Abstract
Leaf color is a highly important agronomic trait, and mutants with altered leaf coloration can serve as excellent models for studies on chloroplast development and chlorophyll biosynthesis, enabling the cloning of genes involved in these processes in rice (Oryza sativa L.). In this study, we isolated a stable genetic rice mutant, oryza sativa albino leaf 50 (osal50), from a breeding population of the japonica cultivar GP50. This mutant exhibited a distinctive albino phenotype, with white-striped leaves in seedlings and white panicles at the heading stage. Compared with wild-type GP50, the osal50 mutant showed lower chlorophyll and carotenoid accumulation, together with abnormal chloroplast ultrastructure. Genetic analysis demonstrated that a recessive nuclear gene was responsible for the albino phenotype of osal50, and a map-based cloning strategy delimited OsAL50 to a 160-kb physical interval on chromosome 1, flanked by two single nucleotide polymorphism (SNP) markers, CAPS-08 and CAPS-37, that included 26 putative open reading frames. Sequence and expression analyses revealed LOC_Os01g20110 as the candidate OsAL50 gene, which was confirmed by knockout using CRISPR/Cas9. Subcellular localization and protein sequence analyses suggested that OsAL50 likely encodes an endoribonuclease E-like protein localized to the chloroplasts. Further investigation indicated that OsAL50 plays a vital role in the regulation of photosynthetic pigment metabolism, photosynthesis, and chloroplast biogenesis. In summary, we identified a novel albino mutant that will serve as useful genetic material for studies of chlorophyll biosynthesis and chloroplast development in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is one of the most important food crops globally and an ideal model for monocotyledonous plants in both genetic and functional genomic research. It is especially useful as it has a relatively small genome, exhibits good co-linearity with other grasses, and is amenable to manipulation using established transformation systems (Han et al. 2007). The primary photosynthetic organs in rice are the leaves. Changes in leaf color are common and tend to occur routinely over the course of growth and development (Su et al. 2017; Chen et al. 2018). Rice leaf-color mutants exhibit various leaf colors and patterns, such as bright green, green-revertible albino, zebra, striped, spotted, albino, yellowing, and light green (Li et al. 2018b). These different phenotypes are caused by variations in the levels and types of pigments, including carotene and chlorophyll, present in the leaves (Wang et al. 2018). The green chlorophyll pigment is present at high levels in plants and algae; it is localized to chloroplasts where it plays a critical role in the capture of light energy and its photosynthetic conversion into chemical energy, thus both controlling plant growth and development and determining the color of the leaves (Yang et al. 2016; Chen et al. 2009). Chlorophyll deficiencies inevitably contribute to reduced photosynthetic efficiency and reduced crop productivity, and may even cause the death of affected plants in severe instances (Wang et al. 2018). Albino mutants are especially associated with chlorophyll deficiency and have been identified in several species, including rice (Oryza sativa L.), soybean (Glycine max), tobacco (Nicotiana tabacum L.), wheat (Triticum aestivum L.), Arabidopsis (Arabidopsis thaliana), barley (Hordeum vulgare L.), and maize (Zea mays L.) (Bae et al. 2001; Dunford and Walden 1991; Hsieh and Goodman 2005; Long et al. 1993). Albino mutants exhibiting distinct coloration can be readily selected and removed, providing an easily recognized phenotypic marker; this has been used to monitor self-pollination during hybrid rice production to ensure the purity of the genetic stock (Chen et al. 2018). While mutants with albino leaves or panicles have been the focus of extensive research owing to their distinctive and easily detectable phenotype and clear relevance to the effective breeding and production of superior rice varieties, the underlying genetic mutations and molecular mechanisms associated with the albino phenotype in rice are not yet fully understood.
At present, over 200 distinctive leaf color mutants in rice have been found, and most of which are under the control of a single recessive nuclear gene. Approximately 175 of these mutants have been characterized, leading to identification of 150 genes associated with leaf color mapped to 12 rice chromosomes. However, at least 53 of these genes, including 14 with more than one allele, have been successfully isolated from different rice cultivars, and their locations on chromosomes have been determined using map-based cloning strategies (Du et al. 2020; Deng et al. 2014). Several genes, such as VAL1, albg, PAPST1, YSA, cisc(t), Gra(t), St1, V3, V2, and V1, have been identified as responsible for the mutant phenotype of albino leaves (Zhang et al. 2018; Jian et al. 2017; Xu et al. 2013; Su et al. 2012; Lan et al. 2010; Chen et al. 2009; Yoo et al. 2009; Sugimoto et al. 2007; Kusumi et al. 2011). The successful cloning of these genes has facilitated substantial progress in clarifying the molecular mechanisms underlying leaf pigmentation alterations. The V1 gene produces an NUS1 protein that localizes to the plastid and contains the bacterial antitermination factor NusB. NUS1 plays a vital role in regulating the transcription and metabolism of ribosomal RNA in the chloroplast. It is imperative during the initial phases of leaf growth and development and is essential for proper chloroplast differentiation when exposed to cold conditions (Kusumi et al. 2011). V2, by contrast, codes for a guanosine kinase that localizes to the mitochondria and plastids, catalyzing (d)GMP phosphorylation to (d)GDP in the guanine nucleotide metabolism pathway and facilitating plastid-to-nucleus signaling that is essential for proper chloroplast differentiation (Sugimoto et al. 2007). St1 and V3 respectively encode RNRS1 and RNRL1, which are the small and large ribonucleotide reductase (RNR) subunits, enabling RNR-mediated regulation of deoxyribonucleotide production rates in the context of DNA repair and biosynthesis while also helping to maintain an appropriate balance between cell division and plastid biogenesis during growth and development of the leaves (Yoo et al. 2009). The chloroplast-localized elongation factor Tu is encoded by Gra(t) and may be functionally related to chloroplast development (Chen et al. 2009). The cisc(t), YSA, and Albg genes code for a pentatricopeptide repeat (PPR) protein, with cisc(t) seedlings exhibiting a cold-sensitive albino phenotype (Lan et al. 2010). Abnormal PPR protein biosynthesis in cisc(t), ysa, and albg mutant plants results in dramatic reductions in total chlorophyll content and impaired chloroplast development (Su et al. 2012; Lan et al. 2010; Jian et al. 2017). The 3’-phosphoadenosine 5’-phosphosulfate transporter encoded by PAPST1 facilitates retrograde signaling between chloroplasts and the nucleus. OsPAPST1 localizes to the chloroplast envelope and outer mitochondrial membrane, and papst1 mutants exhibit defective thylakoid development that results in leaf chlorosis in the initial phases of leaf growth (Xu et al. 2013). VAL1 produced a phosphoribosylamine-glycine ligase that localizes to the chloroplast. It functions as the second enzyme in the de novo purine biosynthesis pathway and plays a crucial role in cell division throughout leaf development, chlorophyll biosynthesis, and in regulating chloroplast biogenesis (Zhang et al. 2018).
