Abstract
Numerous studies have evaluated the association between −159C/T polymorphism of the CD14 gene and asthma risk; however, these studies have yielded inconsistent results. We performed meta-analyses to investigate the association between CD14-159C/T polymorphism and asthma risk. Studies were identified from PubMed, Embase, and two Chinese databases. Odds ratios (ORs) with 95 % confidence interval (CI) were used to evaluate the strength of association. Thirty-six studies were collected for meta-analysis, which involved 6954 cases and 7525 controls. In the overall populations, no significant association between the CD14-159C/T polymorphism and asthma risk was found for the dominant (OR = 0.90, 95 % CI = 0.81–1.01, P = 0.08) or other models; stratified analyses indicated that the CD14-159C/T polymorphism was not associated with asthma risk in Caucasians or Asians or adults or children. Among the atopic asthma populations, no significant results were observed in the all-combined or subgroup analyses. This meta-analysis demonstrates that the CD14-159C/T polymorphism may not be a risk factor for asthma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Asthma is one of the most common chronic respiratory diseases worldwide, characterized by airway inflammation, airway hyper-responsiveness, variable airflow obstruction, and allergy [1]. Although environmental exposures such as allergens may be risk factors for asthma development, both family- and twin-based studies indicated the importance of genetic components [2]. Therefore, the etiology of asthma is considered to be interactions between environmental factors and genetic characteristics of an individual [3]. It is not surprising that numerous studies have investigated the associations between genetic polymorphisms and susceptibility to asthma, and among them, the cluster of differentiation 14 (CD14) gene has been widely studied [3, 4].
The gene-encoding CD14 is located on chromosome 5q31, a region that is linked to asthma in genome-wide linkage studies [5]. Two protein forms are encoded by the CD14 gene: (i) a membrane receptor expressed on the surface of monocytes, macrophages, and neutrophils and (ii) a soluble form in serum [6]. CD14 is the principal receptor for bacterial lipopolysaccharides (LPSs); upon binding, this complex together with co-receptors toll-like receptor 4 can activate the immune system [3]. The subsequent release of pro-inflammatory cytokines can promote Th1 differentiation of naive T cells resulting in a lower prevalence of allergies such as asthma [3, 7].
Many studies have studied the associations between asthma susceptibility and polymorphisms in CD14 gene, especially the −159C/T polymorphism which was first investigated as a candidate one for atopy [8]. However, inconsistent results were observed. This inconsistency may be attributed to the relatively small sample size of the individual studies. A meta-analysis can provide more reliable evidence by combining the data from different studies. Although there have been three meta-analyses concerning the association between the CD14-159C/T polymorphism and asthma risk [1, 9, 10], further investigations are required for the following reasons. The first meta-analysis only included the earlier studies, and no subgroup analyses were presented [10]. The second one had some significant errors in including studies and abstracting data [9]. The last meta-analysis was performed in 2011, and a number of new studies with more data have been published since 2011 [8, 11–19]. In light of this, we performed an updated meta-analysis to re-investigate the association of the CD14-159C/T polymorphism with asthma susceptibility.
MATERIALS AND METHODS
Identification of Eligible Studies
Relevant studies were identified from PubMed, Embase, and two Chinese databases (Wanfang and China National Knowledge Infrastructure (CNKI)) up to 8 May 2015. The search strategy was as follows: “CD14” and “asthma” or “asthmatic” and “mutation” or “polymorphism” or “variant.” There was no restriction on languages or publication date. Also, the references of relevant reviews or meta-analyses were searched manually to identify additional studies. Studies that fulfilled the following criteria were incorporated into the meta-analysis: (1) evaluating CD14-159C/T polymorphism and asthma risk, (2) using a case-control design, and (3) providing enough data for calculating odds ratios (ORs) and 95 % confidence interval (CI). Exclusive criteria in this meta-analysis were (1) studies containing overlapping data; (2) design based on family or sibling pairs; and (3) some publication types such as comments, abstracts, reviews, and proceedings. For the studies with overlapping data, the one with the largest sample was selected.
