Abstract
Several studies have previously assessed the association between interleukin (IL)-10 gene polymorphisms and the risk of asthma, leading to conflicting results. To resolve the incongruent outcomes yielded from different single studies, we conducted the most up-to-date meta-analysis of the IL-10 gene rs1800896, rs1800871, and rs1800872 single-nucleotide polymorphisms (SNPs) and susceptibility to asthma. A systematic literature search performed until April 2020, and the pooled odds ratio (OR) and their corresponding 95% confidence interval (CI) were calculated to determine the association strength. Thirty articles comprising 5678 asthmatic patients and 6079 controls met the inclusion criteria. No significant association was found between rs1800872 SNP and susceptibility to asthma across all genetic models in the overall and subgroup analyses. The rs1800871 SNP had only significant association with a decreased risk of asthma in Europeans (OR 0.66, CI 0.53–0.82, P < 0.001). However, rs1800896 SNP was significantly associated with a decreased risk of asthma by dominant (OR 0.67, CI 0.50–0.90, P < 0.001) and heterozygote (OR 0.66, CI 0.49–0.88, P < 0.001) models in the overall analysis. Subgroup analyses indicated significant association of rs1800896 SNP by dominant (OR 0.45, CI 0.28–0.72, P < 0.001) and heterozygote (OR 0.43, CI 0.26–0.70, P < 0.001) models in the African population. The IL-10 rs1800896 SNP confers protection against the risk of asthma, especially in Africans. Additionally, rs1800871 SNP has a protective role against asthma in Europeans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asthma is a chronic inflammatory disease that is characterized by recurrent attacks of breathlessness, wheezing, chest tightness, airflow obstruction, and airway hyperresponsiveness (AHR). The disease affects children and young adults with varying severity and frequency (Boulet et al. 2012; Makoui et al. 2020). Asthma is the most common allergic disease, and the prevalence of this disease has been increased markedly across the world, with an approximately 250,000–345,000 death cases annually (Bahadori et al. 2009). The pathogenesis of asthma is complex, and several studies implicated that traditional risk factors, including age, allergen, pollution, infections, and smoking interact with susceptibility genetic factors during asthma development (Maddox and Schwartz 2002; Miller and Ho 2008).
A bulk of cytokines and immune cells promote an inflammatory setting in the asthmatic patients (Kim et al. 2010). The disease displays an inflammatory immune response with a noticeable rise in the level of serum immunoglobulin (Ig) E and helper T (Th2) cytokines, including interleukin (IL)-4 and IL-13. These cytokines, in turn, promote class switching of antibody to IgE and further production of this antibody class by B cells (Fahy 2015). Regulatory T (Treg) cells play a pivotal role in coordinating immune responses to maintain and acquire tolerance toward allergens through several mediators, especially IL-10 (Rivas and Chatila 2016). IL-10 is a cytokine that illustrates pleiotropic immunoregulatory and anti-inflammatory effects and contributes to the homeostasis of immune system (Barnes 2008).
To date, scientists have found many candidate genes associated with asthma susceptibility. Among these risk genes, researchers extensively studied IL-10 single-nucleotide polymorphisms (SNPs) (Huang et al. 2016). Studies have reported several polymorphic sites within the promoter region of the IL-10 gene. Growing evidence suggests that three SNPs of the IL-10 gene at positions 1082A/G (rs1800896 or −1087A/G or −1117A/G), 819T/C (rs1800871 or −892C/T or −854C/T), and 592C/A (rs1800872 or −571C/A or −597C/A or −627C/A) located in the transcription initiation site are involved in regulation of the expression of IL-10 gene (Lazarus et al. 2002; Vázquez-Villamar et al. 2016). The A allele of rs1800896 SNP correlates with low IL-10 levels, whereas G allele of this polymorphism associates with increased IL-10 transcription (Barnes 2008; Nie et al. 2012; Turner et al. 1997). Additionally, IL-10 levels in the plasma and peripheral blood mononuclear cells (PBMCs) of the irritable bowel syndrome (IBS) subjects with CT genotype for rs1800896 were significantly higher than those in the TT genotype subjects (Zhu et al. 2019). The −819 C-to-T substitution is a dimorphic polymorphism and may change an estrogen-responsive factor (Lazarus et al. 1997). Also, the C-to-T shift at position −819T/C (rs1800871) was associated with a high level of the total serum IgE by decreasing the expression of IL-10 gene (Zhang et al. 2002). The −592C/A variation is a C-to-A substitution that lies within a region that plays a negative regulatory function in the transcription of IL-10 gene (Kube et al. 1995). This polymorphism was shown to be correlated with increased total serum IgE level in homozygote or heterozygote asthmatics (Hobbs et al. 1998), while the CC genotype was associated with a higher level of serum IL-10 than AA genotype (Jin et al. 2013).
