered early cell cycle parameters (duration of first cytokinesis, t3-t2 and t4-t3), while aneuploid embryos showed much larger standard deviations. Chawla et al. [50] also found differences, though not matching with Chavez and colleagues [49] in early cleavage stage parameters between euploid and aneuploid embryos; while tPNf and t2 happened significantly earlier in euploid embryos, the duration of t5, CC2, CC3, and t5-t2 were significantly prolonged in euploid embryos. Aside from the early cleavage stage parameters, two studies were able to link an early start of cavitation to euploid embryos [31, 51] which is in line with the results presented in this study; embryos that failed to cavitate 118 h post-injection had a higher chance of being aneuploid (47.4% versus 17.1%; p = 0.001). However, the study by Ho and colleagues [31] was unable to show a difference in their day 3 mitoscore values, while we found a significant increase in mitoscore values in cases of failed blastulation (60.22 ± 42.23 versus 50.97 ± 13.37 (p = 0.006)). This discrepancy may be attributed to the use of slow-frozen zygotes and the low number of embryos tested as compared to the high number of fresh cleavage stage embryos in our study. It remains unclear whether the higher mitoscore values in non-cavitating embryos arise due to slower development (fewer cells to split the mitochondria) or can be attributed to early mitochondrial biogenesis initiated by stress [30]. Inappropriate activation of mitochondrial biogenesis can cause the production of oxidants leading to the failure of embryonic development [7].
Concerns have been raised regarding the technique used to measure the mtDNA content of embryos in terms of reproducibility between different centers and the subsequent effect on the treatment outcome [20]. In this study, low coverage NGS combined with an internally validated algorithm for mtDNA content was applied to determine the mitoscore. Corrections for autosomes, normalization of the mtDNA against a multicopy nDNA sequence, or differences in chromosome copy number and sex chromosomes have all been proposed or rejected to reflect a better mtDNA content [15, 19, 31, 44]. To date, there is no agreement on the optimal correction factor; for this reason, no correction factor was applied in this study.
While the current study did not address the implantation potential of embryos based on the mitoscore value, we have found that early cell division timings contribute to differences seen in mtDNA content between embryos. Embryos that fail to cavitate on day 5 have significantly higher mitoscore values on day 3 and a significantly higher chance of being aneuploid. As the dragging effect is visible from tPNf onwards, this would be a good parameter to select the best embryo which potentially develops into a blastocyst with the highest chance of being euploid. The outcome of this study indicates that cleavage stage mtDNA content can predict embryo potential to blastulate and can be used as a valuable tool for fresh embryo transfers. Further studies are required to assess the distribution of mitochondria between the different blastomeres and its biogenesis.
References
Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remohí J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26(10):2658–71.
Montag M, Toth B, Strowitzki T. New approaches to embryo selection. Reprod BioMed Online. 2013;27(5):539–46.
Harton GL, Munne S, Surrey M, Grifo J, Kaplan B, McCulloh DH, et al. Diminished effect of maternal age on implantation after preimplantation genetic diagnosis with array comparative genomic hybridization. Fertil Steril. 2013;100:1695–703.
Ruiz-Alonso M, Galindo N, Pellicer A, Simón C. What a difference two days make: “personalized” embryo transfer (pET) paradigm: a case report and pilot study. Hum Reprod. 2014;29(6):1244–7.
Venetis CA, Kolibianakis EM, Bosdou JK, Tarlatzis BC. Progesterone elevation and probability of pregnancy after IVF: a systematic review and meta-analysis of over 60,000 cycles. Hum Reprod Update. 2013;19(5):433–57.
Van Blerkom J, Davis P, Lee J. ATP content of human oocytes and developmental potential and outcome after in vitro fertilization and embryo transfer. Hum Reprod. 1995;10(2):415–24.
May-Panloup P, Boucret L, Chao de la Barca JM, Desquiret-Dumas V, Ferré-L’Hôttelier V, Morinière C, et al. Ovarian ageing: the role of mitochondria in oocytes and follicles. Hum Reprod Update. 2016;22(6):725–43.
Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, et al. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet. 2015;11:e1005241.