Data Extraction
Two reviewers independently extracted the information from each included publication: author, year of publication, country of origin, ethnicity, age group, atopic status, sample size, asthma definition, and genotype distribution among cases and controls. Any discrepancy was resolved via discussing or consulting two additional authors.
Statistical Analysis
ORs and 95 % CI were employed to evaluate the strength of the associations between the CD14-159C/T polymorphism and asthma risk in the dominant (TT + TC vs. CC), recessive (TT vs. TC + CC), codominant (TT vs. CC), and allelic models (T vs. C). The pooled ORs were calculated using a fixed-effect or random-effect model according to between-study heterogeneity, which was checked by Q test and I 2 statistics [20]. If the P value for the Q test was > 0.10, the pooled OR was calculated with the fixed-effect model; otherwise, the random-effect model was used [9]. I 2 values were used to test the between-study heterogeneity quantitatively. I 2 values range between 0 and 100 %, and the values of 25, 50, and 75 % were nominally assigned as low, moderate, and high heterogeneity, respectively [21]. The statistical significance of the pooled ORs was analyzed by the Z test, and a P value of <0.05 was considered statistically significant. Subgroup analyses were conducted by ethnicity, age, and atopic status. For assessing the age—atopic and ethnicity—atopic associations, we further stratified the atopic asthma populations by age and ethnicity.
Hardy–Weinberg equilibrium (HWE) in controls was assessed with the chi-squared test (significance set at P < 0.05). Sensitivity analysis was performed to check the stability of the results using two methods: (i) removing studies that deviated from HWE in controls and (ii) excluding a single study each time if the statistically significant results were found. The potential publication bias was examined by visual inspection of Begg’s funnel plots and Egger’s test. All statistical tests were conducted with Revman 5.1 (The Nordic Cochrane Centre, The Cochrane Collaboration, Copenhagen, Denmark) and STATA 12.0 (StataCorp LP, College Station, TX, USA) software.
RESULTS
Main Characteristics of Included Studies
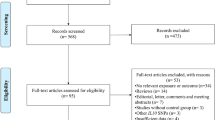
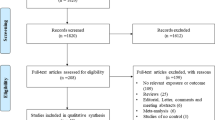
Figure 1 provides a flowchart detailing the procedure for study identification. Finally, 36 case-control studies were included [7, 8, 11–19, 22–46]. The main characteristics of the included studies and genotype distributions of CD14-159C/T polymorphism are presented in Tables 1 and 2, respectively. Except one study without ethnic information [40], 15 studies were performed in Asians, 15 in Caucasians, one in Africans, and four in the populations of mixed ethnicity. Thirteen studies were performed in adults, 16 in children, and seven in undefined or mixed age groups. Eleven studies only included atopic asthma populations; moreover, 18 studies included both non-atopic and atopic asthma populations; however, data could not be extracted separately in nine studies of them. Seven studies did not provide the atopic status of subjects. This information was summarized in Table 1. Based on the same data set, the genotype frequency data for atopic asthma subgroup in one included study [43] were abstracted from another study that had been excluded [47]. The distribution of the CD14-159C/T polymorphism in controls was inconsistent with HWE in three studies [28, 29, 37] (Table 2).
Meta-Analysis of CD14-159C/T Polymorphism and Asthma Risk in the Overall Populations
A total of 6954 cases and 7525 controls in 36 case-control studies were included in meta-analysis. As shown in Fig. 2, the between-study heterogeneity of the dominant model (TT + TC vs. CC) for all 36 studies was tested by Q test and the corresponding P value was 0.007. Thus, a random-effect model was chosen for data synthesis. The pooled OR for all 36 studies was 0.90 (95 % CI = 0.81–1.01, P = 0.08; Fig. 2), indicating no significant association between this polymorphism and asthma risk in the dominant model. Similarly, no statistically significant associations were observed in the recessive, codominant, or allelic model. Summary results of all the genetic comparisons are listed in Table 3.