Several studies examined the association between polymorphisms of the IL-10 gene and susceptibility to asthma. However, the results of these researches were inconclusive due to small sample sizes, clinical heterogeneity, and low statistical power (Hyun et al. 2013; Nie et al. 2012; Zheng et al. 2010). To offset these limitations, we performed the most up-to-date meta-analysis (by including several recent original works) to investigate whether IL-10 gene polymorphisms play a significant role in susceptibility to asthma.
Methods
The present meta-analysis follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Moher et al. 2010). This article does not contain any study involving human participants or animals.
Search Strategy
In order to retrieve all potential publications that consider the association between IL-10 gene polymorphisms (rs1800872, rs1800871, and rs1800896) and susceptibility to asthma, we conducted a comprehensive search through Scopus and Medline. The search covered publications from inception to April 2020. We use combination of the following keywords and medical subject headings (MeSH) terms during search in databases: (“asthma” [MeSH] OR “asthmatic”) AND (“interleukin-10” OR “IL10” OR “IL-10”) AND (“single-nucleotide polymorphism” OR “SNP” OR “polymorphisms” OR “mutation” OR “variation”). Besides, we screened references of eligible studies for additional potentially relevant studies.
Inclusion and Exclusion Criteria
Publications considered eligible if they met the following inclusion criteria: (a) publications with case and control groups (case–control design and cohort design); (b) publications reporting sufficient data to extract or to calculate risk estimates with 95% CI; (c) publications that evaluate the association between IL-10 gene polymorphism and the risk of asthma as the primary outcome; and (d) publications that report genotype or allele distributions of the case and healthy individuals. We excluded duplicated articles as well as those that were reviews, irrelevant, meta-analysis, and book chapters. The application of these criteria results in 22, 11, and18 qualified studies for rs1800872, rs1800871, and rs1800896, respectively.
Data Extraction and Quality Assessment
We exported all retrieved publications to an EndNote library and final eligible studies were selected after duplicates exclusion. Two independent reviewers performed article screening and data extraction. The publications were screened by title, abstract, and full-text examination. Eventually, the data of the eligible studies were extracted as follows: the first author’s last name, journal, year of publication, country of origin, ethnicity, number of subjects in the case and control groups, mean or range of age, genotyping method, and genotype counts in both case and control groups. In case the final extracted file by two reviewers was not similar, they discussed to resolve the discrepancies by consensus. The quality of each study was assessed using the Newcastle–Ottawa Scale (NOS) (Stang 2010). Studies with scores 0–3, 4–6, or 7–9 were considered of low, moderate, or high-quality, respectively.
Statistical Analysis
We conducted Pearson’s χ2 test to examine Hardy–Weinberg equilibrium (HWE) in the healthy control group. The strength of the association between SNPs and asthma risk was assessed by estimating odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) in five genetic models for each SNP: dominant model (AA+AC vs. CC), recessive model (AA vs. CC+CA), allelic model (A vs. C), homozygote model (AA vs. CC), and heterozygotes model (CA vs. CC) for rs1800872 SNP; dominant model (TT+TC vs. CC), recessive model (TT vs. CC+CT), allelic model (T vs. C), homozygote model (TT vs. CC), and heterozygotes model (CT vs. CC) for rs1800871 SNP; dominant model (GG+GA vs. AA), recessive model (GG vs. GA+AA), allelic model (G vs. A), homozygote model (GG vs. AA), and heterozygotes model (AG vs. AA) for rs1800896 SNP. A chi-square-based Q-test and I2 test were used to estimate the between-study heterogeneity. In case of heterogeneity (P-value of < 0.1 for the Q-test or I2 > 50%), the random-effects models (REM) was employed for meta-analysis; otherwise, fixed-effect model (FEM) was applied (DerSimonian and Laird 1986; Mantel and Haenszel 1959). We performed subgroup analysis and meta-regression analysis based on year of publication and ethnicity to assess the pre-defined sources of heterogeneity among included studies. Potential publication bias was estimated using both Begg’s test and Egger’s test (Begg and Mazumdar 1994; Egger et al. 1997). Furthermore, the funnel plot asymmetry was assessed according to the Begg’s test (P-value < 0.05 considered statistically significant). We applied sensitivity analysis in order to evaluate the influence of individual studies on the pooled OR. Stata statistical software (version 14.0; Stata Corporation, College Station, TX, USA) was used for quantitative analysis and preparation of figures and calculation of OR was conducted using SPSS (version 23.0; SPSS, Inc. Chicago, IL, USA).
