Abstract
Bangia fuscopurpurea is a farmed species in the ancient family Bangiales. It inhabits upper intertidal zones and suffers periodical desiccation and osmotic stress. The transcriptomic regulation under dehydration and hyposalinity was investigated. The differentially expressed genes (DEGs) accounted for 18.7% of the unigenes obtained by de novo transcriptome assembly (|log2fold-change| ≥ 1, FDR ≤ 0.001). Over 72% of the DEGs were downregulated under stress. The DEGs were predominantly enriched into the KEGG pathways “metabolic pathways,” “ribosome,” “biosynthesis of secondary metabolites,” “protein processing in endoplasmic reticulum,” and “oxidative phosphorylation.” The optimum photosynthetic efficiency (Fv/Fm) and photochemical quenching (qP) dropped significantly with 89% relative water loss and recovered rapidly after being rehydrated. Most DEGs regarding “photosynthesis” and “C3 carbon fixation” were upregulated in the dehydrated thalli, which may enable the thalli to gain photosynthetic recovery once being rehydrated. Fv/Fm and qP decreased significantly with 1 h of 90% freshwater treatment and then recovered to the control level 1 day later. With 6 h hyposaline treatment, expression of plasma membrane H+-ATPase genes was strongly and predominantly induced while the mRNA abundance of vacuolar, chloroplastic, and mitochondrial H+-ATPase genes decreased or showed no significant change. Some transporter, ion channel, and transmembrane protein genes together with the gene-encoding key enzymes involving in proline and heteroside metabolism were upregulated under hyposalinity. The results indicated that transmembrane exchange of ion and osmolytes was induced under hyposalinity to balance the osmotic fluctuation, which seemed to be triggered by plasma membrane H+-ATPases. These findings will facilitate elucidating the stress acclimation mechanism of B. fuscopurpurea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bangia Lyngbye is a group of widespread red algae in the ancient family Bangiales (Sutherland et al. 2011). The Bangia-like fossil Bangiomorpha pubescens is the oldest taxonomically resolved eukaryotic fossil and seems to represent the oldest known case of sexual reproduction (Butterfield 2000, 2009). Bangia species have been found both in seawater (SW) and freshwater (FW) habitats (Sheath and Cole 1980). Bangia fuscopurpurea is a marine species, distributed along marine shores worldwide (Broom et al. 2004; Wang et al. 2008). It is superior to Pyropia yezoensis (nori) in terms of nutrient value and taste (Li et al. 2003) and has been farmed in China since the 1990s (Wang et al. 2008). Bangia fuscopurpurea inhabits the high intertidal zones and experiences extreme fluctuations in environmental conditions such as desiccation and salinity.
Marine seaweeds growing in intertidal zones are the species most severely challenged by their environment (Lin et al. 2009). Anti-dehydration capacities of intertidal and subtidal seaweeds are coordinated with their vertical distribution and patterns of submersion and emersion (Davison and Pearson 1996). Resistance or adaptation to water deficiency differs among Porphyra/Pyropia species (Smith et al. 1986). Blades of high intertidal Porphyra/Pyropia species can survive 85–95% intercellular water loss during daytime low tide and become crisp sheets against the rocks (Chen et al. 2007; Blouin et al. 2011). The high tolerance of Pyropia allows their survival after routine net desiccation during farming (Chen et al. 2007). Pyropia katadae var. hemiphylla, inhabiting the intertidal marshes, can endure moderate intercellular water loss and is vulnerable to extreme desiccation (Wang et al. 2016). Given its habitat, it is reasonable to speculate that B. fuscopurpurea is resistant to desiccation and the farming practice validates this speculation (Wang et al. 2008). However, the molecular mechanism of desiccation acclimation is poorly understood.
There are ~ 130 species in Bangiales. Except for Bangia atropurpurea, all other Bangiales species live in SW (Sutherland et al. 2011). Although marine Bangia species are phylogenetically clearly separated from B. atropurpurea (Sutherland et al. 2011), they can adapt to 100% freshwater step-by-step in culture (Sheath and Cole 1980). However, little is known about the mechanism for marine Bangia species acclimating to the drastic salinity changes.
The advent of high-throughput RNA-seq technology makes it possible to determine the gene expression profile at transcriptomic level, facilitating the identification of stress-responsive genes in large scale. Transcriptomic reprogramming under abiotic stress has been confirmed in seaweeds such as Chondrus crispus (Collén et al. 2007), Ectocarpus siliculosus (Dittami et al. 2009, 2012), Saccharina latissima (Heinrich et al. 2012), Klebsormidium (Holzinger et al. 2014), and Pyropia sp. (Choi et al. 2013; Im et al. 2015; Sun et al. 2015). To understand the physiological and molecular acclimation mechanism of B. fuscopurpurea under dehydration and hyposalinity, we first determined the photosynthetic response to the stress by PSII chlorophyll fluorescence analysis and then identified the stress-responsive genes by transcriptomic analyses. We hope that the present study would facilitate elucidating the mechanisms of B. fuscopurpurea against dehydration and hyposalinity.
Materials and methods
Sample collection
Gametophytic thalli of Bangia fuscopurpurea were collected from Putian China (24° 59′ N, 118° 48′ E) in mid-November 2013. The thalli were cultured under 12–13 °C, 30 ± 5 μmol photons m−2 s−1 with a photoperiod of 10 h:14 h L/D. Medium was filtered sterilized natural SW enriched with 3 mg L−1 NaNO3–N and 0.3 mg L−1 KH2PO4–P and half-renewed every day. Three days later, the thalli were treated with stress.