Meta-analysis of the association between CD14-159C/T polymorphism and asthma risk among the overall populations in the dominant model, using a random-effect model. The squares and horizontal lines correspond to the odds ratios (ORs) and 95 % confidence interval (CI) in each study. The black diamond represents the pooled OR and 95 % CI.
Stratified Analyses by Ethnicity, Age, and Atopic Status in the Overall Populations
For the subgroup analysis by ethnicity, no association was found between CD14-159C/T polymorphism and asthma risk among Caucasians (OR = 0.91, 95 % CI = 0.77–1.06, P = 0.22) or Asians (OR = 0.87, 95 % CI = 0.68–1.10, P = 0.25) in the dominant model; there were no significant results in the other models (Table 3). Stratification by age indicated no significant associations among adults (TT + TC vs. CC OR = 0.95, 95 % CI = 0.77–1.17, P = 0.63) or children (TT + TC vs. CC OR = 0.99, 95 % CI = 0.88–1.11, P = 0.81) in the dominant or other models (Table 3). The subgroup analysis by atopic status revealed no associations of this polymorphism with asthma risk among the atopic asthma patients, non-atopic asthma patients, or mixed atopic asthma patients in any model (Table 3).
Stratified Analyses in the Atopic Asthma Populations
Twenty eligible studies (3642 cases and 3806 controls) performed among the atopic asthma populations were identified for data analysis. Stratified analyses were conducted according to ethnicity and age. No matter which genetic comparison model we used, there were still no significantly statistical results in any subgroup analysis. Summary of the meta-analysis results is listed in Table 3.
Sensitivity Analysis
In the overall and stratified analyses, the conclusions of non-significance were stable in all the genetic models when removing studies that deviated from HWE. We did not sequentially exclude a single study each time because no statistically significant results were observed.
Heterogeneity and Publication Bias
Between-study heterogeneity was observed during most of the meta-analyses in the overall and subgroup populations, which was indicated by I 2 values (Table 3). We used Begg’s funnel plots to graphically estimate the publication bias, and found the relatively symmetric distribution of funnel plots in the dominant (Fig. 3) and other comparison models (figures not shown), which suggested the absence of publication bias. Moreover, the results of Egger’s test also indicated lack of publication bias in the dominant (P Egger = 0.116), recessive (P Egger = 0.774), codominant (P Egger = 0.524), or allelic model (P Egger = 0.274).
DISCUSSION
Although multifactors contribute to asthma susceptibility, genetic predisposition is regarded as one of the important determinants. Many promising candidate genes for asthma have been identified on chromosome 5q31.1 [48], and CD14 gene is such one. Variants in the promoter region of the CD14 gene may modify the structure of the CD14 protein and influence the CD14-LPS interaction [19]. Baldini et al. first reported the −159C/T polymorphism in the promoter region of CD14 gene [49], which was found to be associated with altered levels of soluble CD14 and IgE in various ethnic patients with asthma [15]. The above facts indicate that the CD14-159C/T polymorphism might be involved in the pathogenesis of asthma.
In order to investigate whether the CD14-159C/T polymorphism is associated with asthma risk, we have performed the current meta-analysis with 11 additional studies compared with previous meta-analyses [1, 9, 10]. The overall analysis and stratified analyses by ethnicity and age revealed no associations between this polymorphism and asthma risk. The findings were consistent with those in the previous meta-analyses. Maybe, the influence of CD14-159C/T polymorphism on soluble CD14 and IgE is not powerful enough so that the relationship between this polymorphism and asthma risk could not be detected. Another plausible explanation is that the solitary CD14-159C/T polymorphism merely influences the production of CD14 protein but does not play a key role in the subsequent pathway to asthma development.