Seli E. Mitochondrial DNA as a biomarker for in-vitro fertilization outcome. Curr Opin Obstet Gynecol. 2016;28(3):158–63.
Reynier P, May-Panloup P, Chretien MF, Morgan CJ, Jean M, Savagner F, et al. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Rep. 2001;7:425–9.
May-Panloup P, Chretien MF, Malthiery Y, Reynier P. Mitochondrial DNA in the oocyte and the developing embryo. Curr Top Dev Biol. 2007;77:51–83.
Babayev E, Seli E. Oocyte mitochondrial function and reproduction. Curr Opin Obstet Gynecol. 2015;27(3):175–81.
Van Blerkom J. Mitochondrial function in the human oocyte and embryo andtheir role indevelopmental competence. Mitochondrion. 2011;11:797–813.
Boucret L, Chao de la Barca JM, Morinière C, Desquiret V, Ferré-L’Hôttelier V, Descamps P, et al. Relationship between diminished ovarian reserve and mitochondrial biogenesis in cumulus cells. Hum Reprod. 2015;30(7):1653–64.
Treff NR, Zhan Y, Tao X, Olcha M, Han M, Rajchel J, et al. Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Hum Reprod. 2017;32(4):954–62.
Desquiret-Dumas Desquiret-Dumas V, Cl_ement A, Seegers V, Boucret L, Ferr_e-L’Hotellier V, Bouet PE, et al. The mitochondrial DNA content of cumulus granulosa cells is linked to embryo quality. Hum Reprod. 2017;32(3):607–14.
de los Santos MJ, Diez-Juan A, Mifsud A, Mercader A, Meseguer M, Rubio C, et al. Variables associated with mitochondrial copy number in human blastocysts: what can we learn from trophectoderm biopsies? Fertil Ster. 2018;109(1):110–7.
Cree LM, Hammond ER, Shelling AN, Berg MC, Peek JC, Green MP. Maternal age and ovarian stimulation independently affect oocyte mtDNA copy number and cumulus cell gene expression in bovine clones. Hum Reprod. 2015;30(6):1410–20.
Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL, et al. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril. 2017;107(1):34–42.
Viotti M, Victor AR, Zouves CG, Barnes FL. Is mitochondrial DNA quantitation in blastocyst trophectoderm cells predictive of developmental competence and outcome in clinical IVF? J Assist Reprod Genet. 2017;34(12):1581–5.
Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril. 2016;85(3):584–91.
Ogino M, Tsubamoto H, Sakata K, Oohama N, Hayakawa HM, Kojima T, et al. Mitochondrial DNA copy number in cumulus cells is a strong predictor of obtaining good-quality embryos after IVF. J Assist Reprod Genet. 2016;33(3):367–71.
Cecchino GN, Garcia-Velasco JA. Mitochondrial DNA copy number as a predictor of embryo viability. Fertil Steril. 2018;0015–0282.
Thundathil J, Filion F, Smith LC. Molecular control of mitochondrial function in preimplantation mouse embryos. Mol Reprod Dev. 2005;71:405–13.
Eichenlaub-Ritter U, Wieczorek M, Lüke S, Seidel T. Age related changes in mitochondrial function and new approaches to study redox regulation in mammalian oocytes in response to age or maturation conditions. Mitochondrion. 2011;11:783–96.
St John J. The control of mtDNA replication during differentiation and development. Biochim Biophys Acta. 1840;2014:1345–54.
St John JC, Facucho-Oliveira J, Jiang Y, et al. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum Reprod Update. 2010;16:488–509.
Motta PM, Nottola SA, Makabe S, Heyn R. Mitochondrial morphology in human fetal and adult female germ cells. Hum Reprod. 2000;15(Suppl 2):129–47.
Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15(Suppl 2):148–59.
Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, et al. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril. 2015;104(3):534–41.
Ho JR, Arrach N, Rhodes-Long K, Salem W, McGinnis LK, Chung K, et al. Blastulation timing is associated with differential mitochondrial content in euploid embryos. J Assist Reprod Genet. 2018;35(4):711–20.
Lin DP, Huang CC, Wu HM, Cheng TC, Chen CI, Lee MS. Comparison of mitochondrial DNA contents in human embryos with good or poor morphology at the 8-cell stage. Fertil Steril. 2014;81(1):73–9.