Stress application
The relative intracellular water loss (RWL; %) of the thalli was calculated as: (W0 − Wt)/W0 × 100, where W0 is the weight of fresh thalli without dehydration and Wt is the weight of the dehydrated thalli. Thalli were air-dried at 12–13 °C for 6 h with the RWL being 89 ± 1.7% and then were subjected to chlorophyll fluorescence measurement. The thalli with 89% RWL were emerged into natural filtered sterilized SW for recovery and the chlorophyll fluorescence measurement was made after 15, 45, and 90 min, respectively.
For hyposalinity treatment, fresh thalli were cultured in medium of filtered sterilized natural SW (salinity 28.5) diluted with 90% sterilized FW. All media were fertilized with 3 mg L−1 NaNO3–N and 0.3 mg L−1 KH2PO4–P. Temperature was 12–13 °C, irradiance was 30 ± 5 μmol photons m−2 s−1, and photoperiod was 10 h:14 h L/D. The chlorophyll fluorescence measurement was made after 1 h, 3 h, 6 h, and 1 day, respectively. Each treatment was performed in triplicate.
Chlorophyll fluorescence measurement
Chlorophyll fluorescence of the thalli was measured using a diving pulse amplitude modulation fluorometer (Diving-PAM, Walz, Germany). The optimum photosynthetic efficiency (Fv/Fm), the photochemical quenching (qP), and the non-photochemical quenching (NPQ) of the samples were recorded after 15 min dark adaptation (Lin et al. 2009). Data were analyzed by ANOVA at a level of p < 0.05 and followed by Tukey’s post hoc test where appropriate. All data are reported as means ± standard deviation (SD) (n = 9).
RNA extraction
Three samples were obtained: fresh thalli without stress treatment (control, salinity 28.5, BC), thalli with 6 h air-drying (approximately 89% RWL, BD), and thalli cultured in 90% FW for 6 h (salinity 3.5, BF). Application of stress was carried out at 12–13 °C and 30 ± 5 μmol photons m−2 s−1. The total RNAs were extracted from each sample and pooled samples, separately, using Trizol protocol (Invitrogen). In sum, ten RNA samples were obtained: one RNA sample from the mixed thalli of equal quantity of BC, BD, and BF, which was used for deep RNA-seq and de novo transcriptome assembly, and three repeats of RNA sample from the BC, BD, and BF, respectively, which were used for global gene expression analysis. The quality and quantity of the RNA samples were assessed using a Nanodrop 2000 spectrophotometer (Thermo, USA) and Agilent 2100 bioanalyzer (Agilent, USA). The RNA samples with OD260/280 1.8–2.2, 28S:18S ≥ 1, and RIN ≥ 8 were used for library construction.
RNA-seq and de novo assembly
cDNA libraries were generated following the manufacturer’s instructions (Illumina). The library products were sequenced via Illumina HiSeq 2000 (Illumina). The sequences of low quality were filtered from the dataset by (a) removing adapter contamination; (b) filtering the reads with ambiguous sequences; (c) discarding the reads with over 4% bases of sequencing quality value (sQ) < 20 (sQ = − 10 lgE, E—sequencing error). The high-quality reads were saved in fastq files and deposited at the NCBI Short Read Archive (SRA) with the accession number SRX759611. The clean reads from the pooled RNA sample were used for de novo assembly of the transcriptome by using the Trinity protocol (Grabherr et al. 2011). The assembled sequences were optimized through sequence splicing and redundancy removing to acquire non-redundant transcripts as long as possible (Pertea et al. 2003).
Annotation and functional classification
Unigenes were first aligned by Blastx to the protein databases, including Nr (NCBI non-redundant protein database), Swissprot, KEGG (Kyoto Encyclopedia of Genes and Genomes), and COG (Cluster of Orthologous Groups of proteins), and those showing the hits with a significance of e value ≤ 10−5 were assigned the same putative function as the corresponding proteins. The unigenes without Blastx hits in the protein databases were predicted using ESTScan (Iseli et al. 1999). The obtained unigenes were further aligned to the draft genome database of Pyropia yezoensis (http://nrifs.fra.affrc.go.jp/ResearchCenter/5_AG/genomes/nori/) and the expressed sequence tags (ESTs) of Porphyra purpurea and Porphyra umbilicalis (http://dbdata.rutgers.edu/nori/) by Blastp with an e value ≤ 10−5. The unigenes were finally assigned to GO (Gene Ontology) functional classification (Conesa et al. 2005; Ye et al. 2006) and KEGG pathway enrichment (Kanehisa and Goto 2000) with an e value cut-off ≤ 1e−5.
Screening of differentially expressed genes (DEGs)
The RNA samples from BC, BD, and BF were separately assigned to RNA-seq via Illumina HiSeq 2000 to find out the DEGs. After sequencing and quality control, the obtained clean reads were mapped to the B. fuscopurpurea transcriptome dataset using SOAPaligner/soap2 (Li et al. 2009). The gene expression level was calculated by using RPKM (reads per kb per million reads) method (Mortazavi et al. 2008). We used |log2fold-change| ≥ 1 and false discovery rate (FDR) ≤ 0.001 as the threshold to assess the significance of gene expression variations. All the DEGs were compared with the whole transcriptome background to search for genes involved in significantly enriched metabolic or signal transduction pathways and assigned to GO and KEG G classification. Validation of DEGs was performed by real-time quantitative PCR (RT-qPCR) according to Wang et al. (2013).