Results
Study Characteristics
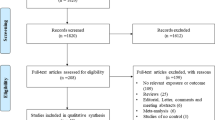
The search and screening process were conducted based on the PRISMA statement depicted in the Fig. 1. In detail, the initial search generated 640 publications. Then, duplicates (n = 72), which were the same among the databases, were omitted, and the remaining articles were excluded either by title and abstracts (n = 473) or full-text (n = 65) evaluations. Ultimately, 30 publications were recognized as eligible and were included in the quantitative analysis (Bo et al. 2011; Cárdaba et al. 2014; Chatterjee et al. 2005; Daley et al. 2009; Gaddam et al. 2012; Hakimizadeh et al. 2012; Hákonarson et al. 2001; Hang et al. 2003; Hsia et al. 2015; Hussein et al. 2011, 2014; Jahromi et al. 2015; Ji et al. 2004; Kadhem and Darweesh 2017; Karjalainen et al. 2003; Kim et al. 2011; Li et al. 2006, 2007; Lim et al. 1998; Movahedi et al. 2008; Park et al. 2004; Schubert et al. 2006; Smolnikova et al. 2013; Trajkov et al. 2008; Undarmaa et al. 2010; Xie 2012; Xu et al. 2014; Yucesoy et al. 2016; Zedan et al. 2008; Zhang et al. 2002). The references of all eligible publications were cross-checked, and no further records were found to meet the inclusion criteria. All eligible studies were published between 2001 and 2017 and had an overall good methodological quality with NOS scores ranging from 6 to 8. Tables 1 and 2 summarize the characteristics and genotype frequency of the included studies, respectively.
Quantitative Synthesis
Meta-analysis of rs1800872 and the Risk of Asthma
Twenty-two studies involving 3629 cases and 4255 controls were included in the final analysis of association between rs1800872 SNP and risk of asthma (Bo et al. 2011; Cárdaba et al. 2014; Chatterjee et al. 2005; Daley et al. 2009; Gaddam et al. 2012; Hakimizadeh et al. 2012; Hákonarson et al. 2001; Hang et al. 2003; Hsia et al. 2015; Ji et al. 2004; Karjalainen et al. 2003; Kim et al. 2011; Li et al. 2006, 2007; Movahedi et al. 2008; Park et al. 2004; Schubert et al. 2006; Smolnikova et al. 2013; Trajkov et al. 2008; Undarmaa et al. 2010; Xie 2012). Among these studies, 15 studies were carried out on Asians, 6 studies on Europeans, and 1 study on Australians. Since there was only one study from Australia, it was omitted from subgroup analysis. The pooled OR did not show association between rs18000872 SNP and asthma risk under all genotype models, including dominant model (OR 1.03, 95% CI 0.81–1.30, P = 0.80; Fig. 2a), recessive model (OR 1.10, 95% CI 0.86–1.40, P = 0.45), allelic model (OR 1.07, 95% CI 0.91–1.26, P = 0.38), homozygote model (OR 1.07, 95% CI 0.79–1.44, P = 0.67), and heterozygote model (OR 0.95, 95% CI 0.73–1.23, P = 0.68). The results also indicated that there were no significant associations between rs18000872 SNP and asthma risk in the subgroup analysis based on atopic status and ethnicity. The results of pooled ORs, heterogeneity tests, and publication bias tests in different analysis models are shown in the Table 3.
Pooled odds OR and 95% confidence interval of individual studies and pooled data for the association between IL-10 gene polymorphism and the risk of asthma in overall populations. The dashed vertical line is the overall OR. a Dominant model (rs1800872), b dominant model (rs1800871), c dominant model (rs1800896)
Meta-analysis of rs1800871 and the Risk of Asthma
Overall, 11 studies containing 1917 cases and 2589 healthy controls were found to be eligible and included for quantitative analysis of association between rs1800871 SNP and asthma risk (Chatterjee et al. 2005; Hsia et al. 2015; Jahromi et al. 2015; Karjalainen et al. 2003; Kim et al. 2010; Li et al. 2006; Movahedi et al. 2008; Trajkov et al. 2008; Xu et al. 2014; Yucesoy et al. 2016; Zhang et al. 2002). Among them, eight studies were on Asians, two studies on Europeans, and only one study on Americans, which was excluded from the subgroup analysis. The pooled OR showed no significant association between rs1800871 SNP and asthma risk across all genotype models, including dominant model (OR 1.23, 95% CI 0.90–1.68, P = 0.19; Fig. 2b), recessive model (OR 1.05, 95% CI 0.90–1.22, P = 0.52), allelic model (OR 1.25, 95% CI 0.67–2.35, P = 0.47), homozygote model (OR 1.07, 95% CI 0.87–1.33, P = 0.50), and heterozygote model (OR 1.26, 95% CI 0.91–1.74, P = 0.17). Additionally, the results of subgroup analysis (based on ethnicity and atopic status) were not statistically significant and rejected the association between rs1800871 SNP and asthma risk, except for significant association of the allelic model of rs1800871 SNP with a decreased risk of asthma in Europeans (OR 0.66, CI 0.53–0.82, P < 0.001). The results of pooled ORs, heterogeneity tests, and publication bias tests in different analytic models are shown in the Table 3.