When performing the stratified analysis by atopic status, there were still no significant results. However, the previous meta-analyses found a protective effect of the T-allele of −159C/T polymorphism for atopic asthma among the overall populations [1], Asians, and children [9]. Those results of those meta-analyses should be interpreted with caution because of a relatively small number of studies. Compared to previous meta-analyses [1, 9], the study inclusion of our meta-analysis is more comprehensive due to the largest sample size, suggesting more reliable results. Probably, this discrepancy reflects spurious relationship caused by the relatively small sample size. We speculate that the CD14-159C/T polymorphism contribute little to risk of atopic asthma. More genetic association studies concerning the relationship between this polymorphism and atopic asthma are warranted to validate our findings.
Heterogeneity is a major issue that needs to be mentioned when interpreting the meta-analysis results. Although we performed a meticulous literature search, precise data extraction, and strict data analysis, there was low to moderate level heterogeneity in the overall populations (Table 3). After performing subgroup analyses, the heterogeneity was not effectively reduced or removed in most of genetic models, indicating that ethnicity, age, or atopic status might not be the source of heterogeneity. The heterogeneity, together with the stable negative results in sensitivity analysis, probably reflects the unclear association of this polymorphism with asthma risk.
Some limitations of the meta-analysis should be considered. First, although no significant publication bias was detected, some relevant studies published in other databases or unpublished negative studies may have been missed. Thus, potential bias could not be excluded. Second, our ethnic-specific meta-analyses were performed in Asians and Caucasians, so our results are applicable only to these ethnic groups. Our findings should be optimized by including other ethnic populations such as Africans and Latinos. Third, data could not be stratified by disease severity, sex, environmental factors, smoking status, or other variables attributing to insufficient original information in the included studies. Fourth, the lack of corresponding data did not allow us to conduct a meta-analysis of the gene-gene or gene-environment interactions. For these reasons, the interpretation of our meta-analysis results should be taken carefully.
In summary, the current study is the most comprehensive and latest meta-analysis concerning the CD14-159C/T polymorphism and asthma risk to date. Our results indicate that there is no evidence of significant association between the CD14-159C/T polymorphism and asthma risk either in Asians or Caucasians or adults or children. Moreover, the negative results are not influenced by atopic status. Due to the limitations showed above, our results should be viewed with caution and future large-scale studies with different ethnic populations and clinical subphenotypes are required for clarifying the role of the CD14-159C/T polymorphism in the pathogenesis of asthma.
References
Zhao, L., and M.B. Bracken. 2011. Association of CD14–260 (-159) C > T and asthma: a systematic review and meta-analysis. BMC Medical Genetics 12: 93.
Los, H., P.E. Postmus, and D.I. Boomsma. 2001. Asthma genetics and intermediate phenotypes: a review from twin studies. Twin Research 4(2): 81–93.
Lau, M.Y., S.C. Dharmage, J.A. Burgess, A.J. Lowe, C.J. Lodge, B. Campbell, et al. 2014. CD14 polymorphisms, microbial exposure and allergic diseases: a systematic review of gene-environment interactions. Allergy 69(11): 1440–1453.
Klaassen, E.M., B.E. Thonissen, G. van Eys, E. Dompeling, and Q. Jobsis. 2013. A systematic review of CD14 and toll-like receptors in relation to asthma in Caucasian children. Allergy, Asthma and Clinical Immunology 9(1): 10.
Bouzigon, E., P. Forabosco, G.H. Koppelman, W.O. Cookson, M.H. Dizier, D.L. Duffy, et al. 2010. Meta-analysis of 20 genome-wide linkage studies evidenced new regions linked to asthma and atopy. European Journal of Human Genetics 18(6): 700–706.
Buckova, D., L.I. Holla, M. Schuller, V. Znojil, and J. Vacha. 2003. Two CD14 promoter polymorphisms and atopic phenotypes in Czech patients with IgE-mediated allergy. Allergy 58(10): 1023–1026.
Kedda, M.A., F. Lose, D. Duffy, E. Bell, P.J. Thompson, and J. Upham. 2005. The CD14 C-159T polymorphism is not associated with asthma or asthma severity in an Australian adult population. Thorax 60(3): 211–214.