Results
Change of chlorophyll fluorescence parameters under dehydration and hyposalinity
With 89% RWL, the Fv/Fm dropped sharply, NPQ increased significantly, and strong fluctuations in the qP values were detected. During rehydration, Fv/Fm and qP increased and NPQ decreased to the control level during 45–90 min (Table 1). The Fv/Fm and qP values decreased while NPQ increased significantly after 1 h of hyposaline treatment (p < 0.05). The Fv/Fm and qP values increased and NPQs decreased to the control level with 6–24 h treatment (Table 1).
Deep sequencing and de novo assembly of transcriptome
A total of 77.6 M paired-end qualified clean reads with a total length of 7.84 G bp were obtained. The GC percent of the clean reads was 64.25%. The clean reads were assembled into 35,421 unigenes with mean length of 537.5 bp and N50 length of 673 bp, ranging from 201 bp to 14,567 bp (Fig. 1). The mean GC content of the unigenes was 66.24%, a little higher than that of the assembled Pyropia transcriptome and genome (Yang et al. 2011; Nakamura et al. 2013; Xie et al. 2013; Im et al. 2015; Sun et al. 2015; Brawley et al. 2017).
Transcriptome annotation
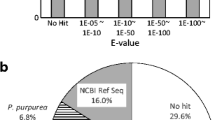
There were 14,534 unigenes with homologous protein sequences in at least one of the Nr, Swissprot, COG, and KEGG databases (Fig. 2). A total of 9186 unigenes had both hits in Nr and Swissprot database with e value ≤ 1e−10 (Table 2). Among the unigenes without hits in the protein databases, 8399 unigenes were predicted to be possible novel or highly diverged transcripts in B. fuscopurpurea based on ESTScan analysis.
The unigenes were assigned to GO classification and got 3419 returns, which were sorted into 57 functional groups and 1993 GO terms. In each of the three main GO categories (biological process, cellular component, and molecular function), “metabolic process,” “cell,” and “metabolic process” predominated, respectively (Fig. 3). Searching against COG database, 8508 unigenes were classified into 24 functional categories. The top largest three categories were “translation, ribosomal, and biogenesis”; “general function prediction only”; and “posttranslational modification, protein turnover, and chaperones” (Fig. 4). By KEGG pathway enrichment, 7619 unigenes were mapped to 117 pathways that were sorted into 20 groups, including the primary metabolism, energy metabolism, and regulatory systems (Supplementary Table S1). Except phosphoketolase (EC 4.1.2.9) in C3-pathway and malate dehydrogenase (NADP+) (EC 1.1.1.82) and pyruvate orthophosphate dikinase (PPDK, EC 2.7.9.1) in C4-pathway, all the other genes involved in eukaryotic C3 and C4 photosynthetic carbon fixation were identified (Supplementary Table S2). Phosphoketolase and PPDK were not identified in P. yezoensis (Yang et al. 2011) and Porphyra haitanensis (Xie et al. 2013) either. Malate dehydrogenase was present in P. yezoensis (Yang et al. 2011) while absent in P. haitanensis (Xie et al. 2013).
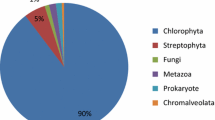
About 34.57% of the Blastx hits in Nr belonged to three red algae Chondrus crispus, Galdieria sulphuraria, and Cyanidioschyzon merolae whose complete genome has been published (Fig. 5). Only hundreds of unigenes were identified to be homologous to the putative proteins of Pyropia species by Blastx search against the four public databases. Some of the recent research regarding Pyropia genome or transcriptome has been deposited in local databases other than Nr and Swissprot. In order to compare B. fuscopurpurea and Pyropia species, the obtained unigenes were aligned to the draft genome database of P. yezoensis (http://nrifs.fra.affrc.go.jp/ResearchCenter/5_AG/genomes/ nori/) and the transcriptome database of P. purpurea and P. umbilicalis (http://dbdata.rutgers.edu/nori/). A total of 5338 B. fuscopurpurea unigenes matched to the sequences of three Pyropia species (Supplementary Table S3). Approximately, 90% of the matches had an e value ≤ 1e−50.
Determination of stress application
Experiments carried out on land plants (Seki et al. 2002), red alga (Collén et al. 2007), and brown alga (Dittami et al. 2009, 2012) indicate that application of stressors for 6 h induces marked transcriptional changes. Six hours of air-drying resulted in ca. 90% RWL in B. fuscopurpurea. There were significant changes in the PAM parameters of the thalli with 89% RWL, which fully recovered after rehydration (Table 1). The PAM parameters of the thalli cultured under 90% FW were affected in short period of treatment (< 6 h) while acclimated after 1 day of treatment (Table 1). Therefore, air-drying for 6 h (90% RWL) and 6 h treatment of 90% freshwater (salinity 3.5) were applied in this study.
Differential gene expression under dehydration and hyposalinity
Over 1.5 billion clean reads were obtained for each sample, which were mapped to at least 26,420 unigenes from B. fuscopurpurea transcriptome dataset (Supplementary Table S4). Bangia fuscopurpurea gametophytes are small in size and the sample used for RNA-seq is composed of a large population. In order to eliminate artificial deviation and to get a more general view of B. fuscopupurea transcriptomic response to stress, DEGs were screened based on three replicates (Fig. 6).