Meta-analysis of rs1800896 and the Risk of Asthma
Herein, 18 studies were included for analysis of association between rs1800896 SNP and risk of asthma (Cárdaba et al. 2014; Chatterjee et al. 2005; Daley et al. 2009; Hsia et al. 2015; Hussein et al. 2014; Hussein et al. 2011; Jahromi et al. 2015; Kadhem and Darweesh 2017; Karjalainen et al. 2003; Kim et al. 2011; Li et al. 2007; Movahedi et al. 2008; Park et al. 2004; Trajkov et al. 2008; Xu et al. 2014; Yucesoy et al. 2016; Zedan et al. 2008; Zhang et al. 2002). A total of 3894 cases and 3884 controls were included in the quantitative analysis. Of eligible studies, 11 studies were on Asians, 2 studies on Africans, 3 studies on Europeans, and one study was on each Australian and American populations. Since there was only one study from Australia and America, they were omitted from subgroup analysis. Our results showed that rs1800896 SNP was associated with decrease risk of asthma across dominant model (OR 0.67, 95% CI 0.50–0.90, P < 0.001; Fig. 2c) and heterozygote model (OR 0.66, 95% CI 0.49–0.88, P < 0.001), but not recessive model (OR 1.03, 95% CI 0.79–1.40, P = 0.19), allelic model (OR 0.85, 95% CI 0.69–1.03, P = 0.09), and homozygote model (OR 0.93, 95% CI 0.77–1.11, P = 0.39). Additionally, the results of subgroup analysis based on ethnicity and atopic status reject association between rs1800896 and asthma risk except for Africans in dominant model (OR 0.45, 95% CI 0.28–0.72, P < 0.001) and heterozygote model (OR 0.43, 95% CI 0.26–0.70, P < 0.001). The results of pooled ORs, heterogeneity tests, and publication bias tests in different analysis models are shown in Table 3.
Publication Bias
We estimate publication bias using funnel plot, Begg’s and Egger’s tests. Except in some models, no evidence of publication bias was observed for the overall population and subgroup analysis under all genetic models (Table 3). Additionally, the shape of the funnel plot (just for dominant models of three SNPs) appeared to be symmetrical, demonstrating no significant publication bias (Fig. 3).
Sensitivity Analysis
The leave-one-out method was used in the sensitivity analysis to explore the effect of individual data on the pooled ORs. The significance level of ORs was not altered through omitting any single study in the dominant model for rs1800872, rs1800871, and rs1800896, indicating that our results were statistically robust (Fig. 4).
Meta-regression Analyses
Meta-regression analysis was performed to explore potential sources of heterogeneity among the included studies (Table 4). The findings indicated that none of the potential parameters, including publication year and ethnicity, were a source of heterogeneity (Fig. 5).
Discussion
Asthma is a chronic inflammatory disease and AHR and airway remodeling are the main hallmarks of the disease (Murphy and O’Byrne 2010). Although the pathogenesis of asthma is unknown, many studies have demonstrated that genetic background is involved in susceptibility to asthma (Kim et al. 2010). Our previous studies also indicated association of genetic polymorphisms in the critical cytokines involved in asthma pathogenesis, namely IL-4 (Kousha et al. 2020) and IL-13 (Omraninava et al. 2020) with the risk of asthma. Moreover, inflammation plays a significant role in the incidence and development of the disease (Noutsios and Floros 2014). On the other hand, IL-10 is a critical anti-inflammatory cytokine that suppresses the immune responses (Kurowski et al. 2018). A significant reduction of IL-10 expression in asthmatics suggests that IL-10 plays a crucial role in the pathogenesis of asthma (Raeiszadeh Jahromi et al. 2014). The results of previous studies on the association between IL-10 polymorphisms and asthma risk were inconsistent. The reasons for this contradiction may underly in the demographic differences and small sample size the low statistical power of the individual studies. Consequently, meta-analysis can help to obtain a more accurate finding of the association of IL-10 gene polymorphisms with risk of asthma.
Nie et al. (2012) performed the first meta-analysis in 2012 by including 17 studies (containing 4478 cases and 4803 controls) and indicated that rs1800896 and rs1800872, but not rs1800871, were associated with asthma risk. Similarly, the meta-analysis by Zheng et al. (2014) in 2014 on a total of 23 studies (22 articles) involving 4716 asthmatic patients and 5093 controls reported that rs1800896 and rs1800872 (but not rs1800871) had significant association with asthma risk. However, Hyun et al.'s (2013) meta-analysis in 2013 on 11 studies (containing 2215 asthma patients and 2170 controls) revealed an association between rs1800896 (but not rs1800871 and rs1800872) and asthma in adults. The meta-analysis by Huang et al. (2016) in 2016 on 16 studies (comprising 2494 cases and 2160 controls) showed no significant association between these three SNPs and pediatric asthma risk.