Wells D. Mitochondria DNA quantity as a biomarker for blastocyst implantation potential. Fertil Steril. 2017;108(5):742–7.
Decoster J, Gallucci M, Iselin AR. Best practices for using median splits, artificialcategorization, and their continuous alternatives. J Exp Psychopathal. 2011;5.
La Marca A, Sunkara SK. Individualization of controlled ovarian stimulation in IVF using ovarian reserve markers: from theory to practice. Hum Reprod Update. 2014;20(1):124–40.
Papanikolaou EG, Pozzobon C, Kolibianakis EM, Camus M, Tournaye H, Fatemi HM, et al. Incidence and prediction of ovarian hyperstimulation syndrome in women undergoing gonadotropin-releasing hormone antagonist in vitro fertilization cycles. Fertil Steril. 2006;85(1):112–20.
Steirteghem V, Nagy Z, Joris H, Liu J, Staessen C, Smitz J, et al. High fertilization and implantation rates after intracytoplasmic sperm injection. Hum Reprod. 1993;8(7):1061–6.
Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26(6):1270–83.
Harton GL, Magli MC, Lundin K, Montag M, Lemmen J, Harper JC. European Society for Human Reproduction and Embryology (ESHRE) PGD Consortium/Embryology Special Interest Group. ESHRE PGD Consortium/Embryology Special Interest Group--best practice guidelines for polar body and embryo biopsy for preimplantation genetic diagnosis/screening (PGD/PGS). Hum Reprod. 2011;26(1):41–622.
Phillips NR, Sprouse ML, Roby RK. Simultaneous quantification of mitochondrial DNA copy number deletion ratio: a multiplex real-time PCR assay. SciRep. 2014;4:3887.
Capalbo A, Ubaldi FM, Rienzi L, Scott R, Treff N. Detecting mosaicism in trophectoderm biopsies: current challenges and future possibilities. Hum Reprod. 2017;32(3):492–8.
Vera-Rodriguez M, Rubio C. Assessing the true incidence of mosaicism in preimplantation embryos. Fertil Steril. 2017;107(5):1107–12.
Ravichandran K, McCaffrey C, Grifo J, Morales A, Perloe M, Munne S, et al. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Hum Reprod. 2017;32(6):1282–92.
Fragouli E, McCaffrey C, Ravichandran K, Spath K, Grifo J, Munne S, et al. Clinical implications of mitochondrial DNA quantification on pregnancy outcomes: a blinded prospective non-selection study. Hum Reprod. 2017;32(11):2340–7.
Klimczak AM, Pacheco LE, Lewis KE, Massahi N, Richards JP, Kearns WG, et al. Embryonal mitochondrial DNA: relationship to embryo quality and transfer outcomes. J Assist Reprod Genet. 2018;35(5):871–7.
Gualtieri R, Mollo V, Barbato V, Fiorentino I, Iaccarino M, Talevi R. Ultrastructure and intracellular calcium response during activation in vitrified and slow-frozen human oocytes. Hum Reprod. 2011;26(9):2452–60.
Murakoshi Y, Sueoka K, Takahashi K, Sato S, Sakurai T, Tajima H, et al. Embryo developmental capability and pregnancy outcome are related to the mitochondrial DNA copy number and ooplasmic volume. J Assist Reprod Genet. 2013;30:1367–75.
Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, et al. Dynamic blastomere behavior reflects human embryo ploidy by the four-cell stage. Nat Commun. 2012;3:1251.
Chawla M, Fakih M, Shunnar A, Bayram A, Hellani A, Perumai V, et al. Morphokinetic analysis of cleavage stage embryos and its relationship to aneuploidy in a retrospective time-lapse imaging study. J Assist Reprod Genet. 2015;32(1):69–75.
Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod BioMed Online. 2013;26(5):477–85.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bayram, A., De Munck, N., Elkhatib, I. et al. Cleavage stage mitochondrial DNA is correlated with preimplantation human embryo development and ploidy status. J Assist Reprod Genet 36, 1847–1854 (2019). https://doi.org/10.1007/s10815-019-01520-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-019-01520-y