Abstract
The −159C/T polymorphism in the CD14 gene has been implicated in susceptibility to asthma, but a large number of studies have reported inconclusive results. The aim of this study is to investigate the association between the −159C/T polymorphism in the CD14 gene and the risk of asthma by meta-analysis. We searched Pubmed, Embase, CNKI database, Wanfang database, Weipu database, and Chinese Biomedical database, covering all publications (last search been performed on April 20, 2010). Statistical analysis was performed by using the softwares Revman 4.2 and STATA 10.0. A total of 17 case–control studies in 17 articles (4,246 cases and 3,631 controls) were included in this meta-analysis. There was no association between this polymorphism and asthma risk in combined analyses (odds ratio (OR) = 0.86 and 95% confidence interval (95% CI) = 0.72–1.02, P = 0.09 for TC + TT vs. CC). In the subgroup analysis by age, ethnicity, and atopic status, no significant associations of asthma risks were obtained from age groups, ethnic groups, and atopic groups for TC + TT vs. CC comparison. For atopic population, significant decreased atopic asthma risks were found among Asian population (OR = 0.69, 95% CI 0.52–0.92, P = 0.01) and children population (OR = 0.69, 95% CI 0.54–0.89, P = 0.0004) for TC + TT vs. CC comparison. This meta-analysis suggests that CD14 is a candidate gene for atopic asthma susceptibility. The −159C/T polymorphism may be a protective factor for atopic asthma in Asian and children. More studies are needed to validate these associations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asthma is a complex inflammatory disease affecting nearly 300 million individuals worldwide (Gupta et al. 2009). It is characterized by airway hyperresponsiveness, hypertrophy, and hyperplasia of smooth muscle, hypersecretion of mucus, bronchial epithelial desquamation, and structural remodeling (Mahn et al. 2010; Zhang et al. 2010). Previous studies have indicated that asthma is a complex disorder with multiple determinants that include genetic variations, environmental exposures, and gene–environment interactions (Carroll et al. 2009; Weiss et al. 2009). Numerous published studies have focused on the association of genetic variants with asthma susceptibility (Hattori et al. 2009; Jung et al. 2009; Kabesch 2010; Møller-Larsen et al. 2008), and among them, CD14 gene has been extensively studied (Bernstein et al. 2006; Chen et al. 2009; Cui et al. 2003; de Faria et al. 2008; Heinzmann et al. 2003; Hong et al. 2007; Kedda et al. 2005; Kowal et al. 2008; Leung et al. 2003; Sharma et al. 2004; Smit et al. 2007, 2009; Tan et al. 2006; Wang et al. 2009; Woo et al. 2003; Wu et al. 2010; Zambelli-Weiner et al. 2005).
The CD14 gene is localized on chromosome 5q31.1, a region that is linked to both asthma and total serum IgE concentration. It encodes two protein isoforms: a membrane molecule (mCD14) on the surface of monocytes, macrophages, and neutrophils and a soluble form in serum (sCD14; Buckova et al. 2003). CD14 is a membrane recognition factor involved in the binding of bacterial components or inhaled endotoxin, a potent inducer of lung inflammation which may activate immune pathways that promote Th1 differentiation and Th2-dependent immunoglobulin E responses (Kedda et al. 2005). As previously shown in genetic studies, a polymorphism in the CD14 gene, −159C/T (rs2569190), might interact with environmental factors in the development of asthma and asthma phenotypes (Bernstein et al. 2006; Chen et al. 2009; Cui et al. 2003; de Faria et al. 2008; Heinzmann et al. 2003; Hong et al. 2007; Kedda et al. 2005; Kowal et al. 2008; Leung et al. 2003; Sharma et al. 2004; Smit et al. 2007, 2009; Tan et al. 2006; Wang et al. 2009; Woo et al. 2003; Wu et al. 2010; Zambelli-Weiner et al. 2005). This polymorphism is in the promoter region of CD14 gene and has been associated with altered CD14 and IgE levels (Zambelli-Weiner et al. 2005). There have been a large number of studies investigating this polymorphism with asthma risk; however, the results were inconsistent and inconclusive. Considering a single study may lack the power to provide reliable conclusion, we performed a meta-analysis to investigate these associations. This is, to our knowledge, the most comprehensive meta-analysis of genetics studies on the association between asthma susceptibility and the −159C/T polymorphism in CD14 gene.