In order to update and provide a more precise evaluation of the association between IL-10 promoter rs1800896, rs1800871, and rs1800872 SNPs and asthma susceptibility, we conducted a meta-analysis of the studies on all articles published up to April 2020. Our meta-analysis consisted of 30 eligible case–control studies comprising 5678 asthmatic patients and 6079 controls. We observed that there was no association between rs1800872 and susceptibility to asthma in all genetic models both in the overall analysis and in thesubgroup analyses based on ethnicity and atopic status. Although rs1800871 had no significant association with asthma risk in the overall analysis, the results of subgroup analysis indicated a significant association of the allelic model of this SNP with a decreased risk of asthma in Europeans. However, analyses indicated that rs1800896 was associated with a decreased risk of asthma among dominant and heterozygote genetic models with no association across recessive, allelic, and homozygote models. The subgroup analysis based on ethnicity revealed that the dominant and heterozygote models had a significant association with decreased susceptibility to asthma in Africans.
The demonstrated association between rs1800896 polymorphism and asthma risk in Africans should be interpreted with caution. Although we applied sensitivity analysis for studies in HWE to explore the effect of individual data on pooled results and revealed significant associations, there were only two studies in HWE, which contained small sample sizes. Furthermore, we found no evidence of a significant association between rs1800871 (11 eligible studies) and rs1800872 (22 eligible studies) polymorphisms and risk of asthma under different genetic models with subgroup analysis based on atopic status and ethnicity. Our result disagrees with four previous meta-analysis studies (Huang et al. 2016; Hyun et al. 2013; Nie et al. 2012; Zheng et al. 2014), which may due to inclusion of more studies in the analysis. However, inclusion of more studies implies to higher sample size with more statistical power, suggesting that our findings are more valid. Compared to the previous meta-analysis studies, we identified thirty relevant papers using a robust retrieval method, far more than the number of studies included in previous analyses. Hence our analyses’ statistical power is higher.
Some limitations have to be taken into account when interpreting our findings. First, the heterogeneity of asthma can influence the analysis. Various criteria for inclusion and diagnosis of asthmatic patients and selection of the control group without screening test for asthma may distort the results. Second, the limited number of available studies possibly accounts for the observed results, implying that the polymorphisms of IL-10 may be associated with asthma susceptibility if more original data become available in the future. Although the subgroup analysis performed based on ethnicity, a small number of studies were found in the Europeans and African populations. Third, although we estimated publication bias using a funnel plot, Begg’s and Egger’s tests, their results in most of groups had P-value higher than < 0.05. However, the results for some subgroups showed significant publication bias and we think possible unpublished studies or many unknown reasons can be the source.
Conclusion
Our findings showed that rs1800896 SNP (by dominant and heterozygote genetic models) plays a protective role against asthma risk. Also, the rs1800896 SNP was associated with decreased risk of asthma in Africans. We found no association between rs1800872 SNP and asthma risk. Despite rs1800871 had no significant association with asthma risk in the overall analysis, this SNP had a protective role against asthma risk in Europeans. Nevertheless, due to the limited number of available studies in the subgroup analyses, the findings should be interpreted cautiously. Hopefully, further original works will contribute to our understanding of the association between IL-10 gene polymorphisms and risk of asthma.
Data Availability
All data generated or analyzed during this study are included in this article.
Abbreviations
- IL-10 :
-
Interleukin-10
- SNP:
-
Single-nucleotide polymorphism
- IL:
-
Interleukin
- PRISMA:
-
Preferred Reporting Items for Systematic reviews and Meta-Analyses
- NOS:
-
Newcastle–Ottawa Scale
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- AHR:
-
Airway hyperresponsiveness
- Ig:
-
Immunoglobulin
- Th2:
-
Helper T
- Treg:
-
Regulatory T
- IBS:
-
Irritable bowel syndrome
- HWE:
-
Hardy–Weinberg equilibrium
- FEM:
-
Fixed-effect model
- REM:
-
Random-effects models
References
Bahadori K, Doyle-Waters MM, Marra C, Lynd L, Alasaly K, Swiston J, FitzGerald JM (2009) Economic burden of asthma: a systematic review. BMC Pulm Med 9:24. https://doi.org/10.1186/1471-2466-9-24
Barnes PJ (2008) The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Investig 118:3546–3556. https://doi.org/10.1172/JCI36130
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50:1088–1101. https://doi.org/10.2307/2533446
Bo W, LiLi Z, DingFang D (2011) Association between polymorphisms in promoter region of interleukin-10 and interleukin-17 gene and childhood asthma. Prog Mod Biomed 11:307–313
Boulet L-P, Turcotte H, Plante S, Chakir J (2012) Airway function, inflammation and regulatory T cell function in subjects in asthma remission. Can Respir J 19:19–25. https://doi.org/10.1155/2012/347989
Cárdaba B, Calzada D, Baos S, Aguerri M, Quiralte J, Lahoz C (2014) Polymorphisms of tumor necrosis factor-α, transforming growth factor-β, and interleukin-10 in asthma associated with olive pollen sensitization. J Immunol Res. https://doi.org/10.1155/2014/276345
Chatterjee R, Batra J, Kumar A, Mabalirajan U, Nahid S, Niphadkar P, Ghosh B (2005) Interleukin-10 promoter polymorphisms and atopic asthma in North Indians. Clin Exp Allergy 35:914–919. https://doi.org/10.1111/j.1365-2222.2005.02273.x
Daley D et al (2009) Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet 125:445–459. https://doi.org/10.1007/s00439-009-0643-8
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188. https://doi.org/10.1016/0197-2456(86)90046-2
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634. https://doi.org/10.1136/bmj.315.7109.629
Fahy JV (2015) Type 2 inflammation in asthma—present in most, absent in many. Nat Rev Immunol 15:57–65. https://doi.org/10.1038/nri3786
Gaddam SL, Priya VHS, Srikanth Babu B, Joshi L, Venkatasubramanian S, Valluri V (2012) Association of interleukin-10 gene promoter polymorphism in allergic patients. Genet Test Mol Biomark 16:632–635. https://doi.org/10.1089/gtmb.2011.0255
Hakimizadeh E et al (2012) Association of-592 region of IL-10 polymorphisms with asthma in south-eastern Iranian patients. Clin Lab 58:267–271
Hákonarson H et al (2001) Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am J Respir Crit Care Med 164:2036–2044. https://doi.org/10.1164/ajrccm.164.11.2101086
Hang LW, Hsia TC, Chen WC, Chen HY, Tsai JJ, Tsai FJ (2003) Interleukin-10 gene–627 allele variants, not interleukin-I beta gene and receptor antagonist gene polymorphisms, are associated with atopic bronchial asthma. J Clin Lab Anal 17:168–173. https://doi.org/10.1002/jcla.10088
Hobbs K, Negri J, Klinnert M, Rosenwasser LJ, Borish L (1998) Interleukin-10 and transforming growth factor-β promoter polymorphisms in allergies and asthma. Am J Respir Crit Care Med 158:1958–1962. https://doi.org/10.1164/ajrccm.158.6.9804011
Hsia T-C et al (2015) The contribution of interleukin-10 promoter genotypes to susceptibility to asthma in adults. In Vivo 29:695–699
Huang Z, Cheng B, Wan Y, Zhou C (2016) Meta-analysis of the IL-10 promoter polymorphisms and pediatric asthma susceptibility. Genet Mol Res 15:gmr.15028320. https://doi.org/10.4238/gmr.15028320
Hussein YM, Shalaby SM, Mohamed RH, Hassan TH (2011) Association between genes encoding components of the IL-10/IL-0 receptor pathway and asthma in children. Ann Allergy Asthma Immunol 106:474–480. https://doi.org/10.1016/j.anai.2011.02.021
Hussein Y, Alzahrani S, Alharthi A, Ghonaim M, Alhazmi A, Eed E, Shalaby S (2014) Association of serum cytokines levels, interleukin 10– 1082G/A and interferon-γ+ 874T/A polymorphisms with atopic asthma children from Saudi Arabia. Cell Immunol 289:21–26. https://doi.org/10.1016/j.cellimm.2014.03.006
Hyun M-H, Lee C-H, Kang M-H, Park B-K, Lee YH (2013) Interleukin-10 promoter gene polymorphisms and susceptibility to asthma: a meta-analysis. PLoS One 8:e53758. https://doi.org/10.1371/journal.pone.0053758
Jahromi SR, Mahesh P, Jayaraj B, Holla AD, Vishweswaraiah S, Ramachandra NB (2015) IL-10 and IL-17F promoter single nucleotide polymorphism and asthma: a case-control study in South India. Lung 193:739–747. https://doi.org/10.1007/s00408-015-9753-3