In rice, white panicle mutants have rarely been reported. Map-based cloning strategies have identified just eight genes related to this mutant phenotype to date: WLP1 (Song et al. 2014), WLP2 (Lv et al. 2017), St-wp (Chen et al. 2015), SLWP (Zhou et al. 2018), WP1 (Wang et al. 2016), WP2 (Wang et al. 2021), WP3 (Li et al. 2018a), and WSP1 (Zhang et al. 2017). WLP1 codes for a chloroplast 50S ribosome L13 protein, a primary plastidial ribosome component necessary for chloroplast development in rice, particularly at lower temperatures (Song et al. 2014). WLP2 encodes a plastid-encoded RNA polymerase (PEP)-associated protein that localizes to chloroplasts and is primarily expressed within green tissues. Both WLP2 and its paralog OsFLN2 are capable of directly interacting with the thioredoxin OsTRXz to form a TRX-FLN regulatory module that facilitates appropriate chloroplast development under conditions of heat stress (Lv et al. 2017). Various mutations in the St1 allelic genes St-wp and SLWP are responsible for different phenotypic characteristics, with St1 encoding RNRS1 as discussed above (Zhou et al. 2018; Chen et al. 2015). The Val-tRNA synthetase OsvalRS2 encoded by WP1 is an essential regulator of the biogenesis of chloroplast ribosomes and overall chloroplast development, with a single base change in the WP1 gene resulting in rice seedlings exhibiting virescent to albino phenotypes with white panicles at the heading stage (Wang et al. 2016). WP2 encodes OsTRXz, which regulates RNA editing by controlling the redox status of multiple organellar RNA editing factors, serving as an essential regulator of chloroplast development under high-temperature conditions (Wang et al. 2021). Appropriate interactions and coordination between chloroplasts and mitochondria are crucial for the growth and development of plants. The nuclear WP3 gene is expressed specifically in rice panicles and codes for a product that is located in the mitochondria. WP3 is crucial in maintaining proper mitochondrial function, which regulates chloroplast development in rice panicles. Mutations in WP3 lead to significant developmental abnormalities in both mitochondria and chloroplasts (Li et al. 2018a). WSP1 encodes a chloroplast-localized RNA editing factor, and a point mutation in this gene affects the editing of multiple organellar RNA sites such that wsp1 mutants exhibit impaired biogenesis of chloroplast ribosomes, impacting the development of chloroplasts in panicles and leaf tissues during the flowering stage (Zhang et al. 2017).
While a growing number of genes controlling leaf and panicle coloration in rice have been identified, the specific mechanisms underlying these changes in coloration remain an area of active research interest. In the present study, a naturally occurring rice albino mutant designated osal50 (oryza sativa albino leaf 50), derived from a breeding population of the japonica cultivar GP50 was identified and characterized. The mutant plants exhibit white-striped leaves as seedlings and white panicles at the heading stage owing to severe defects in both chloroplast development and chlorophyll biosynthesis. Moreover, this mutant has numerous white-striped leaves during the tillering stage, which is the period of most active growth. Notably, the albino phenotype exhibited by the osal50 mutant is distinct from that of any other variegated mutants reported to date in rice. We determined that the osal50 mutant phenotype is controlled by a recessive nuclear gene tentatively named OsAL50 and confined to a 160-kb physical interval on chromosome 1 between two single nucleotide polymorphism (SNP) markers, CAPS-08 and CAPS-37. Sequence and expression analyses revealed LOC_Os01g20110 as the candidate OsAL50 gene, which was confirmed by knockout using CRISPR/Cas9. Subcellular localization and protein sequence analyses indicated that the OsAL50 gene likely encodes an endoribonuclease E (RNase E)-like protein localized to the chloroplasts, where it is involved in RNA processing within the plastids and is an essential regulator of the biogenesis and development of chloroplasts. Our results offer valuable insights into the function of OsAL50 and provide genetic resources for studying the molecular determinants of changes in leaf and panicle coloration.
Materials and methods
Plant materials, mapping populations, and growth conditions
The osal50 is a spontaneous rice albino mutant derived from an advanced-generation breeding population of the japonica cultivar GP50 grown under normal field conditions and originating from the Sanming Academy of Agricultural Sciences (SAAS). This osal50 mutant and its progeny exhibited distinctive albinism affecting their leaves and panicles, and the mutant phenotype was stably inherited over several generations. In this study, segregating populations used for genetic analysis and gene mapping were developed by crossing the osal50 mutant with the japonica cultivars GP50 and Ningjing44 (NJ44) as well as the indica cultivar 9311. In total, 526 and 685 recessive F2 individuals exhibiting the albino phenotype were obtained from the respective NJ44 × osal50 and 9311 × osal50 crosses and used for genetic mapping of OsAL50. The rice plants utilized in the present study were cultivated using standard management practices in a paddy field at the SAAS in Shaxian, Fujian Province, China.
Analysis of agronomic traits
During the summer of 2022, the identical crop fields at the SAAS were planted with wild-type GP50 and osal50 mutant rice plants. The plants were cultivated under the same environmental conditions and management techniques. Three biological replicates were used to investigate the following key agronomic traits after the plants reached the mature stage: plant height, seed setting rate, 1000-grain weight, panicle length, number of grains per panicle, effective panicle number, grain length, grain width, flag leaf length, and flag leaf width.