Perin, P., V. Berce, and U. Potocnik. 2011. CD14 gene polymorphism is not associated with asthma but rather with bronchial obstruction and hyperreactivity in Slovenian children with non-atopic asthma. Respiratory Medicine 105(Suppl 1): S54–S59.
Zhang, Y., C. Tian, J. Zhang, X. Li, H. Wan, C. He, et al. 2011. The -159C/T polymorphism in the CD14 gene and the risk of asthma: a meta-analysis. Immunogenetics 63(1): 23–32.
Nishimura, F., M. Shibasaki, K. Ichikawa, T. Arinami, and E. Noguchi. 2006. Failure to find an association between CD14-159C/T polymorphism and asthma: a family-based association test and meta-analysis. Allergology International 55(1): 55–58.
Wang, J., M. Simayi, Q. Wushouer, Y. Xia, Y. He, F. Yan, et al. 2014. Association between polymorphisms in ADAM33, CD14, and TLR4 with asthma in the Uygur population in China. Genetics and Molecular Research 13(2): 4680–4690.
Sahin, F., P. Yildiz, A. Kuskucu, M.A. Kuskucu, N. Karaca, and K. Midilli. 2014. The effect of CD14 and TLR4 gene polymorphisms on asthma phenotypes in adult Turkish asthma patients: a genetic study. BMC Pulmonary Medicine 14: 20.
Hussein, Y.M., S.M. Shalaby, H.E. Zidan, N.A. Sabbah, N.A. Karam, and S.S. Alzahrani. 2013. CD14 tobacco gene-environment interaction in atopic children. Cellular Immunology 285(1-2): 31–37.
Bose, P., R. Bathri, S. De, and K.K. Maudar. 2013. CD14 C-159T polymorphism and its association with chronic lung diseases: A pilot study on isocyanate exposed population of Central India. Indian Journal of Human Genetics 19(2): 188–195.
Yazdani, N., M.M. Amoli, M. Naraghi, A. Mersaghian, F. Firouzi, F. Sayyahpour, et al. 2012. Association between the functional polymorphism C-159T in the CD14 promoter gene and nasal polyposis: potential role in asthma. Journal of Investigational Allergology & Clinical Immunology 22(6): 406–411.
Micheal, S., K. Minhas, M. Ishaque, F. Ahmed, and A. Ahmed. 2011. Promoter polymorphisms of the CD14 gene are associated with atopy in Pakistani adults. Journal of Investigational Allergology & Clinical Immunology 21(5): 394–397.
Wu, S.Q., K.B. Zhang, H.Q. Chen, Z.G. Zhao, G.L. Wang, and C. Chen. 2011. Association between gene polymorphism of CD14-159 (C/T) and allergic asthma. Molecular Medicine Reports 4(6): 1127–1130.
Zaborowski, T., K. Wojas-Krawczyk, P. Krawczyk, O. Jankowska, J. Siwiec, T. Kucharczyk, et al. 2011. The effect of CD14 and TLR4 gene polymorphisms on the occurrence of atopic and non-atopic asthma. Advances in Clinical and Experimental Medicine 20(4): 413–421.
Zhang, Y.N., Y.J. Li, H. Li, H. Zhou, and X.J. Shao. 2015. Association of CD14 C159T polymorphism with atopic asthma susceptibility in children from Southeastern China: a case-control study. Genetics and Molecular Research 14(2): 4311–4317.
Higgins, J.P., and S.G. Thompson. 2002. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine 21(11): 1539–1558.
Higgins, J.P., S.G. Thompson, J.J. Deeks, and D.G. Altman. 2003. Measuring inconsistency in meta-analyses. BMJ 327(7414): 557–560.
Kuo Chou, T.N., Y.S. Li, K.H. Lue, C.F. Liao, C.Y. Lin, P.R. Tzeng, et al. 2010. Genetic polymorphism of manganese superoxide dismutase is associated with childhood asthma. The Journal of Asthma 47(5): 532–538.