On the whole, 6623 DEGs above the threshold |log2fold-change| ≥ 1 and FDR ≤ 0.001 were identified under dehydration and hyposalinity, accounting for 18.7% of the transcriptome profile. Most DEGs were significantly downregulated under either stress (Fig. 7, Table 3). A larger number of DEGs was identified in BF than in BD (Table 3). Nevertheless, more drastic variations occurred in BD than in BF. The DEGs can be grouped into eight clusters based on their regulation pattern among samples (Table 3).
Annotation and classification of DEGs
A total of 3854 DEGs had significant Blastx hits in Nr, Swissprot, COG, and/or KEGG protein databases (e value < 1e−5). Under both stress, DEGs from the GO items “catalytic activity” and “regulation of molecular function” were upregulated while those from “developmental process,” “reproduction,” “reproductive process,” “multicellular organismal process,” “growth,” “negative regulation of biological process,” and “immune system process” were downregulated (Fig. 8). Approximately, 20% DEGs were mapped to 109 KEGG pathways (e value < 1e−5). The top significantly enriched pathways were in accordance with the KEGG enrichment of the transcriptome profile (Supplementary Table S1). The KEGG items with top minimum Q value indicated that these KO items were predominantly affected by dehydration or hyposalinity. The difference in the distribution of the KEGG items indicated that different response and acclimation mechanism occurred between dehydration and hyposalinity (Table 4).
The DEGs that play vital roles in stress signaling in plants are summarized in Fig. 9. Except for the respiratory burst oxidase genes, genes involved in “plant circadian rhythm,” and photosynthesis-related genes, it was found that the DEGs regarding the other categories were mostly downregulated under both stresses. Especially, all responsive hydrolase ubiquitin-mediated proteolysis genes were downregulated. However, the DEGs involved in “circadian rhythm-plant,” “phosynthesis-antenna protein,” and “photosynthesis” were mostly upregulated in BD and DEGs regarding “chloroplastic components” and “porphyrin and chlorophyll metabolism” were mostly upregulated in BF (Fig. 9).
Validation by RT-qPCR
Twelve DEGs in response to stress were analyzed by RT-qPCR. Although the fold-changes of the DEGs varied between RNA-seq and RT-qPCR, the expression patterns of 12 genes were similar under the same conditions, with the Pearson correlation coefficients ranging from 0.71–0.99 (Table 5). It was found that expression of four plasma membrane-type H+-ATPase genes was induced in dozens to hundreds of folds under hyposalinity while showing no significant difference under dehydration by RT-qPCR, which were among the DEGs with top ten highest transcriptional abundance in BF.
Discussion
Transcriptome characteristics of B. fuscopurpurea
There are several reports about the global transcriptomic characteristics of Pyropia sp. (Yang et al. 2011; Chan et al. 2012; Choi et al. 2013; Xie et al. 2013; Im et al. 2015; Sun et al. 2015). A large set of Porphyra/Pyropia ESTs have been deposited in public databases that facilitate investigation of these algae. However, mere 168 Bangia sp. ESTs deposited in NCBI GenBank and 92.26% are redundant sequences encoding two proteins. Additional 1252 ESTs have been recently deposited in the NCBI EST database but lack functional annotation. The present study identified 35,421 ESTs of B. fuscopurpurea and 41% of them were homologous to the published putative proteins. Over one third of the Blastx hits in the Nr database belonged to the red algae Chondrus crispus, Galdieria sulphuraria, and Cyanidioschyzon merolae (Fig. 5). Nakamura et al. (2013) have reported the draft genome of P. yezoensis that shows a low genome similarity with C. merolae. Making a comparison between B. fuscopurpurea and Porphyra/Pyropia, only hundreds of unigenes returned above cut-off Blastx results (e value < 10−5). We further compared B. fuscropurpurea unigenes to the dataset of P. yezoensis draft genome and P. purpurea and P. umbilicalis transcriptomes, which has been deposited in local databases. A total of 15% unigenes of B. fuscropurpurea matched to sequences of three Pyropia species. The findings are consistent with that these red algae are divergent from each other (Yoon et al. 2004). The large set of ESTs deciphered here would facilitate further investigation of B. fuscropurpurea.
Global transcriptomic reprogramming under abiotic stress
Approximate 18.7% of the unigenes were significantly affected by 90% RWL and salinity 3.5 based on the screening threshold |log2ratio| ≥ 1 and FDR ≤ 0.001. Statistical method significantly affects the number of DEGs, such as FDR (see Fig. 3 in Dittami et al. 2009). Using the present threshold, DEGs identified in stress-applied E. siliculosus was < 1% (Dittami et al. 2009, 2012). Four to 5% of C. crispus genes were significantly regulated by osmotic stresses (Collén et al. 2007). The Arabidopsis transcriptome varied from 1 to 30% under drought, osmotic, or cold stress (Kreps et al. 2002; Seki et al. 2002). The global transcriptomic reprogramming identified here helped to reveal the endurance mechanism of B. fuscopurpurea to dehydration and hyposalinity.
The number of downregulated genes was 2.6–3.2-fold of that of the upregulated genes under dehydration and hyposalinity (Fig. 7; Table 3). The pathways regarding translation, transcription, metabolism of carbohydrate and lipid, reproduction, and growth were all downregulated under both stresses. Transcription of ribosomal proteins (RPs) has shown to be one of the least affected by abiotic stress in Ectocarpus siliculosus (Dittami et al. 2009). It seems very different in Bangiales. The number of the RP DEGs accounted for 25% of the total RP unigenes identified in B. fuscopurpurea and ranked the second highest among all the KEGG classification. Most RP DEGs were downregulated under both stress while the chloroplastic RP genes were all upregulated, indicating that photosynthetic regulation may play vital roles against dehydration and hyposalinity.