Analysis of pigment levels
Fresh leaves, glumes, and branches from wild-type GP50 and osal50 mutant plants were sampled (approximately 0.2 g) during different periods of growth. The samples were cut into segments and incubated for 24 h in 10 mL of 100% (v/v) alcohol at 26 °C in the dark until the segments were fully discolored. Using a multifunctional enzyme-labeling instrument (TECAN infinite M200PRO), absorbances at 470, 649, and 665 nm were determined after the supernatants were collected. For every sample, three biological duplicates were examined. The levels of carotenoid (Car) and chlorophyll (Chl a and Chl b) in the samples were determined as described by Lichtenthaler (1987).
Transmission electron microscopic (TEM) analysis of chloroplast ultrastructure
White sections of fresh leaves harvested from osal50 plants at the four-leaf stage and white sections of fresh branches and glumes obtained separately from young panicles at the flowering stage were collected for analysis, as were the corresponding green tissue samples from wild-type plants. The samples were separated into small pieces, and fixed with 2.5% (v/v) glutaraldehyde (diluted with phosphate buffer, pH 7.2) under vacuum until the specimens were completely submerged. Standard protocols were used to fix, dehydrate, embed, section, and stain the specimens, and the processed specimens were then examined under a TEM (HITACHI HT-7800, Tokyo, Japan) to analyze chloroplast ultrastructure (Wu et al. 2016; Rao et al. 2015).
DNA extraction, gene pool construction, and development of molecular markers
The genomic DNA was extracted from fresh leaves using a slightly modified version of the cetyltrimethyl ammonium bromide-based (CTAB) approach (Murray and Thompson 1980). The multifunctional enzyme-labeling equipment was used to quantify the genomic DNA's OD value. DNA samples from each plant were diluted to a final concentration of 50 ng/μL, and diluted DNA of randomly selected wild-type or mutant individuals from the F2 population (n = 20 each) were mixed to produce two extreme DNA bulk-pools (Michelmore et al. 1991). Simple sequence repeat (SSR) primers uniformly distributed over the 12 rice chromosomes were collected from the International Rice Microsatellite Initiative (http://archive.gramene.org/markers/microsat). Genome sequences of the japonica and indica cultivars Nipponbare and 9311 were obtained by referring to the National Center for Biotechnology Information (NCBI) GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) and the Rice Genome Annotation Project (RGAP; http://rice.plantbiology.msu.edu/). Sequences were searched for SSR repeats using the SSRIT program (http://archive.gramene.org/db/markers/ssrtool). Furthermore, Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and DNAMAN (http://www.lynnon.com/) were used to align the genomic sequences of Nipponbare and 9311 to find any loci for insertion/deletion (InDels) or single nucleotide polymorphisms (SNPs). Using Primer Premier 5.0 (Premier Biosoft International, CA, USA), site-specific primer pairs flanking SSR, SNP, and InDel sequences were developed. These were subsequently used to examine the polymorphism in both the parental lines and the extreme pools and to genotype F2 individuals with mutant phenotypes using 8% nondenaturing polyacrylamide gel electrophoresis. This analysis uncovered SSR, InDel, and SNP molecular markers closely associated with the OsAL50 locus, facilitating fine mapping to be carried out (Wei et al. 2019). Online Resource 1 lists the enzymes utilized, PCR product sizes, and primer sequences.
Genetic analysis and fine mapping of OsAL50
Reciprocal crosses were made between osal50 and indica cultivar 9311 as well as osal50 and the japonica cultivars GP50 and NJ44 to determine whether the osal50 mutant phenotype was under the control of a single recessive nuclear gene. After that, the χ2 test was used to examine and assess the phenotypes of the F1 and F2 plants that resulted from these crosses.
For the genetic mapping of OsAL50, recessive class analysis (RCA) and bulk segregation analysis (BSA) were employed (Michelmore et al. 1991). F2 populations developed from crossings between the osal50 mutant and the japonica cultivar NJ44 or the indica cultivar 9311 were used for fine mapping. A standard paddy field was used for growing parental, F1, and F2 plants. Employing Mapmaker/Exp 3.0, a linkage analysis for molecular markers and the OsAL50 locus was carried out based on genotyping data for 1,211 recessive F2 individuals displaying the mutant phenotype (Lander et al. 1987). After converting recombination rates into genetic distances using the Kosambi operating function, a genetic linkage map was produced using the MapDraw program (Liu and Meng 2003). Based on the contig map, a physical map spanning the OsAL50 locus was generated using chromosomal walking (International Rice Genome Sequencing Project 2005).
Sequencing and phylogenetic analysis
Gene prediction and structural analysis were performed using the RAP-DB (http://rapdb.dan.affrc.go.jp/) and RGAP (http://rice.plantbiology.msu.edu/) databases. Candidate genes were amplified using genomic DNA from the wild-type GP50 and the osal50 mutant as templates. The products amplified from these DNA sequences were separated using 1% (w/v) agarose gel electrophoresis, purified using a DNA Gel Extraction Kit (Takara, Dalian, China), and sequenced by Takara Biotech (Dalian, China). Clustal Omega and DNAMAN software were used to assemble and align the sequences.
The BLASTP Search Program of the NCBI database (https://www.ncbi.nlm.nih.gov/) was utilized to find homologous sequences of OsAL50, and the DNAMAN program was then utilized for multiple sequence alignments. Employing MEGA 7.0, a phylogenetic tree was constructed using the neighbor-joining method with 1000 replicates. PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to search for cis-elements in the approximately 1300 bp of DNA sequence that came before the ATG start codon in the OsAL50 promoter, which was received via RGAP.