Methods
Publication search
Two reviewers independently searched Pubmed, Embase, China National Knowledge Infrastructure (CNKI), Wanfang Database, Weipu Database, and Chinese Biomedical (CBM) database, to identify studies that had investigated the association between asthma susceptibility and CD14 polymorphisms, with the last updated search being performed on 20 April 2010. The search terms were used as follows: “asthma or asthmatic” and “CD14” in combination with “polymorphism or variant or mutation”. Studies published in English and Chinese were all included. Articles reporting on the association between asthma and CD14 polymorphism were identified. Studies included in our meta-analysis had to meet the following inclusion criteria: (1) evaluation of the −159C/T polymorphism in CD14 gene and asthma risk, (2) using a case–control design, (3) genotype distributions in both cases and controls should be available for estimating an odds ratio (OR) with 95% confidence interval (CI), and (4) genotype distribution of control population must be consistent with Hardy–Weinberg equilibrium (HWE). Accordingly, the following exclusion criteria were also used: (1) abstracts, reviews, and repeat studies; (2) genotype frequency not reported; and (3) genotype distribution in the control population not consistent with HWE.
Data extraction
Two independent reviewers checked all potentially relevant studies and reached a consensus on all studies. In case of disagreement, a third author also assessed those articles. A standardized data form was used and included first author, year of publication, country of origin, ethnicity of the study population, definition of cases, source of control, genotyping methods, total number of cases and controls, and genotype distribution in cases and controls.
Statistical analysis
The strength of the association between the −159C/T polymorphism and asthma risk was measured by OR and 95% CI. The statistical significance of summary OR was determined with Z test. We first estimated with the dominant model (TT + TC vs. CC) and recessive model (TT vs. TC + CC) and then evaluated variant genotype TT and compared with the wild-type CC homozygote (TT vs. CC). We also estimated the risks of T vs. C and TC vs. CC. Heterogeneity was evaluated by a χ 2-based Q statistic and was considered statistically significant at P value less than 0.10. When the P value is greater than 0.10, the pooled OR of each study was calculated by the fixed-effects model; otherwise, a random-effects model was used, as this was more appropriate when there was significant heterogeneity. The significance of the pooled OR was determined by the Z test, and P less than 0.05 was considered as statistically significant. To evaluate the ethnicity-specific, age-specific, and atopic-specific effects, subgroup analyses were performed by ethnic, age, and atopic status. To evaluate the ethnicity–atopic status and age–atopic status associations, the atopic asthma populations were stratified analyzed by ethnicity and age.
Sensitivity analysis was performed through sequentially excluding individual studies to assess the stability of the results (Zhang et al. 2010). Asymmetry funnel plots were used to assess potential publication bias. The Begg’s test and Egger’s test were also used to assess publication bias statistically (Zhang et al. 2010).
Hardy–Weinberg equilibrium was tested by using a web-based program (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl). All statistical tests were performed by using the Revman 4.2 software and STATA 10.0 software.
Results
Study included in the meta-analysis
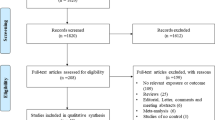
Figure 1 outlines the process of selecting studies. Briefly, a total of 254 articles were identified after an initial search from the Pubmed, Embase, CNKI database, Wanfang database, Weipu database, and CBM database. After reading the titles and abstracts, 217 articles were excluded for being not relevant to CD14 polymorphisms and asthma risk, or abstracts and reviews. After reading full texts of the remaining 38 articles, two studies which did not concern −159C/T polymorphism were excluded. Thus, 36 articles were left for data extraction. Of the 36 articles, 15 did not report the genotype frequencies and were excluded. Thus, a total of 21 case–control studies were extracted from 21 articles. Among 21 case–control studies, genotype frequencies for control group in two studies were not consistent with HWE, and data in two studies were overlapped. So these four case–control studies were excluded. Finally, a total of 17 case–controls in 17 articles were identified for meta-analysis (Bernstein et al. 2006; Chen et al. 2009; Cui et al. 2003; de Faria et al. 2008; Heinzmann et al. 2003; Hong et al. 2007; Kedda et al. 2005; Kowal et al. 2008; Leung et al. 2003; Sharma et al. 2004; Smit et al. 2007, 2009; Tan et al. 2006; Wang et al. 2009; Woo et al. 2003; Wu et al. 2010; Zambelli-Weiner et al. 2005). The characteristics of each case–control studies are shown in Table 1. Genotype and allele distributions for each case–control studies are listed in Table 2. There were eight case–controls of Asians (Chen et al. 2009; Cui et al. 2003; Hong et al. 2007; Leung et al. 2003; Sharma et al. 2004; Tan et al. 2006; Wang et al. 2009; Wu et al. 2010), seven of Caucasians (Bernstein et al. 2006; Heinzmann et al. 2003; Kedda et al. 2005; Kowal et al. 2008; Smit et al. 2007, 2009; Woo et al. 2003), and two were mixed (de Faria et al. 2008; Zambelli-Weiner et al. 2005). Nine studies were performed in adults (Bernstein et al. 2006; Chen et al. 2009; Kedda et al. 2005; Kowal et al. 2008; Sharma et al. 2004; Smit et al. 2007, 2009; Woo et al. 2003; Zambelli-Weiner et al. 2005), while eight were performed in children (Cui et al. 2003; de Faria et al. 2008; Heinzmann et al. 2003; Hong et al. 2007; Leung et al. 2003; Tan et al. 2006; Wang et al. 2009; Wu et al. 2010).