Wu, X., Y. Li, Q. Chen, F. Chen, P. Cai, L. Wang, et al. 2010. Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle China. The Journal of Asthma 47(3): 238–244.
Bjornvold, M., M.C. Munthe-Kaas, T. Egeland, G. Joner, K. Dahl-Jorgensen, P.R. Njolstad, et al. 2009. A TLR2 polymorphism is associated with type 1 diabetes and allergic asthma. Genes and Immunity 10(2): 181–187.
Smit, L.A., V. Siroux, E. Bouzigon, M.P. Oryszczyn, M. Lathrop, F. Demenais, et al. 2009. CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. American Journal of Respiratory and Critical Care Medicine 179(5): 363–368.
Wang, J.Y., Y.H. Liou, Y.J. Wu, Y.H. Hsiao, and L.S. Wu. 2009. An association study of 13 SNPs from seven candidate genes with pediatric asthma and a preliminary study for genetic testing by multiple variants in Taiwanese population. Journal of Clinical Immunology 29(2): 205–209.
Kowal, K., A. Bodzenta-Lukaszyk, A. Pampuch, M. Szmitkowski, S. Zukowski, M.B. Donati, et al. 2008. Analysis of -675 4G/5G SERPINE1 and C-159T CD14 polymorphisms in house dust mite-allergic asthma patients. Journal of Investigational Allergology & Clinical Immunology 18(4): 284–292.
de Faria, I.C., E.J. de Faria, A.A. Toro, J.D. Ribeiro, and C.S. Bertuzzo. 2008. Association of TGF-beta1, CD14, IL-4, IL-4R and ADAM33 gene polymorphisms with asthma severity in children and adolescents. The Journal of Pediatrics 84(3): 203–210.
Lachheb, J., I.B. Dhifallah, H. Chelbi, K. Hamzaoui, and A. Hamzaoui. 2008. Toll-like receptors and CD14 genes polymorphisms and susceptibility to asthma in Tunisian children. Tissue Antigens 71(5): 417–425.
Smit, L.A., S.I. Bongers, H.J. Ruven, G.T. Rijkers, I.M. Wouters, D. Heederik, et al. 2007. Atopy and new-onset asthma in young Danish farmers and CD14, TLR2, and TLR4 genetic polymorphisms: a nested case-control study. Clinical and Experimental Allergy 37(11): 1602–1608.
Bernstein, D.I., N. Wang, P. Campo, R. Chakraborty, A. Smith, A. Cartier, et al. 2006. Diisocyanate asthma and gene-environment interactions with IL4RA, CD-14, and IL-13 genes. Annals of Allergy, Asthma & Immunology 97(6): 800–806.
Hong, S.J., H.B. Kim, M.J. Kang, S.Y. Lee, J.H. Kim, B.S. Kim, et al. 2007. TNF-alpha (-308 G/A) and CD14 (-159T/C) polymorphisms in the bronchial responsiveness of Korean children with asthma. The Journal of Allergy and Clinical Immunology 119(2): 398–404.
Zambelli-Weiner, A., E. Ehrlich, M.L. Stockton, A.V. Grant, S. Zhang, P.N. Levett, et al. 2005. Evaluation of the CD14/-260 polymorphism and house dust endotoxin exposure in the Barbados Asthma Genetics Study. The Journal of Allergy and Clinical Immunology 115(6): 1203–1209.
Sharma, M., J. Batra, U. Mabalirajan, S. Goswami, D. Ganguly, B. Lahkar, et al. 2004. Suggestive evidence of association of C-159T functional polymorphism of the CD14 gene with atopic asthma in northern and northwestern Indian populations. Immunogenetics 56(7): 544–547.
Heinzmann, A., H. Dietrich, S.P. Jerkic, T. Kurz, and K.A. Deichmann. 2003. Promoter polymorphisms of the CD14 gene are not associated with bronchial asthma in Caucasian children. European Journal of Immuno Genetics 30(5): 345–348.