RNA extraction and gene expression analysis
Using a MiniBEST Universal RNA Extraction Kit (Takara, Dalian, China), total RNA was isolated from the fourth leaves (four-leaf stage), seventh leaves (seven-leaf stage), and immature panicles (comprising young branches and glumes; heading stage) of wild-type and osal50 plants. After undergoing RNase-free DNase treatment, the purity of the RNA was evaluated using 1% (w/v) agarose gel electrophoresis. Following this, 2 μg of RNA per sample was used to prepare cDNA with HiScript Reverse Transcriptase SuperMix (Vazyme, Nanjing, China). ChamQ SYBR Green qPCR Master Mix (Vazyme, Nanjing, China) and an ABI QuantStudio 3 instrument were used for real-time fluorogenic quantitative PCR analysis, with three biological replicates per sample. Relative gene expression was calculated using the comparative 2−△△CT method. Sixteen target candidate genes were analyzed, with OsActin serving as a reference control. Online Resources 2 and 3 contain the primers utilized in sequencing analysis and for determining the expression levels of candidate genes, respectively. The transcript levels of genes associated with photosynthetic pigment metabolism (OsCAO1, OsCHLI, OsCHLM, OsCHLD, OsCHLa/b, OsDVR, OsHEMA1, OsPSY1, OsPSY2, and OsPSY3), chloroplast development (OsPOLP, Ftsz, OsRPOTP, OsRPOB, V2, and V3), and photosynthesis (OsCAB1R, OsCAB2R, OsPETA, OsPETB, OsPSAA, OsPSBA, OsRBCL, and OsRBCS) were further examined in various tissues and at different growth stages of wild-type and osal50 mutant plants. Specific primers used for analyzing the expression levels of these genes were designed according to previous reports (Zhang et al. 2019; Shang et al. 2019) and are listed in Online Resource 4.
Vector construction for the CRIPSR/Cas9 system
OsAL50 was knocked out in wild-type GP50 using a modified tRNA-processing method based on the CRISPR/Cas9 system. The methods employed have been described previously (Wang et al. 2018). Two sgRNA target sites (CAATTGATGCATGCTGTTAGAGG, TCTGGTGGACATCAATGGCTTGG) in exon 1 were designed to construct the OsAL50-Cas9 vector. This vector was then transformed into wild-type calli by an Agrobacterium tumefaciens-mediated process (Lv et al. 2020). Positive transgenic plants from gene-edited lines of the T0 generation were identified using the specific functional markers FMHPT-F/R (5′-ATTTGTGTACGCCCGACAGT-3′ and 5′-GTGCTTGACATTGGGGAGTT-3′) and FMCas9-F/R (5′-AGAACCTCTCCGATGCTATCC-3′ and 5′-AGCAAGAGGACAACGTAG-3′) (Wei et al. 2023).
Subcellular localization of OsAL50
The pOsAL50-GFP vector was constructed by fusing the full-length coding sequence of OsAL50 to the N-terminus of GFP in the pAN580 vector. As previously described, the pOsAL50-GFP vector and the empty vector pAN580 were transformed into rice protoplasts (Lv et al. 2020). Employing a confocal laser scanning microscope (LSM700, Jena, Germany), images of the GFP signals were acquired.
Statistical analysis
The data is presented as the average value ± standard deviation (SD) derived from three biological replicates. Comparison of results was conducted using unpaired Student’s t-tests. A significance level of P < 0.05 was used to determine statistical significance. The *P < 0.05 and **P < 0.01 indicated significance levels. The statistical analyses in this study followed the techniques mentioned above.
Results
Phenotypic and agronomic characteristics of the osal50 mutant
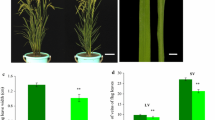
We initially subjected osal50 mutant rice plants to comprehensive phenotypic characterization. As shown in Fig. 1, both leaves and panicles of the mutant plants displayed significant albino characteristics in a growth stage-dependent manner when cultivated under normal field conditions. Specifically, in seedlings, it was observed that the leaves of this mutant developed white-green stripes running parallel to the mid-veins, with some leaves appearing almost fully white. However, we noticed that the branches of young panicles were completely white, while the glumes were white with some yellow and green coloration at the flowering stage. To fully explore the development of the albino character in the osal50 mutant, we next investigated osal50 mutant plants during different stages of growth, finding that the seedlings of this mutant produced a white-striped leaf phenotype starting from the three-leaf stage, which was particularly obvious at the four-leaf stage, whereafter they gradually became increasingly green from the five-leaf stage onward (Fig. 1a–e). When reaching the seven-leaf stage, the osal50 mutant seedlings exhibited an almost completely normal green leaf phenotype consistent with that of the wild type (Fig. 1f). Moreover, it is interesting that these mutant plants also had numerous white-striped or white leaves intermixed with normal leaves at the tillering stage, which is the period of most active growth (Fig. 1g). The osal50 mutant plants thus showed inconsistent leaf coloration at the tillering stage, with varying patterns and degrees of variegation even within a single plant, ranging from leaves that were completely white to those that were green, heavily striped, and slightly striped (Fig. 1h). In contrast to the light green glumes and branches exhibited by immature panicles of wild-type plants at the heading stage, young panicles of osal50 showed distinctive albino features, as described above (Fig. 1i–k). While osal50 mutant branches remained white when reaching the mature stage, the glumes of the panicles gradually turned black-brown in color (Fig. 1l). We were then cultivated wild-type and osal50 plants at a range of temperatures (20, 25, 30, and 35 °C) to determine whether the albino phenotype was temperature-sensitive. Based our examination, no pronounced phenotypic differences were observed in the osal50 mutant plants compared with the wild type at any of the tested temperatures, suggesting that generation of the albino phenotype in osal50 is not influenced by temperature changes (data not shown). To evaluate the link between the mutant phenotype and yields, we next investigated the following agronomic traits: plant height, 1000-grain weight, panicle length, number of grains per panicle, effective panicle number, seed setting rate, grain length, grain width, flag leaf length, and flag leaf width. Key yield-related traits, including 1000-grain weight, seed setting rate, and effective panicle number were lower in osal50 mutant plants than in the wild type (Fig. 2). This suggests that changes in leaf and panicle color may lead to a reduction in photosynthetic efficiency, thereby decreasing the accumulation of substances required for effective growth in rice.