Ji X, Zhang WZ, Zhang QY, Liu XD, Ge YJ, Huang JQ (2004) Gene polymorphism of interleukin-10 and its association with asthma in east China. Zhonghua Yi Xue Shi Jian Za Zhi 3:769–771
Jin H, Wang Y, Xu L (2013) Association of interleukin 10 gene-592C/A polymorphism with coronary artery disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 30:724–728. https://doi.org/10.3760/cma.j.issn.1003-9406.2013.06.020
Kadhem EJ, Darweesh MF (2017) Association of Il-101082g/A gene polymorphism with its serum levels in asthma patients. Biochem Cell Arch 17:709–713
Karjalainen J, Hulkkonen J, Nieminen M, Huhtala H, Aromaa A, Klaukka T, Hurme M (2003) Interleukin-10 gene promoter region polymorphism is associated with eosinophil count and circulating immunoglobulin E in adult asthma. Clin Exp Allergy 33:78–83. https://doi.org/10.1046/j.1365-2222.2003.01577.x
Kim HY, DeKruyff RH, Umetsu DT (2010) The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol 11:577–584. https://doi.org/10.1038/ni.1892
Kim KW, Lee KE, Hong JY, Kim MN, Heo WI, Sohn MH, Kim K-E (2011) Involvement of IL-10 gene promoter polymorphisms in the susceptibility for childhood asthma. Lung 189:417. https://doi.org/10.1007/s00408-011-9312-5
Kousha A et al (2020) Interleukin 4 gene polymorphism (− 589C/T) and the risk of asthma: a meta-analysis and met-regression based on 55 studies. BMC Immunol 21:1–16. https://doi.org/10.1186/s12865-020-00384-7
Kube D, Platzer C, Von Knethen A, Straub H, Bohlen H, Hafner M, Tesch H (1995) Isolation of the human interleukin 10 promoter. Characterization of the promoter activity in Burkitt’s lymphoma cell lines. Cytokine 7:1–7. https://doi.org/10.1006/cyto.1995.1001
Kurowski M, Jurczyk J, Olszewska-Ziąber A, Jarzębska M, Krysztofiak H, Kowalski ML (2018) A similar pro/anti-inflammatory cytokine balance is present in the airways of competitive athletes and non-exercising asthmatics. Adv Med Sci 63:79–86. https://doi.org/10.1016/j.advms.2017.07.004
Lazarus M, Hajeer A, Turner D, Sinnott P, Worthington J, Ollier W, Hutchinson I (1997) Genetic variation in the interleukin 10 gene promoter and systemic lupus erythematosus. J Rheumatol 24:2314–2317
Lazarus R et al (2002) Single-nucleotide polymorphisms in the interleukin-10 gene: differences in frequencies, linkage disequilibrium patterns, and haplotypes in three United States ethnic groups. Genomics 80:223–228. https://doi.org/10.1006/geno.2002.6820
Li HL, Zhao XF, Huang YK (2006) A study on association between the interleukin-10 _627 (C/A) polymorphism and childhood asthma. Zhongguo You Sheng Yu Yi Chuan Za Zhi 14:15–17. https://doi.org/10.1016/zhon.2006.0003
Li ZF, Lu JH, Li JR, Liao BP (2007) Lack of association between childhood asthma and interleukin-10 polymorphism gene. Zhongyuan Yi Kan 34:34–35
Lim S, Crawley E, Woo P, Barnes PJ (1998) Haplotype associated with low interleukin-10 production in patients with severe asthma. Lancet 352:113. https://doi.org/10.1016/S0140-6736(98)85018-6
Maddox L, Schwartz DA (2002) The pathophysiology of asthma. Annu Rev Med 53:477–498. https://doi.org/10.1146/annurev.med.53.082901.103921
Makoui MH, Imani D, Motallebnezhad M, Azimi M, Razi B (2020) Vitamin D receptor gene polymorphism and susceptibility to asthma: meta-analysis based on 17 case-control studies. Ann Allergy Asthma Immunol 124:57–69. https://doi.org/10.1016/j.anai.2019.10.014
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748. https://doi.org/10.1093/jnci/22.4.719
Miller RL, Ho S-m (2008) Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med 177:567–573. https://doi.org/10.1164/rccm.200710-1511PP
Moher D, Liberati A, Tetzlaff J, Altman DG (2010) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 8:336–341. https://doi.org/10.1016/j.ijsu.2010.02.007
Movahedi M, Mahdaviani SA, Rezaei N, Moradi B, Dorkhosh S, Amirzargar AA (2008) IL-10, TGF-ß, IL-2, IL-12, and IFN-γ cytokine gene polymorphisms in asthma. J Asthma 45:790–794. https://doi.org/10.1080/02770900802207261
Murphy DM, O’Byrne PM (2010) Recent advances in the pathophysiology of asthma. Chest 137:1417–1426. https://doi.org/10.1378/chest.09-1895
Nie W, Fang Z, Li B, Xiu Q-y (2012) Interleukin-10 promoter polymorphisms and asthma risk: a meta-analysis. Cytokine 60:849–855. https://doi.org/10.1016/j.cyto.2012.08.023
Noutsios GT, Floros J (2014) Childhood asthma: causes, risks, and protective factors; a role of innate immunity. Swiss Med Wkly. https://doi.org/10.4414/smw.2014.14036