Quantitative data synthesis
All studies
As shown in Fig. 2, we analyzed the heterogeneity of TT + TC vs. CC for all 17 studies and the value of χ 2 was 44.78 with 16° of freedom and P = 0.0002 in a random-effects model. Additionally, I-square value is another index of the test of heterogeneity. In Fig. 2, the I-square was 64.3%, suggesting a present of heterogeneity. Thus, we chose the random-effects model to synthesize the data. Overall, OR was 0.88 (95% CI = 0.73–1.07) and the test for overall effect Z value was 1.26 (P = 0.21). The results suggested that the TT homozygote and TC heterozygote carriers did not have an increased risk of asthma compared with those individuals with the CC homozygote. Summary of the results of other genetic comparisons are listed in Table 3.
Subgroup analyses
In the subgroup analysis by ethnicity (Supplement Fig. 1), no associations were found among Asians (OR 0.80, 95% CI 0.56–1.15, P = 0.23) and Caucasians (OR 1.01, 95% CI 0.77–1.34, P = 0.94) in dominant model (TT + TC vs. CC). Similarly, in the subgroup analysis by age (Supplement Fig. 2), no associations were found among adults (OR 0.84, 95% CI 0.63–1.10, P = 0.21) and children (OR 0.94, 95% CI 0.71–1.25, P = 0.67). Subgroup analyses were also performed by atopic status (Supplement Fig. 3); no associations were found among atopic asthma patients, non-atopic asthma patients, and mixed atopic status asthma patients in dominant model (Table 3). Summary of the results of other genetic comparisons are listed in Table 3.
Atopic asthma patients
A total of eight case–control studies performed among atopic asthma patients were identified in the meta-analysis (Cui et al. 2003; de Faria et al. 2008; Hong et al. 2007; Kedda et al. 2005; Kowal et al. 2008; Leung et al. 2003; Sharma et al. 2004; Woo et al. 2003). Four studies were performed among Asians (Cui et al. 2003; Hong et al. 2007; Leung et al. 2003; Sharma et al. 2004), three among Caucasians (Kedda et al. 2005; Kowal et al. 2008; Woo et al. 2003), and one among mixed ethnicities (de Faria et al. 2008). Five studies were performed in children (Cui et al. 2003; de Faria et al. 2008; Hong et al. 2007; Leung et al. 2003; Sharma et al. 2004), while three were performed in adults (Kedda et al. 2005; Kowal et al. 2008; Woo et al. 2003). Subgroup analyses were also performed according to ethnicity and age. As shown in Supplement Fig. 4, significant decreased risk of asthma was found among children (OR = 0.69, 95% CI 0.54–0.89, P = 0.0004). The results suggested that the T allele carriers (TT + TC) have a 31% deceased risk of atopic asthma compared with those individuals with the CC homozygote in children population. In the subgroup analysis by ethnicity (Supplement Fig. 5), significant decreased risks of atopic asthma were found among Asians (OR = 0.69, 95% CI 0.52–0.92, P = 0.01). The results suggested that the T allele carriers (TT + TC) have a 31% deceased risk of atopic asthma compared with those individuals with the CC homozygote in Asian population. Summary of the results of other genetic comparisons are listed in Table 3.