Phenotypic characterization of wild-type GP50 and osal50 mutant plants. a–d, f Phenotypic comparisons of wild-type and osal50 seedlings at the one- (a), two- (b), three- (c), four- (d), and seven-leaf (f) stages. e White-striped leaves from osal50 mutant plants at the four-leaf stage. g-h Wild-type and osal50 leaf phenotypes at the tillering stage. i–j Wild-type and osal50 plant (i) and panicle (j) phenotypes at the heading stage. k–l Wild-type and osal50 grain phenotypes at the heading (k) and mature (l) stages. osal50-1, -2, and -3 correspond to three different types of white-striped leaves from osal50 mutant plants at the four-leaf stage (e) and tillering stage (h), respectively. Bars: 1 cm (a–f, k, l); 5 cm (g, h, j); 10 cm (i)
Comparisons of agronomic traits in wild-type GP50 and osal50 mutant rice plants. a Plant height (cm); b Effective panicle number; c Panicle length (cm); d Grain length (cm); e Grain width (cm); f Seed setting rate (%); g Number of grains per panicle; h 1000-grain weight (g); i Flag leaf length (cm); j Flag leaf width (cm). Data represent the mean ± SD of three biological replicates. Student’s t-test was used for statistical analysis
Photosynthetic pigment levels and chloroplast ultrastructural characteristics of osal50 mutant plants
The white-striped leaf phenotypes exhibited by osal50 mutant plants were most pronounced at the four-leaf and tillering stages, and marked differences in the extent of leaf albinism were seen even within a given plant during these growth stages. Therefore, to determine the levels of photosynthetic pigments in osal50 mutant plants over the course of growth and development, we analyzed chlorophyll a, chlorophyll b, and carotenoid levels in three different types of white-striped leaves, osal50-1 (slightly striped), osal50-2 (heavily striped), and osal50-3 (almost fully white), collected from osal50 mutant plants at the four-leaf and tillering stages, respectively (Fig. 1e, h), together with green leaves harvested from wild-type plants at these two stages. As expected, osal50 leaves had significantly lower pigment levels relative to those in wild-type leaves, with 26.01‒94.91%, 33.56‒96.87%, and 40.67‒98.19% less chlorophyll a, chlorophyll b, and carotenoid in these white-striped leaves at the four-leaf stage, respectively, and 29.96‒89.03%, 35.43‒91.89%, and 42.93‒96.33% less chlorophyll a, chlorophyll b, and carotenoid, respectively, at the tillering stage (Fig. 3a, b). Similarly, the chlorophyll a, chlorophyll b, and carotenoid levels in glumes and branches harvested at the heading stage from young panicles of osal50 were 54.06%, 49.09%, and 61.79% lower and 95.81%, 97.4%, and 92.07% lower, respectively, than those of the wild type (Fig. 3c, d).
Comparisons of photosynthetic pigment levels and chloroplast ultrastructures in wild-type GP50 and osal50 mutant plants. a, b Photosynthetic pigment levels in leaves harvested at the four-leaf stage (a) and tillering stage (b) of wild-type and osal50 mutant plants. c-d Photosynthetic pigment levels in glumes (c) and branches (d) from young panicles of wild-type and osal50 mutant plants at the heading stage. e-j Chloroplast ultrastructural characteristics in green sections of leaves (e, f), branches (g, h), and glumes (i, j) from wild-type plants. k–p Chloroplast ultrastructural characteristics in white sections of leaves (k, l), branches (m, n), and glumes (o, p) from osal50 mutant plants. Bars: 1 μm (f, h, j, l, n, p); 5 μm (e, g, i, k, m, o). FW fresh weight; osal50-1, osal50-2, and osal50-3 three different types of white-striped leaves from osal50 mutant plants as shown in e and h; Ch chloroplast; N nucleus; G grana stacks; A amyloplast; OG osmophilic plastoglobuli; CW cell wall; M mitochondria. Data represent the mean ± SD of three biological replicates. Student’s t-test was used for statistical analysis, *P < 0.05, **P < 0.01
The underlying physiological basis for lower photosynthetic pigment levels in osal50 mutant plants was explored by investigating differences in chloroplast morphology and development in various tissues and at different growth stages of wild-type and osal50 mutant plants via TEM. Leaves collected at the four-leaf stage, as well as glumes and branches harvested at the heading stage, were examined. As expected, organelles in the green sections of various tissue samples from wild-type plants were intact and evenly distributed within appropriately developed cells, with fully developed chloroplasts showing tightly stacked grana lamellae (Fig. 3e–j). By contrast, most cells in the white sections of leaves, glumes, and branches from osal50 mutant plants showed extremely simple internal structures; the chloroplasts were severely degraded and lacked any organized thylakoid structures. In addition to overall reductions in chloroplast numbers and volumes, there was a clear absence of lamellae stacking within the disordered grana (Fig. 3k–p). The findings demonstrate that the mutant phenotype and decreased chlorophyll and carotenoid levels of osal50 result from abnormal chloroplast development under natural conditions.
Genetic analysis and preliminary mapping of OsAL50
The F1 and F2 populations derived from reciprocal crosses between the osal50 and the japonica cultivars GP50 or NJ44 or the indica cultivar 9311 were used for genetic analysis to determine whether a single recessive nuclear gene was responsible for the osal50 mutant phenotype. Leaves from all F1 plants exhibited normal phenotypes consistent with those of wild-type GP50 plants, confirming that the osal50 mutant phenotype is recessive. Moreover, no intermediate phenotypes were identified, and the mutant phenotype was segregated in the F2 populations. The segregation ratio of the wild-type to the mutant phenotype in these four populations was approximately 3:1 as determined through a χ2 test (χ2 < χ20.05 = 3.84; P > 0.05; Table 1), suggesting that the osal50 mutant albino phenotype is under the control of a single recessive nuclear gene.
We next performed OsAL50 mapping using the F2 population generated from the cross between osal50 and NJ44. Both parental lines and two extreme DNA bulk-pools were initially screened for polymorphisms using 336 pairs of SSR markers released by the International Rice Microsatellite Initiative, distributed evenly across all 12 rice chromosomes. Polymorphic SSR markers associated with the albino phenotype in both the parental lines and the extreme pools were confirmed by analysis of F2 mutant individuals. An SSR marker pair, Rm562, on the short arm of chromosome 1 that was linked to the OsAL50 locus was successfully identified (Fig. 4a). Analysis of 115 pairs of SSR markers densely distributed in regions adjacent to Rm562 revealed four additional pairs linked to OsAL50 (Rm6642, Rm10701, Rm10710, and Rm10747) (Fig. 4b). PCR genotyping of 526 F2 individuals exhibiting the mutant phenotype derived from the cross between osal50 and NJ44 parental lines was next performed, leading to the identification of 35, 5, 4, and 15 recombinant events, respectively. The recombinant positions of the Rm6642, Rm10701, and Rm10710 markers were identical and were located on the same side of the OsAL50 locus, whereas Rm10747 was on the other side of the locus along with Rm562. These results were used to preliminarily map OsAL50 to chromosome 1 between the Rm10710 and Rm10747 markers at respective genetic distances of 0.4 cM and 1.8 cM (Fig. 4b). Further review of the chromosomal location of OsAL50 suggested that it was distinct from other genes previously associated with changes in leaf or panicle coloration, so we speculate that OsAL50 is a newly identified gene.