Omraninava M, Eslami MM, Aslani S, Razi B, Imani D, Feyzinia S (2020) Interleukin 13 gene polymorphism and susceptibility to asthma: a meta-regression and meta-analysis. Eur Ann Allergy Clin Immunol. https://doi.org/10.23822/EurAnnACI.1764-1489.180
Park BL et al (2004) Interleukin 3 (IL3) polymorphisms associated with decreased risk of asthma and atopy. J Hum Genet 49:517–527. https://doi.org/10.1007/s10038-004-0184-x
Raeiszadeh Jahromi S, Mahesh PA, Jayaraj BS, Madhunapantula SR, Holla AD, Vishweswaraiah S, Ramachandra NB (2014) Serum levels of IL-10, IL-17F and IL-33 in patients with asthma: a case-control study. J Asthma 51:1004–1013. https://doi.org/10.3109/02770903.2014.938353
Rivas MN, Chatila TA (2016) Regulatory T cells in allergic diseases. J Allergy Clin Immunol 138:639–652. https://doi.org/10.1016/j.jaci.2016.06.003
Schubert K et al (2006) A comprehensive candidate gene study on bronchial asthma and juvenile idiopathic arthritis. Dis Markers 22:127–132
Smolnikova MV, Smirnova SV, Freidin MB, Tyutina OS (2013) Immunological parameters and gene polymorphisms (C-590T IL4, C-597A IL10) in severe bronchial asthma in children from the Krasnoyarsk region, West Siberia. Int J Circumpolar Health 72:21159. https://doi.org/10.3402/ijch.v72i0.21159
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25:603–605. https://doi.org/10.1007/s10654-010-9491-z
Trajkov D et al (2008) Association of cytokine gene polymorphisms with bronchial asthma in Macedonians. Iran J Allergy Asthma Immunol 7:143–156
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV (1997) An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 24:1–8. https://doi.org/10.1111/j.1365-2370.1997.tb00001.x
Undarmaa S et al (2010) Replication of genetic association studies in asthma and related phenotypes. J Hum Genet 55:342–349. https://doi.org/10.1038/jhg.2010.32
Vázquez-Villamar M et al (2016) Frequency distribution of interleukin-10 haplotypes (-1082 A> G,-819 C> T, and-592 C> A) in a Mexican population. Genet Mol Res 15:1–11. https://doi.org/10.4238/gmr15048411
Xie Q (2012) Study of the correlation between IL-10 gene-627C/A polymorphism and childhood asthma. Mod Med J 40:51–53. https://doi.org/10.1022/Mod.10.1111.2012
Xu J, Shi F, Li J (2014) Polymorphisms and susceptibility to asthma in children study of Interleukin-10 gene single nucleotide. China J Chin Med 29:639–641
Yucesoy B et al (2016) Genetic variants in TNF α, TGFB1, PTGS1 and PTGS2 genes are associated with diisocyanate-induced asthma. J Immunotoxicol 13:119–126. https://doi.org/10.3109/1547691X.2015.1017061
Zedan M, Settin A, Farag MK, El-Bayoumi M, El Regal ME, El Baz R, Osman E (2008) Gene polymorphisms of tumor necrosis factor alpha-308 and interleukin-10-1082 among asthmatic Egyptian children. In: Allergy and asthma proceedings, vol 3. OceanSide Publications, p 268
Zhang J, Chen H, Hu L, Fu J, Zhang H, Chen Y (2002) Correlation between polymorphism of IL-4 and IL-10 gene promoter and childhood asthma and their impact upon cytokine expression. Zhonghua Yi Xue Za Zhi 82:114–118
Zheng X-Q, Li C-C, Xu D-P, Lin A, Bao W-G, Yang G-S, Yan W-H (2010) Analysis of the plasma soluble human leukocyte antigen–G and interleukin-10 levels in childhood atopic asthma. Hum Immunol 71:982–987. https://doi.org/10.1016/j.humimm.2010.06.018
Zheng X-y et al (2014) Interleukin-10 promoter 1082/− 819/− 592 polymorphisms are associated with asthma susceptibility in Asians and atopic asthma: a meta-analysis. Lung 192:65–73. https://doi.org/10.1007/s00408-013-9519-8
Zhu SW, Liu ZJ, Sun QH, Duan LP (2019) Effect of the interleukin 10 polymorphisms on interleukin 10 production and visceral hypersensitivity in Chinese patients with diarrhea-predominant irritable bowel syndrome. Chin Med J 132:1524–1532. https://doi.org/10.1097/cm9.0000000000000306
Acknowledgements
The authors thank Mr. Karim Imani for all his support.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
DI and ND generated the idea. AP, SH, and BR searched the literature and analyzed and interpreted the data. FA, SS, and MHH prepared the original draft. MA, ND, BR, MM, and SA wrote and critically revised the paper. DI supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Imani, D., Dashti, N., Parvari, A. et al. Interleukin-10 Gene Promoter Polymorphisms and Susceptibility to Asthma: Systematic Review and Meta-analysis. Biochem Genet 59, 1089–1115 (2021). https://doi.org/10.1007/s10528-021-10056-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10056-9