Protein kinases (PKs) and transcription factors (TFs) are classical stress signaling components in plants and shown to be affected at the transcriptional level by abiotic stress (Xu et al. 2013). Here, about 25% PK unigenes and 15% TFs unigenes were significantly differentially expressed in BD and/or BF, similar to that reported in higher plants (Seki et al. 2002; Xu et al. 2013). Heat shock proteins (HSPs) play vital roles in protecting plants against stress by stabilizing proteins and membranes and their transcription is induced under abiotic stress in many organisms (Dittami et al. 2009; Sun et al. 2015). Nearly 24% HSP/chaperon unigenes were affected by dehydration and/or hyposalinity, which is higher than the findings in stress-applied C. crispus (Collén et al. 2007), E. siliculosus (Dittami et al. 2009), and P. yezoensis (Sun et al. 2015). Most HSP DEGs were downregulated, similar to the hyposaline response of E. siliculosus (Dittami et al. 2009). Plant hormones are involved in response to various stresses. Seventeen transcripts were identified in the “plant hormone signal” pathway and five of them coding for PP2C, SnRK2, and PR-1 were downregulated under both stress, different from the stress response in some plants (Dinakar et al. 2012; Xu et al. 2013). Three ATP-dependent Clp protease adapter protein genes were upregulated in BF, which is hypothesized to mediate an abscisic acid-independent signaling pathway (Contreras-Porcia et al. 2013). Cytochrome P450s are another stress signaling protein family that are induced by stress and catalyze oxygenation of many substrates (Narusaka et al. 2004). Four DEGs of cytochrome P450s were downregulated in BD or BF and only one cytochrome P450 gene was upregulated in BF (log2fold-change 1.103). Sixty unigenes of protease and proteasome and 45 ubiquitin-related unigenes changed significantly under dehydration and hyposalinity. Only two E3 ubiquitin-protein ligase genes were upregulated in BD. Cyclophilin has been shown to be involved in stress signaling in many plants and algae, including P. haitanensis (Jia et al. 2013). Seven DEG-encoding cyclophilins were identified in BD or BF and only one were upregulated under hyposalinity (Fig. 9). Downregulation of these functional genes at transcriptional level indicated that the corresponding proteins may be downregulated, mediating downregulation of the most metabolic pathways under dehydration and hyposalinity and protecting the organism against stress.
Response of reactive oxygen species (ROS)-redox system
ROS usually accumulates under extreme environmental conditions. ROS-redox balance is essential for plants to maintain normal energy metabolism, control signaling pathways, and activate acclimation responses (Suzuki et al., 2012). Carotenoids are supposed to act mainly against potential ROS-induced damage to photosynthetic apparatus in phycobilisome-containing species (Sampath-Wiley et al. 2008). Several DEGs regarding carotenoid biosynthesis were identified while only one gene-encoding zeaxanthin epoxides was upregulated in BD and a gene-encoding (+)-abscisic acid 8′-hydroxylase was upregulated in BF. There was a DEG identified in “flavonoid biosynthesis” pathway that was upregulated in BF. Dozens of transcripts regarding the ascorbate-glutathione cycle were affected under dehydration and six of them was upregulated in BD or BF. Thirty DEGs of antioxidant enzymes including superoxide dismutase, peroxidise, catalase, and thioredoxin reductase were identified in B. fuscopurpurea and six were upregulated under either stress (Fig. 9). The result that the genes regarding the classic ROS scavenging components were not predominantly induced in dehydration- or hyposalinity-applied B. fuscopurpurea was in accordance with the findings in Chondrus crispus (Collén et al. 2007) and E. siliculosus (Dittami et al. 2009). In flowering plants, these proteins are known to carry out important functions against ROS. We further identified 30 DEGs coding for kinds of oxidases and found that only four were upregulated in BD. ROS may not burst in BD and BF-applied B. fuscopurpurea; thus, there was no need for extensive induction of the redox system.