Genetic and physical maps of the region including the OsAL50 locus. a Molecular marker-based linkage maps of rice chromosome 1, with the short arm of the chromosome at the top. Marker names and corresponding map distances (centimorgan, cM) are shown, with the centromere (CEN) and approximate OsAL50 locus locations represented by black and red rectangles, respectively. b Fine genetic linkage map of the OsAL50 locus on chromosome 1. Markers tightly linked to OsAL50 are named on the right of the map, while genetic distances are shown to the left in cM. c Physical contig map of the region spanning the OsAL50 locus, generated using 1,211 F2 mutant individuals. Chromosomal orientation is as noted, and numbers in parentheses beneath each molecular marker denote the numbers of recombinants identified from 526 (NJ44 × osal50) and 685 (9311 × osal50) F2 mutant individuals. Physical locations on chromosome 1 of the corresponding linkage markers are denoted by numbers below the map that were determined using the GRAMENE database (http://www.garmene.org). Relative positions of linkage markers in Nipponbare BAC clones are designated with vertical and dashed lines. d The locus spanning OsAL50 was narrowed down to a 160-kb region of the genome between the CAPS-08 and CAPS-37 markers. e Twenty-six predicted open reading frames identified as candidate OsAL50 genes according to genomic information of the japonica rice cultivar Nipponbare (http://rice.plantbiology.mus.edu/)
Fine mapping of OsAL50
The preliminary mapping results above were next used to perform additional fine mapping of OsAL50 using a new mapping population developed by crossing the osal50 mutant with the indica rice cultivar 9311, which showed significant polymorphism in the target mapping region. To facilitate the localization of OsAL50 to a smaller region on chromosome 1 and develop tightly linked molecular markers, the publicly available genome sequences of the preliminary mapping regions in Nipponbare and 9311 were obtained from the GRAMENE and NCBI databases. These sequences were then analyzed using the SSRIT program, which allowed for the development of 80 pairs of new SSR markers within the preliminary mapping region. Alignment of the Nipponbare and 9311 sequences led to the development of 95 pairs of de novo InDel and CAPS markers that were used to narrow the genomic region encompassing the OsAL50 locus between the flanking Rm10710 and Rm10747 markers. Following screening of polymorphic markers in the two parental lines and the two extreme DNA bulk-pools, and confirmation by analysis of F2 mutant individuals, Mapmaker/Exp 3.0 was employed to conduct a linkage analysis, leading to the identification of 14 molecular markers closely associated with the OsAL50 locus (InDel-01, CAPS-08, CAPS-40, CAPS-37, InDel-07, InDel-08, ZZY-01, ZZY-04, InDel-11, InDel-13, InDel-15, InDel-18, InDel-20, and InDel-29) (Fig. 4b). These markers were used for genotyping 685 F2 individuals with the mutant phenotype derived from the cross between osal50 and 9311. Three recombinant events were detected with the InDel-01 marker, four with the ZZY-01 or ZZY-04 markers, five with the InDel-11, InDel-13, InDel-15, InDel-18, InDel-20, or InDel-29 markers, and just one with the CAPS-08 or CAPS-37 markers; the CAPS-40 marker co-segregated with OsAL50. We thus determined that the OsAL50 locus is delimited by the closely flanking CAPS-08 and CAPS-37 SNP markers, with a genetic distance of 0.1 cM from each of them. Finally, a high-resolution genetic linkage map of the OsAL50 locus was constructed using a combination of Mapmaker/Exp 3.0 and MapDraw software (Fig. 4b). Moreover, DNA sequences of bacterial artificial chromosome (BAC) clones and their corresponding physical locations within the Nipponbare (japonica) genome allowed anchorage of the molecular markers linked to OsAL50 to appropriate positions of the BAC clones, enabling the construction of a physical contig map spanning the OsAL50 locus based on the genotyping results of 1211 F2 mutant individuals (Fig. 4c). This demonstrated that the OsAL50 locus is located within the AP002873 and AP003567 regions, and that the CAPS-40 marker co-segregating with OsAL50 is located on the AP002873 BAC clone (Fig. 4c).