Publication bias
Publication bias was assessed by Begg’s funnel plot and Egger’s test. The shape of the funnel plots seemed symmetrical in the TT + TC vs. CC comparison genetic model, suggesting the absence of publication bias (Fig. 3). Then, the Egger’s test was performed to provide statistical evidence of funnel plots asymmetry. The results indicated a lack of publication bias of the current meta-analysis (t = −1.31, P = 0.211).
Sensitivity analysis
As describe previously (Zhang et al. 2010), in order to assess the stability of the results, sensitivity analysis was performed by sequentially excluding each study. Statistically similar results were obtained after sequentially excluding each case–control study, suggesting the stability of the results.
Discussion
Common variants in genes in the pathway of pathogenesis may alter protein function and individual’s susceptibility to disease (Contopoulos-Ioannidis et al. 2005; Gao et al. 2006; Ohar et al. 2010; Pabst et al. 2010). CD14 is a membrane recognition factor involved in binding of bacterial components and plays a central role in innate immunity. One polymorphism in the CD14 gene, 159C/T (rs2569190), might interact with environmental factors in the development of asthma and asthma phenotypes. Correlation of this polymorphism and asthma risk has been studied, but the results remain controversial. Therefore, we performed a meta-analysis to clarify the relationship between this polymorphism and susceptibility to asthma.
A total of 17 case–control studies were included in the meta-analysis. The strength of the present analysis was based on the accumulation of published data giving information to detect more precise conclusion. In this study, the effect of allele frequency and the effect of dominant and recessive models were all estimated. In addition, the consistency of genetic effects across populations from different ethnicities and ages were investigated. The consistency of atopic status was also investigated. Main and subgroup analyses for ethnicity, age, and atopic status for all genetic contrasts produced no significant results, and the heterogeneity ranged from none to high. For the atopic asthma patients, subgroup analysis was performed by ethnicity and age. Significant decreased risks of asthma were found among Asian and children groups.
In this study, we did not find a clear association between the −159C/T polymorphism and risk of asthma in the total combined analysis. This may be related to the fact that the functionality of the polymorphism, at least with respect to the extent of the influence on CD14 plasma levels among asthma, has not been definitively established. Although an effect on transcription and CD14 levels was described (Zambelli-Weiner et al. 2005) and the potential effect on asthma was also described in some studies (Fageras et al. 2004; Woo et al. 2003), it is possible that that effect may be affected by several polymorphisms, rather than one individual polymorphism. This implies that the solitary −159C/T polymorphism might influence plasma levels but not in a substantial way.
In this study, subgroup analysis was also performed among atopic status patients. Significant decreased risk of asthma was found among atopic status patients in Asian and children, but not in Caucasians and adults, suggesting a possible role of ethnic differences in genetic backgrounds and etiology in different age. In addition, there is no reported study using Asian adults and Caucasian children in atopic asthma patients. So it is likely that the observed ethnic and age differences may be due to chance because studies with small sample size may have insufficient statistical power to detect a slight effect. Therefore, additional studies are warranted to further validate ethnic and age differences in the effect of this polymorphism on asthma susceptibility, especially in atopic status patients.
We have to mention two studies which were based on Hong Kong Chinese. These two studies were from almost the same investigators based on the same ethnic population. The first publication was based on atopic population, and the second publication was based on atopic and non-atopic population. The second publication included more cases and controls than the first publication. There may be some potential overlapped for cases or controls. Thus, we excluded one study. In order to include more cases and controls for atopic population, we just included the first publication in this meta-analysis, which is not according with our previous study. However, if we included this later study instead of the previous study for data analysis, the same no associations were found for overall analysis (data not shown).