Plasma membrane H+-ATPase and osmolytic regulation under hyposalinity
H+-ATPases (proton pumps) play vital roles in osmotic regulation on many organisms. There are three types of proton pumps based on the localization: vacuolar membrane (V-type) H+-ATPases, chloroplastic or mitochondrial inner membrane (F-type) H+-ATPases, and plasma membrane (P-type) H+-ATPases. Five DEG-encoding P-type proton pumps were strongly induced under hyposalinity while were unaffected under dehydration. These DEGs were among the most significantly upregulated genes (123–8019-fold). By contrast, expression of nine V-type proton pump DEGs was downregulated in BF and seven of them were also downregulated in BD. No F-type proton pump genes were identified to be significantly differentially expressed under both stresses. The results indicated that P-type proton pumps of B. fuscopurpurea gametophytes play special roles in response to hyposalinity. P-type proton pumps generally cooperate with 14-3-3 proteins (Hanstein et al. 2011), which is activated by kinases (Yu et al. 2006), phosphorylase (Fuglsang et al. 2007), and chaperons (Yang et al. 2010), et al. Thirteen unigenes were identified to be homologous to 14-3-3 proteins and none of them showed significant differential transcription in BF. Piette et al. (2011) reported that P-type proton pump functions in other ways without cooperation with 14-3-3 proteins in tobacco. In B. fuscopurpurea, three HSP/chaperon genes, four serine/threonine PK genes, and one uncharacterized aarF domain-containing PK gene were upregulated in BF, which may cooperates with P-type proton pump. The HSP gene (K04079, e value = 3.00e−35) was upregulated over 500-fold but showed no changes in mRNA abundance in BD. The result indicated that this HSP protein is critical in hyposalinity acclimation of B. fuscopurpurea. The activity of P-type proton pumps is reported to be dependent on unsaturated fatty acids (UFAs) in some species (Martz et al. 2006; Janicka-Russak and Kabala 2015). UFA content accounts for over 80% of the total fatty acids in B. fuscopurpurea (Li et al. 2003), providing a favorable intracellular environment for activation of P-type proton pumps. The plant P-type proton pump generates an electrochemical potential difference across the plasma membrane, which is essential for transporting of many ions and metabolites (Janicka-Russak and Kabala 2015). Aquaporins are transmembrane channel proteins involved in the transportation of water and some small molecules. An aquaporin gene was found to be upregulated in P. yezoensis under desiccation, hyposalinity, and high-temperature stress (Kong et al. 2017). Five aquaporin unigenes of B. fuscopurpurea were identified in this study and two of them were specifically upregulated under dehydration while none was significantly up- or downregulated under hyposalinity. Six transporter genes were upregulated in BF, including ABC transporters, band 3 anion transport protein, sodium/sulfate cotransporter, and K+ efflux antiporter. We further found that two chloride channel unigenes, one sodium/potassium/calcium exchanger unigene, a potassium voltage-gated channel unigene, and two two-pore potassium channel unigenes together with two transmembrane protein unigenes were upregulated in BF. Almost all these unigenes were specially upregulated in BF and unaffected in BD. Upregulation of these transporter and channel genes suggested an induction of transmembrane exchange under hyposalinity. Proline is a compatible osmolyte in higher plants and algae (Dittami et al. 2009). A proline synthetase gene was upregulated in BF. In addition, a nitrate reductase gene and a DEG coding for glutamate-cysteine ligase and a DEG-encoding asparagine synthase were upregulated in BF. Except amino acids, heterosides play an important role in osmotic adjustment in this species. It has been shown that floridoside increases with increasing osmotic pressure in B. atropurpurea (Sheath and Cole 1980). Here, two DEGs coding for glycosyl hydrolase and glycoside hydrolase were upregulated in BF. These results indicated that transmembrane exchange of ion and osmolytes (such as amino acids and heterosides) may be induced to reduce the osmotic pressure during hypoosmotic adaptation by upregulating the expression of P-type proton pumps, transporters, and channels.
Photosynthetic regulation under dehydration
The chlorophyll fluorescence results indicated a high photo-physiological tolerance to dehydration in B. fuscopurpurea gametophytes (Table 1), similar to the other stress-resistant Bangiales (Smith et al. 1986; Chen et al. 2007). It was found that the Fv/Fm and qP values dropped significantly under 90% RWL while 43.6% of the chloroplastic DEGs and 91% DEGs relating to “photosynthesis” were upregulated by dehydration (Fig. 9). The dehydrated thalli were capable of regaining the normal Fv/Fm and qP levels soon after rehydration (Table 1). Thus, we suggest that upregulation of these genes enabled the thalli to recover rapidly once being rehydrated. On the one hand, the excessive electrons generated in dehydrated B. fuscopurpurea thalli were dissipated as heat as indicated by the strong increase in NPQs (Table 1). On the other hand, 90% of the DEGs relating to “porphyrin and chlorophyll metabolism” were downregulated in BD, indicating downregulation of light absorbability. Thus, the ROS stress was diminished, helping to explain the less induction of the classical ROS scavenging genes identified here. Most DEGs encoding the key enzymes involved in C3 pathway were upregulated in BD while the responsive ESTs except phosphoenolpyruvate carboxykinase (PEPCK) in C4 pathway and three DEG-encoding carbonic anhydrase (CA) were downregulated in BD. CA is responsible for dissolved inorganic carbon uptake. During dehydration, free CO2 is predominant. It has been reported that during moderate desiccation, P. yezoensis can utilize CO2 in ambient air (Zhou et al. 2014) and photosynthetic oxygen evolution is enhanced (Gao and Aruga 1987). An oxygen-evolving complex component gene was upregulated upon 9-fold under dehydration. This indicated that B. fuscopurpurea thalli may be capable to take up CO2 directly from the air resulting in the enhanced C3 carbon fixation independent of CAs and C4 pathway CO2 concentrating mechanism during moderate dehydration.
Conclusion
The transcriptome of the primitive red alga B. fuscopurpurea is first reported. A total of 35,421 ESTs were identified and 41% of them were homologous to published putative proteins. About 15% identified ESTs matched sequences of Pyropia/Porphyra species based on currently published transcriptome or genome data, indicating that B. fuscropurpurea is divergent from Pyropia/Porphyra. Approximately, 18.7% of the ESTs were significantly affected by dehydration and hyposalinity based on the threshold |log2ratio| ≥ 1 and FDR ≤ 0.001, with more DEGs present under hyposalinity. About 43.6% of the chloroplastic DEGs and 91% DEGs relating to “photosynthesis” were upregulated by dehydration and expression of plasma membrane H+-ATPase genes was strongly and predominantly induced under hyposaline stress. The results suggest distinct regulatory pathways of B. fuscopurpurea towards desiccation and hyposalinity stress. The large set of ESTs and DEGs deciphered here would facilitate further investigation of this important farmed species.