Candidate genes for OsAL50
Fine mapping of the OsAL50 locus successfully delimited it to a 160-kb region flanked by the CAPS-08 and CAPS-37 markers, covering portions of the AP002873 and AP003567 BAC clones (Fig. 4d). Using the RAP-DB and RGAP genomic annotation databases, 26 predicted open reading frames were identified within this 160-kb region as candidate OsAL50 genes (Fig. 4e). These included LOC_Os01g19970, LOC_Os0120140, and LOC_Os01g20160, which, respectively, encode a member of the MYB transcription factor family, a latency-associated nuclear antigen, and an OsHKT1;5-Na+ transporter. Of these genes, 13 (LOC_Os01g19910, LOC_Os01g19940, LOC_Os01g19950, LOC_Os01g19980, LOC_Os01g19990, LOC_Os01g20000, LOC_Os01g20010, LOC_Os01g20020, LOC_Os01g20030, LOC_Os01g20060, LOC_Os01g20100, LOC_Os01g20110, and LOC_Os01g20120) encode hypothetical or expressed proteins, whereas 10 (LOC_Os01g19920, LOC_Os01g19930, LOC_Os01g19960, LOC_Os01g20040, LOC_Os01g20050, LOC_Os01g20070, LOC_Os01g20080, LOC_Os01g20090, LOC_Os01g20130, and LOC_Os01g20150) encode transposons or retrotransposons (Online Resource 5). These latter 10 genes were excluded, considering the potential relationships between their predicted functions and the mutant phenotypes observed in osal50 mutant plants. Next, we performed a series of semi-quantitative reverse-transcription PCR (RT-PCR) analyses to investigate the transcript levels of the remaining 16 genes in wild-type GP50 and osal50 mutant leaves at the four-leaf stage, as this was the stage at which the albino phenotype was most pronounced. The transcript levels of LOC_Os01g20110 were markedly lower in the osal50 mutant compared with those in the wild type (Fig. 5a, b). Of the remaining 15 candidate genes, LOC_Os01g19980 and LOC_Os01g20160 exhibited extremely poor expression in both the wild type and the osal50 mutant, while the other 13 genes were expressed at comparable levels in both lines (Fig. 5a, b). Considering these findings, we further examined the accumulation of LOC_Os01g20110 mRNA transcripts in young panicles of wild type and osal50, including both glumes and branches, at the heading stage using a combination of semi-quantitative RT-PCR and quantitative RT-PCR (qRT-PCR), with additional qRT-PCR analysis used to confirm the previously detected differences in LOC_Os01g20110 expression in leaves at the four-leaf stage between the two parental lines. Consistent with earlier results, expression levels of LOC_Os01g20110 were markedly lower in osal50 mutant glumes and branches at the heading stage and leaves at the four-leaf stage, with 51.9%, 58.5%, and 71.5% lower expression levels, respectively, relative to wild-type GP50 plant tissues (Fig. 5b, c). Overall, these results indicated that LOC_Os01g20110 is the most likely candidate for the OsAL50 gene.
OsAL50 candidate gene analysis. a Expression levels of 15 candidate genes in wild-type GP50 and osal50 mutant leaves at the four-leaf stage, determined by semi-quantitative RT-PCR. b–c Expression levels of LOC_Os01g20110 in wild-type GP50 and osal50 mutant plants, determined by semi-quantitative RT-PCR (b) and qRT-PCR (c). Leaves were collected at the four-leaf stage (left) and glumes (middle) and branches (right) were harvested separately from young panicles at the heading stage for cDNA preparation. d Mutation analysis in the LOC_Os01g20110 promoter region, where “–” indicates base deletion. e Mutation types and locations in the defined promoter region of LOC_Os01g20110, with specific positional information being provided in brackets. Relative gene expression was calculated using the comparative 2−△△CT method, OsActin served as a normalization control. Error bars indicate SD (n = 3). Student’s t-test was used for statistical analysis, *P < 0.05, **P < 0.01
Sequence analysis and functional confirmation of OsAL50
To confirm the identity of LOC_Os01g20110 as the OsAL50 gene, the coding sequences (CDS) of LOC_Os01g20110 were amplified using genomic DNA from the wild-type GP50 and the osal50 mutant as templates, followed by sequencing. The DNA sequences were then aligned to compare differences in the LOC_Os01g20110 sequences between the two parental lines. The RGAP database (http://rice.plantbiology.msu.edu/) indicates that LOC_Os01g20110 is a single-copy gene consisting of seven exons and six introns, with a 1338-bp CDS encoding a 445-aa polypeptide. However, we did not identify any differences in the coding regions between the allelic LOC_Os01g20110 gene sequences from the wild type and the osal50 mutant. Accordingly, we amplified the respective promoter regions upstream of the gene sequences from the two parental genomes and identified 20 mutation sites within the promoter of LOC_Os01g20110 in the osal50 mutant comprising mostly single-base substitution and deletion mutations. Two small fragments, namely, ‘TATTTACTTGTA’ and ‘ATGTACCG’, were inserted into adjacent positions 2813 and 2808 bp upstream of the ATG start codon, respectively (Fig. 5d, e). Further investigation of the promoter and coding sequences of LOC_Os01g20110 in Nipponbare, 9311, and NJ44 revealed that these sequences were identical to those of the wild-type GP50 (Fig. 5d, e). Moreover, to confirm that these mutations were exclusive to the osal50 mutant, we examined another 20 rice varieties and found that only the osal50 mutant, and not even one of the wild-type rice varieties, contained these specific mutations in the promoter region of LOC_Os01g20110 (Online Resource 6). These results further supported the tentative identification of LOC_Os01g20110 as the candidate OsAL50 gene. To confirm this conclusion, we used the CRISPR/Cas9 system to knock out LOC_Os01g20110 in the wild-type GP50 (Fig. 6a). Two homozygous positive transgenic plants were identified among gene-edited lines of the T0 generation, and seedlings of these independent transgenic lines displayed an obvious white-striped leaf phenotype consistent with that of the osal50 mutant (Fig. 6b, c). Specifically, 3-bp and 11-bp deletions were detected at the target site of LOC_Os01g20110 in the two transgenic lines, respectively (Fig. 6d). These deletions resulted in frameshift mutations. Thus, we verified that LOC_Os01g20110 was the candidate OsAL50 gene, with mutations in the associated promoter region potentially contributing to the observed albino phenotype in osal50 mutant plants.
CRISPR/Cas9 verification of the OsAL50 gene. a Vector map of Cas9/gRNA. LB, UBI, Cas9, gRNA, rU6, 35S, Hygro, and RB represent the vector left border, UBI promoter, Cas9 gene, guide RNA, rice U6 promoter, 35S promoter, hygromycin, and vector right border, respectively. b PCR identification of positive transgenic plants in gene-edited lines of the T0 generation using the functional markers FMHPT-F/R (left) and FMCas9-F/R (right), respectively, where “+” and “−” indicate the positive control and negative control, respectively; M represents a 2000-bp DNA marker. c Phenotype of T0 mutant seedlings generated using the CRISPR/Cas9 system. Bar: 5 cm. d Mutation analysis at the target site of LOC_Os01g20110 in transgenic lines cas9-1# and -2#, where “-” indicates base deletion and PAM represents the protospacer adjacent motif
The non-coding promoter region contains many different sites for transcription-factor binding, including the basal core promoter (BCP) as well as regions involved in transcriptional regulation. The BCP facilitates the basal transcriptional level and serves as the minimum genomic unit necessary for proper transcription. Other transcriptional regulatory regions are associated with adjustment of the expression levels of specific genes in response to changes in environmental conditions (Du et al. 2020). Given that OsAL50 mutations were only detected in the promoter region in the osal50 mutant, a 1300-bp region located 2100‒3400 bp upstream of the OsAL50 start codon and including the primary locations of these mutation sites was selected for in-depth analysis to understand the regulatory mechanism of OsAL50 expression. Sequences of this region were analyzed using PlantCARE, which revealed the presence of a cluster of light-responsive G-box cis-acting elements, meristem expression-related CAT-box elements, MeJA-responsive CGTCA- and TGACG-motifs, light-responsive TCT-motifs and Sp1 elements, salicylic acid-responsive TCA-elements, and auxin-responsive TGA-elements, in addition to abundant core promoter elements including TATA- and CAAT-boxes (Online Resource 7). Thus, it appears likely that OsAL50 serves as a regulator of photosynthesis in rice, given the abundant light-responsive elements in its promoter region, with the promoter sequence covering these mutations being essential for OsAL50 function.