Since 1990s, host genetic variants and their associations with susceptibility to asthma have been extensively studied, and many variants have been suggested as determinants of susceptibility to asthma. In recently years, the genome-wide association studies (GWAS) has become an important scientific focus in asthma genetic studies (Barnes 2010; DeWan et al. 2010; Kabesch 2010; Moffatt et al. 2010; Weidinger et al. 2010). Up to now, several loci have been suggested as asthma risk factors by GWAS (Barnes 2010; DeWan et al. 2010; Kabesch 2010; Moffatt et al. 2010; Weidinger et al. 2010), such as DENND1B, RAD50/IL13, HLA-DQB1, ORMDL3, PDE11A, SMAD3, and et al. (Barnes 2010; DeWan et al. 2010; Moffatt et al. 2010; Weidinger et al. 2010), and a few have also been replicated in different ethnic populations (Barnes 2010; Kabesch 2010; Weidinger et al. 2010). Although a few GWAS reported some loci which were associated with total IgE such as RAD50/IL13 and HLA-DQB1 (Barnes 2010; Weidinger et al. 2010), clearly none of these GWAS reported the significant association with −159C/T (rs2569190) polymorphism. This can be partly explained by the slight effects of this polymorphism on asthma susceptibility. In the future, more GWAS should be performed in atopic asthma patients to resolve whether the −159C/T polymorphism shows significant association with asthma susceptibility.
There are several limitations in this study that need to be addressed. First, only publications in Chinese and English included by the selected electronic databases were identified in this meta-analysis; it is possible that some relevant published studies in other languages or unpublished studies which had null results were not included, which might bias the results, while our statistical test might not show it clearly. Second, the number of studies and the number of subjects in the studies included in the meta-analysis by specific subgroups were small. This may not have enough power to explore the association between the −159C/T polymorphism and the disease; thus, caution should be adopted when explaining our results. Third, there is heterogeneity among studies, maybe owing to ages, ethnicity, atopic status, or environmental factors and so on. Fourth, most of the studies were performed in Asian and Caucasian populations; further studies are needed in other ethnic population because of possible ethnic differences of the −159C/T polymorphism.
To our knowledge, this is the most comprehensive meta-analysis to date to have assessed the relationship between the −159C/T polymorphism in CD14 gene and asthma risk. Our results revealed that the −159C/T polymorphism was significantly associated with decreased risk of asthma among Asian atopic population. This finding suggests that the −159C/T polymorphism may be a low-penetrance susceptibility marker of asthma. In the future, more studies in different ethnic population or ages should be performed to assess these associations.
Abbreviations
- HWE:
-
Hardy–Weinberg equilibrium
- OR:
-
Odds ratio
- CI:
-
Confidence interval
References
Barnes KC (2010) Genomewide association studies in allergy and the influence of ethnicity. Curr Opin Allergy Clin Immunol 10:427–433
Bernstein DI, Wang N, Campo P, Chakraborty R, Smith A, Cartier A, Boulet LP, Malo JL, Yucesoy B, Luster M, Tarlo SM, Hershey GK (2006) Diisocyanate asthma and gene–environment interactions with IL4RA, CD-14, and IL-13 genes. Ann Allergy Asthma Immunol 97:800–806
Buckova D, Holla LI, Schuller M, Znojil V, Vacha J (2003) Two CD14 promoter polymorphisms and atopic phenotypes in Czech patients with IgE-mediated allergy. Allergy 58:1023–1026
Carroll CL, Stoltz P, Schramm CM, Zucker AR (2009) Beta2-adrenergic receptor polymorphisms affect response to treatment in children with severe asthma exacerbations. Chest 135:1186–1192
Chan IH, Tang NL, Leung TF, Huang W, Lam YY, Li CY, Wong CK, Wong GW, Lam CW (2008) Study of gene-gene interactions for endophenotypic quantitative traits in Chinese asthmatic children. Allergy 63:1031–1039