References
Blouin NA, Brodie JA, Grossman AC, Xu P, Brawley SH (2011) Porphyra: a marine crop shaped by stress. Trends Plant Sci 16:29–37
Brawley SH, Blouin NA, Fickoblean E, Wheeler GL, Lohr M, Goodson HV, et al. (2017) Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc Natl Acad Sci 114(31):E6361–E6370
Broom JES, Farr TJ, Nelson WA (2004) Phylogeny of the Bangia flora of New Zealand suggests a southern origin for Porphyra and Bangia (Bangiales, Rhodophyta). Mol Phylogenet Evol 31:1197–1207
Butterfield NI (2000) Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 26:86–404
Butterfield NJ (2009) Modes of pre-Ediacaran multicellularity. Precamb Res 173:201–211
Chan CX, Blouin NA, Zhuang Y, Zäuner S, Prochnik SE, Lindquist E, Lin S, Benning C, Lohr M, Yarish C, Gantt E, Grossman AR, Lu S, Müller K, W. Stiller J, Brawley SH, Bhattacharya D (2012) Porphyra (Bangiophyceae) transcriptomes provide insights into red algal development and metabolism. J Phycol 48:1328–1342
Chen CS, Wang L, Ji DH, Xie CT, Xu Y (2007) Influence of desiccation and cold preservation on the survival and growth of Porphyra haitanensis and unwanted alga. Acta Oceanol Sinica 29(2):131–136
Choi S, Hwang MS, Im S, Kim N, Jeong WJ, Park EJ, Gong YG, Choi DW (2013) Transcriptome sequence and comparative analysis of the gametophyte thalli of Pyropia tenera under normal and high temperature conditions. J Appl Phycol 25:1237–1246
Collén J, Guisle-Marsollier I, Léger JJ, Boyen C (2007) Response of the transcriptome of the intertidal red seaweed Chondrus crispus to controlled and natural stresses. New Phytol 176:45–55
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Contreras-Porcia L, López-Cristoffanini C, Lovazzano C, Flores-Molina MR, Thomas D, Núñez A, Fierro C, Guajardo E, Correa JA, Kube M, Reinhardt R (2013) Differential gene expression in Pyropia columbina (Bangiales, Rhodophyta) under natural hydration and desiccation conditions. Lat Am J Aquat Res 41:933–958
Davison IR, Pearson GA (1996) Stress tolerance in intertidal seaweeds. J Phycol 32:197–211
Dinakar C, Djilianov D, Bartels D (2012) Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci 182:29–41
Dittami SM, Gravot A, Goulitquer S, Rousvoal S, Peters AF, Bouchereau A, Boyen C, Tonon T (2012) Towards deciphering dynamic changes and evolutionary mechanisms involved in the adaptation to low salinities in Ectocarpus (brown algae). Plant J 71:366–377
Dittami SM, Scornet D, Petit JL, Ségurens B, da Silva C, Corre E, Dondrup M, Glatting KH, König R, Sterck L, Rouzé P, van de Peer Y, Cock JM, Boyen C, Tonon T (2009) Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol 10:R66
Fuglsang AT, Guo Y, Cuin TA, Qiu QS, Song CP, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19:1617–1634
Gao K, Aruga Y (1987) Preliminary studies on the photosynthesis and respiration of Porphyra yezoensis under emersed conditions. J Tokyo Univ Fish 47:51–65
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotech 29:644–652
Hanstein S, Wang X, Qian X, Friedhoff P, Fatima A, Shan Y, Feng K, Schubert S (2011) Changes in cytosolic Mg2+ levels can regulate the activity of the plasma membrane H+-ATPase in maize. Biochem J 435:93–101
Heinrich S, Valentin K, Frichenhaus S, John U, Wiencke C (2012) Transcriptomic analysis of acclimation to temperature and light stress in Saccharina latissima (Phaeophyceae). PLoS One 7(8):e44342
Holzinger A, Kaplan F, Blaas K, Zechmann B, Komsic-Buchmann K, Becker B (2014) Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. PLoS One 9(10):e110630
Im S, Choi S, Hwang MS, Park E, Jeong W, Choi D (2015) De novo assembly of transcriptome from the gametophyte of the marine red algae Pyropia seriata and identification of abiotic stress response genes. J Appl Phycol 27:1343–1353
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 1:138–148
Janicka-Russak M, Kabala K (2015) The role of plasma membrane H+-ATPase in salinity stress of plants. Progr Bot 76:77–92
Jia Z, Niu J, Huan L, Wu X, Wang G, Hou Z (2013) Cyclophilin participates in responding to stress situations in Porphyra haitanensis (Bangiales, Rhodophyta). J Phycol 49:194–201
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30
Kong F, Yang J, Li N, Zhao H, Mao Y (2017) Identification and characterization of PyAQPs from Pyropia yezoensis, which are involved in tolerance to abiotic stress. J Appl Phycol 29:1695–1706
Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Li R, Yu C, Li Y, Lam T, Yiu S, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Li SJ, Ma JH, Ji HH, Li EY (2003) Evaluation of nutrient components of Bangia sp. Acta Oceanol Sinica 22:89–95
Lin AP, Wang GC, Yang F, Pan GH (2009) Photosynthetic parameters of sexually different parts of Porphyra katadai var. hemiphylla (Bangiales, Rhodophyta) during dehydration and rehydration. Planta 229:803–810
Martz F, Sutinen ML, Kiviniemi S, Palta JP (2006) Changes in freezing tolerance, plasma membrane H+-ATPase activity and fatty acid composition in Pinus resinosa needles during cold acclimation and de-acclimation. Tree Physiol 26:783–790