Woo, J.G., A. Assa’ad, A.B. Heizer, J.A. Bernstein, and G.K. Hershey. 2003. The -159 C-->T polymorphism of CD14 is associated with nonatopic asthma and food allergy. The Journal of Allergy and Clinical Immunology 112(2): 438–444.
Sengler, C., A. Haider, C. Sommerfeld, S. Lau, M. Baldini, F. Martinez, et al. 2003. Evaluation of the CD14 C-159 T polymorphism in the German Multicenter Allergy Study cohort. Clinical and Experimental Allergy 33(2): 166–169.
Lis, G., E. Kostyk, M. Sanak, and J.J. Pietrzyk. 2001. Molecular studies in a population of children with bronchial asthma. I. Polymorphism in the promotor region of gene CD14. Pneumonologia i Alergologia Polska 69(5-6): 265–272.
Koppelman, G.H., N.E. Reijmerink, Stine O. Colin, T.D. Howard, P.A. Whittaker, D.A. Meyers, et al. 2001. Association of a promoter polymorphism of the CD14 gene and atopy. American Journal of Respiratory and Critical Care Medicine 163(4): 965–969.
Alexis, N., M. Eldridge, W. Reed, P. Bromberg, and D.B. Peden. 2001. CD14-dependent airway neutrophil response to inhaled LPS: role of atopy. The Journal of Allergy and Clinical Immunology 107(1): 31–35.
Murk, W., K. Walsh, L.I. Hsu, L. Zhao, M.B. Bracken, and A.T. Dewan. 2011. Attempted replication of 50 reported asthma risk genes identifies a SNP in RAD50 as associated with childhood atopic asthma. Human Heredity 71(2): 97–105.
Hakonarson, H., U.S. Bjornsdottir, E. Ostermann, T. Arnason, A.E. Adalsteinsdottir, E. Halapi, et al. 2001. Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. American Journal of Respiratory and Critical Care Medicine 164(11): 2036–2044.
Chan, I.H., N.L. Tang, T.F. Leung, W. Huang, Y.Y. Lam, C.Y. Li, et al. 2008. Study of gene-gene interactions for endophenotypic quantitative traits in Chinese asthmatic children. Allergy 63(8): 1031–1039.
Park, S.O., M.Y. Jo, S.S. Jung, and G.S. Ahn. 2006. No association of polymorphisms in RANTES and CD14 genes with asthma in the Korean population. Korean Journal of Genetics 28(1): 75–81.
Cui, T.P., W.C. Jiang, and J.M. Wu. 2003. Genetic polymorphism of CD14 and allergic asthma in children. Central China Medical Journal 27(5): 235–236.
Chen, M., B. Wu, and W. Li. 2009. Influence of CD14 gene -159C/T polymorphism on IL-5 level in patients with asthma. Shang Dong Yi Yao 49(5): 13–15.
Leung, T.F., N.L. Tang, Y.M. Sung, A.M. Li, G.W. Wong, I.H. Chan, et al. 2003. The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatric Allergy and Immunology 14(4): 255–260.
Marsh, D.G., J.D. Neely, D.R. Breazeale, B. Ghosh, L.R. Freidhoff, E. Ehrlich-Kautzky, et al. 1994. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science 264(5162): 1152–1156.
Baldini, M., I.C. Lohman, M. Halonen, R.P. Erickson, P.G. Holt, and F.D. Martinez. 1999. A polymorphism* in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. American Journal of Respiratory Cell and Molecular Biology 20(5): 976–983.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Compliance with Ethical Standard
ᅟ
Conflict of Interest
The authors declare no conflicts of interest.
Additional information
Rui Zhang and Rui Deng contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, R., Deng, R., Li, H. et al. No Association Between −159C/T Polymorphism of the CD14 Gene and Asthma Risk: a Meta-Analysis of 36 Case-Control Studies. Inflammation 39, 457–466 (2016). https://doi.org/10.1007/s10753-015-0269-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-015-0269-z