Chen M, Wu B, Li W (2009) Influence of CD14 gene −159C/T polymorphism on IL5 level in patients with asthma (Chinese). Shan Dong Yi Yao 49:13–14
Contopoulos-Ioannidis DG, Manoli EN, Ioannidis JP (2005) Meta-analysis of the association of beta2-adrenergic receptor polymorphisms with asthma phenotypes. J Allergy Clin Immunol 115:963–972
Cui TP, Jiang WC, Wu JM (2003) Genetic polymorphism of CD14 and allergic asthma in children (Chinese). Hua Zhong Yi Xue Za Zhi 27:235–236
de Faria IC, de Faria EJ, Toro AA, Ribeiro JD, Bertuzzo CS (2008) Association of TGF-beta1, CD14, IL-4, IL-4R and ADAM33 gene polymorphisms with asthma severity in children and adolescents. J Pediatr (Rio J) 84:203–210
DeWan AT, Triche EW, Xu X, Hsu LI, Zhao C, Belanger K, Hellenbrand K, Willis-Owen SA, Moffatt M, Cookson WO, Himes BE, Weiss ST, Gauderman WJ, Baurley JW, Gilliland F, Wilk JB, O’Connor GT, Strachan DP, Hoh J, Bracken MB (2010) PDE11A associations with asthma: results of a genome-wide association scan. J Allergy Clin Immunol 126:871–873.e9
Fageras BM, Hmani-Aifa M, Lindstrom A, Jenmalm MC, Mai XM, Nilsson L, Zdolsek HA, Bjorksten B, Soderkvist P, Vaarala O (2004) A TLR4 polymorphism is associated with asthma and reduced lipopolysaccharide-induced interleukin-12(p70) responses in Swedish children. J Allergy Clin Immunol 114:561–567
Gao J, Shan G, Sun B, Thompson PJ, Gao X (2006) Association between polymorphism of tumour necrosis factor alpha-308 gene promoter and asthma: a meta-analysis. Thorax 61:466–471
Gupta S, Siddiqui S, Haldar P, Raj JV, Entwisle JJ, Wardlaw AJ, Bradding P, Pavord ID, Green RH, Brightling CE (2009) Qualitative analysis of high-resolution CT scans in severe asthma. Chest 136:1521–1528
Hattori T, Konno S, Hizawa N, Isada A, Takahashi A, Shimizu K, Shimizu K, Gao P, Beaty TH, Barnes KC, Huang SK, Nishimura M (2009) Genetic variants in the mannose receptor gene (MRC1) are associated with asthma in two independent populations. Immunogenetics 61:731–738
Heinzmann A, Dietrich H, Jerkic SP, Kurz T, Deichmann KA (2003) Promoter polymorphisms of the CD14 gene are not associated with bronchial asthma in Caucasian children. Eur J Immunogenet 30:345–348
Hong SJ, Kim HB, Kang MJ, Lee SY, Kim JH, Kim BS, Jang SO, Shin HD, Park CS (2007) TNF-alpha (−308G/A) and CD14 (−159T/C) polymorphisms in the bronchial responsiveness of Korean children with asthma. J Allergy Clin Immunol 119:398–404
Jung JS, Park BL, Cheong HS, Bae JS, Kim JH, Chang HS, Rhim T, Park JS, Jang AS, Lee YM, Kim KU, Uh ST, Na JO, Kim YH, Park CS, Shin HD (2009) Association of IL-17RB gene polymorphism with asthma. Chest 135:1173–1180
Kabesch M (2010) Novel asthma-associated genes from genome-wide association studies: what is their significance? Chest 137:909–915
Kedda MA, Lose F, Duffy D, Bell E, Thompson PJ, Upham J (2005) The CD14 C-159T polymorphism is not associated with asthma or asthma severity in an Australian adult population. Thorax 60:211–214
Kowal K, Bodzenta-Lukaszyk A, Pampuch A, Szmitkowski M, Zukowski S, Donati MB, Iacoviello L (2008) Analysis of −675 4g/5G SERPINE1 and C-159T CD14 polymorphisms in house dust mite-allergic asthma patients. J Investig Allergol Clin Immunol 18:284–292
Leung TF, Tang NL, Sung YM, Li AM, Wong GW, Chan IH, Lam CW (2003) The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol 14:255–260
Li X, Zhang Y, Zhang J, Xiao Y, Huang J, Tian C, He C, Deng Y, Yang Y, Fan H (2010) Asthma susceptible genes in Chinese population: a meta-analysis. Respir Res 11:129
Mahn K, Ojo OO, Chadwick G, Aaronson PI, Ward JP, Lee TH (2010) Ca2+ homeostasis and structural and functional remodelling of airway smooth muscle in asthma. Thorax 65:547–552
Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO (2010) A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 363:1211–1221
Møller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, Børglum AD (2008) Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax 63:1064–1069
Ohar JA, Hamilton RJ, Zheng S, Sadeghnejad A, Sterling DA, Xu J, Meyers DA, Bleecker ER, Holian A (2010) COPD is associated with a macrophage scavenger receptor-1 gene sequence variation. Chest 137:1098–1107