Mortazavi A, Williams BA, McCue K, SchaeVer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Meth 5:621–628
Nakamura Y, Sasaki N, Kobayashi M, Ojima N, Yasuike M, Shigenobu Y, Satomi M, Fukuma Y, Shiwaku K, Tsujimoto A, Kobayashi T, Nakayama I, Ito F, Nakajima K, Sano M, Wada T, Kuhara S, Inouye K, Gojobori T, Ikeo K (2013) The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS One 8:e57122
Narusaka Y, Narusaka M, Seki M, Umezawa T, Ishida J, Nakajima M, Enju A, Shinozaki K (2004) Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol 55:327–342
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652
Piette AS, Derua R, Waelkens E, Boutry M, Duby G (2011) A phosphorylation in the C-terminal auto-inhibitory domain of the plant plasma membrane H+-ATPase activates the enzyme with no requirement for regulatory 14-3-3 proteins. J Biol Chem 286:18474–18482
Sampath-Wiley P, Neefus CD, Jahnke LS (2008) Seasonal effects of sun exposure and emersion on intertidal seaweed physiology: fluctuations in antioxidant contents, photosynthetic pigments and photosynthetic efficiency in the red alga Porphyra umbilicalis Kützing (Rhodophyta, Bangiales). J Exp Mar Biol Ecol 361:83–91
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Sheath RG, Cole KM (1980) Distribution and salinity adaptations of Bangia atropurpurea (Rhodophyta), a putative migrant into the Laurentian Great Lakes. J Phycol 16:412–420
Smith CM, Satoh K, Fork DC (1986) The effects of osmotic tissue dehydration and air drying on morphology and energy transfer in two species of Porphyra. Plant Physiol 80:843–847
Sun P, Mao Y, Li G, Cao M, Kong F, Wang L, Bi G (2015) Comparative transcriptome profiling of Pyropia yezoensis (Ueda) M.S.Hwang & H.G. Choi in response to temperature stresses. BMC Genomics 16:463
Sutherland JE, Lindstrom SC, Nelson WA, Brodie J, Lynch MDJ, Hwang MS, Choi HG, Miyata M, Kikuchi N, Oliveira MC, Farr T, Neefus C, Mols-Mortensen A, Milstein D, Müller KM (2011) A new look at an ancient order: generic revision of the Bangiales (Rhodophyta). J Phycol 47:1131–1151
Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signaling in the response of plants to abiotic stress. Plant Cell Environ 35:259–270
Wang WJ, Sun XT, Liu FL, Liang ZR, Zhang JH, Wang FJ (2016) Effect of abiotic stress on the gameophyte of Pyropia katadae var. hemiphylla (Bangiales, Rhodophyta). J Appl Phycol 28:469–479
Wang WJ, Wang FJ, Sun XT, Liu FL, Liang ZR (2013) Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae). Planta 237:1123–1133
Wang WJ, Zhu JY, Xu P, Xu JR, Lin XZ, Huang CK, Song W, Peng G, Wang GC (2008) Characterization of the life history of Bangia fuscopurpurea (Bangiaceae, Rhodophyta) in connection with its artificial cultivation in China. Aquaculture 278:101–109
Xie C, Li B, Ji D, Chen C (2013) Characterization of the global transcriptome for Pyropia haitanensis (Bangiales, Rhodophyta) and development of cSSR markers. BMC Genomics 14:107
Xu Y, Gao S, Yang Y, Huang M, Cheng L, Wei Q, Fei Z, Hong B (2013) Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genomics 14:662
Yang Y, Qin Y, Xie C, Zhao F, Zhao J, Liu D, Chen S, Fuglsang AT, Palmgren MG, Schumaker KS, Deng XW, Guo Y (2010) The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22:1313–1332
Yang H, Mao YX, Kong FN, Yang GP, Ma F, Wang L (2011) Profiling of the transcriptome of Porphyra yezoensis with Solexa sequencing technology. Chin Sci Bull 20:2119–2130
Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34(Web Server issue):W293–W297
Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21:809–818
Yu X, Li M, Gao G, Feng HZ, Geng XQ, Peng CC, Zhu SY, Wang XJ, Shen YY, Zhang DP (2006) Abscisic acid stimulates a calcium-dependent protein kinase in grape berry. Plant Physiol 140:558–579
Zhou W, He L, Yang F, Lin A, Zhang B, Niu J, Wang G (2014) Pyropia yezoensis can utilize CO2 in the air during moderate dehydration. Chin J Oceanol Limnol 32:358–364
Acknowledgements
We are grateful to the anonymous reviewers for their constructive comments and suggestions on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31672630), Primary Research & Development Plan of Shandong Province (2016GSF115038), Special Scientific Research Funds for Central Non-profit Institutes, Chinese Academy of Fishery Sciences (2015A02XK01), National Science and Technology Infrastructure Project (2012), and the Open Funds of Seaweed Genetics and Germplasm Key Laboratory, Changshu Institute of Technology (2014-2016).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, W., Shen, Z., Sun, X. et al. De novo transcriptomics analysis revealed a global reprogramming towards dehydration and hyposalinity in Bangia fuscopurpurea gametophytes (Rhodophyta). J Appl Phycol 31, 637–651 (2019). https://doi.org/10.1007/s10811-018-1501-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-018-1501-7