Pabst S, Karpushova A, Diaz-Lacava A, Herms S, Walier M, Zimmer S, Cichon S, Nickenig G, Nothen MM, Wienker TF, Grohe C (2010) VEGF gene haplotypes are associated with sarcoidosis. Chest 137:156–163
Sharma M, Batra J, Mabalirajan U, Goswami S, Ganguly D, Lahkar B, Bhatia NK, Kumar A, Ghosh B (2004) Suggestive evidence of association of C-159T functional polymorphism of the CD14 gene with atopic asthma in northern and northwestern Indian populations. Immunogenetics 56:544–547
Smit LA, Bongers SI, Ruven HJ, Rijkers GT, Wouters IM, Heederik D, Omland O, Sigsgaard T (2007) Atopy and new-onset asthma in young Danish farmers and CD14, TLR2, and TLR4 genetic polymorphisms: a nested case–control study. Clin Exp Allergy 37:1602–1608
Smit LA, Siroux V, Bouzigon E, Oryszczyn MP, Lathrop M, Demenais F, Kauffmann F (2009) CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med 179:363–368
Tan CY, Chen YL, Wu LS, Liu CF, Chang WT, Wang JY (2006) Association of CD14 promoter polymorphisms and soluble CD14 levels in mite allergen sensitization of children in Taiwan. J Hum Genet 51:59–67
Wang JY, Liou YH, Wu YJ, Hsiao YH, Wu LS (2009) An association study of 13 SNPs from seven candidate genes with pediatric asthma and a preliminary study for genetic testing by multiple variants in Taiwanese population. J Clin Immunol 29:205–209
Weidinger S, Baurecht H, Naumann A, Novak N (2010) Genome-wide association studies on IgE regulation: are genetics of IgE also genetics of atopic disease? Curr Opin Allergy Clin Immunol 10:408–417
Weiss ST, Raby BA, Rogers A (2009) Asthma genetics and genomics 2009. Curr Opin Genet Dev 19:279–282
Woo JG, Assa’Ad A, Heizer AB, Bernstein JA, Hershey GK (2003) The −159C-->T polymorphism of CD14 is associated with nonatopic asthma and food allergy. J Allergy Clin Immunol 112:438–444
Wu X, Li Y, Chen Q, Chen F, Cai P, Wang L, Hu L (2010) Association and gene–gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle China. J Asthma 47:238–244
Zambelli-Weiner A, Ehrlich E, Stockton ML, Grant AV, Zhang S, Levett PN, Beaty TH, Barnes KC (2005) Evaluation of the CD14/−260 polymorphism and house dust endotoxin exposure in the Barbados Asthma Genetics Study. J Allergy Clin Immunol 115:1203–1209
Zhang YG, Huang J, Zhang J, Li XB, He C, Xiao YL, Tian C, Wan H, Zhao YL, Tsewang YG, Fan H (2010) RANTES gene polymorphisms and asthma risk: a meta-analysis. Arch Med Res 41:50–58
Acknowledgments
The authors thank Prof. Lidwien A. M. Smit for providing original data of their study.
Funding
This study was supported by grants #30470761 and 30871117 from National Natural Science Foundation of China.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yonggang Zhang and Can Tian contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Fig. 1
Meta-analysis with a random-effects model for the association between asthma risk and the CD14 −159C/T polymorphism (TT + TC vs. CC): subgroup analysis by ethnicity (DOC 41 kb)
Supplement Fig. 2
Meta-analysis with a random-effects model for the association between asthma risk and the CD14 −159C/T polymorphism (TT + TC vs. CC): subgroup analysis by age (DOC 39 kb)
Supplement Fig. 3
Meta-analysis with a random-effects model for the association between asthma risk and the CD14 −159C/T polymorphism (TT + TC vs. CC): subgroup analysis by atopic status. (DOC 43 kb)
Supplement Fig. 4
Meta-analysis with a random-effects model for the association between atopic asthma risk and the CD14 −159C/T polymorphism (TT + TC vs. CC): subgroup analysis by age (DOC 35 kb)
Supplement Fig. 5
Meta-analysis with a random-effects model for the association between atopic asthma risk and the CD14 −159C/T polymorphism (TT + TC vs. CC): subgroup analysis by ethnicity (DOC 37 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Tian, C., Zhang, J. et al. The −159C/T polymorphism in the CD14 gene and the risk of asthma: a meta-analysis. Immunogenetics 63, 23–32 (2011). https://doi.org/10.1007/s00251-010-0489-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-010-0489-1