Multiple sequence alignment revealed that OsAL50 shares high sequence similarity with homologous proteins from Zea mays, Brachypodium distachyon, Sorghum bicolor, Triticum aestivum, Hordeum vulgare, and Setaria italica, with identities of 74.45%, 85%, 86.35%, 86.57%, 87.41%, and 88.32%, respectively (Fig. 7a). Notably, genes homologous to OsAL50 in these plant species were predicted to encode an RNase E-like protein according to annotations in the NCBI and Phytomozome databases (https://phytozome-next.jgi.doe.gov/). Moreover, this protein is localized to the thylakoids, where it plays a crucial role in the processing of plastid RNA and is required for chloroplast development and biogenesis (Schein et al. 2008; Mudd et al. 2008; Walter et al. 2010). Furthermore, a phylogenetic tree was constructed to investigate the possible relationships between OsAL50 homologs in evolutionary history. As shown in Fig. 7b, they could be divided into two subgroups: the proteins from monocotyledonous plants were placed clearly into one subgroup, while the proteins from dicotyledonous plants formed another subgroup (Fig. 7b). These data indicate that OsAL50 is highly conserved in higher plants, particularly among the Gramineae. To determine whether OsAL50 was localized to the chloroplasts, the pOsAL50-GFP vector was introduced into rice protoplasts. The green fluorescent signals from the OsAL50-GFP fusion proteins overlapped with the chloroplast autofluorescence (Online Resource 8). Considering these findings, we inferred that OsAL50 most likely encodes an RNase E-like protein localized to the chloroplasts that is essential for chlorophyll biosynthesis and chloroplast development in rice.
Sequence analysis of the OsAL50 protein. a Alignment of derived amino acid sequences of OsAL50 and its homologous proteins. Dark blue highlights identical residues, while similar residues are marked in pink or light blue. b Phylogenetic tree representing the alignment of OsAL50 and homologous protein sequences constructed by MEGA 7.0 using the neighbor-joining method with 1000 replications. All protein sequences were obtained from the NCBI database, with corresponding GenBank accession numbers shown in parentheses. The scale indicates the percentage substitution rate per site
Expression patterns of genes associated with photosynthetic pigment metabolism, chloroplast development, and photosynthesis in osal50
Both the leaves and panicles of osal50 mutant plants exhibited distinctive growth stage-dependent albino characteristics during the regular growing season. Changes in the coloration of these tissues and organs are associated with photosynthetic pigment metabolism, chloroplast development, and photosynthesis. We therefore investigated the transcript levels of 25 genes involved in these processes in various tissues of wild-type and osal50 mutant plants during different stages of development using qRT-PCR analysis. These included 11 genes associated with chlorophyll and carotenoid biosynthesis, encoding glutamyl-tRNA reductase (OsHEMA1; Schmied et al. 2011), chlorophyllide a oxygenase 1 (OsCAO1; Yang et al. 2016), chlorophyll synthase (OsCHLa/b; Karlin-Neumann 1988), magnesium chelatase subunits I and D (OsCHLI and OsCHLD; Zhang et al. 2006), Mg-protoporphyrin IX methyltransferase (OsCHLM; Tomiyama et al. 2014), uroporphyrinogen-III synthase (OsHEMD), 3,8-divinyl reductase (OsDVR; Wang et al. 2010), and phytoene synthases 1, 2, and 3 (OsPSY1, OsPSY2, and OsPSY3; Yang et al. 2021). Further analysis was conducted on six chloroplast development-related genes encoding guanylate kinase (V2 and V3; Sugimoto et al. 2007; Yoo et al. 2009), plastid division protein (Ftsz; Gong et al. 2014), RNA polymerase β subunit (OsRPOB; Inada et al. 1996), plastid RNA polymerase (OsRPOTP; Kusumi et al. 2004), and plastid polI-like DNA polymerase (OsPOLP; Takeuchi et al. 2007), as well as eight genes related to photosynthesis, encoding the light-harvesting chlorophyll a/b-binding protein 1/2R of PSII (OsCAB1R and OsCAB2R; Matsuoka 1990), photosystem I/II reaction-center protein A/B (OsPSAA and OsPSBA; Kapoor et al. 1994), cytochrome subunits f and B6 (OsPETA and OsPETB; Shang et al. 2019; Zhang et al. 2019), and the small and large subunits of soluble Rubisco proteins (OsRBCS and OsRBCL; Kyozuka et al. 1993). All of these genes were significantly down-regulated in osal50 mutant leaves compared with wild-type leaves at the four-leaf stage (Fig. 8a), with consistent expression patterns of transcriptional suppression in young panicles, including glumes and branches, of osal50 at the heading stage, except for the chloroplast development-related FtsZ, OsRPOB, and V3 genes (Fig. 8c, d). Moreover, the expression levels of all of these genes were markedly increased in osal50 mutant leaves at the seven-leaf stage (when the leaves turn green) compared with those at the four-leaf stage (Fig. 8b), with OsCHLI, OsCHLD, OsDVR, OsPSAA, OsPSBA, OsRBCL, OsPOLP, and V2 expression restored to wild-type levels and OsHEMA1, OsCAB1R, OsPETA, OsPETB, and OsRBCS expression levels in the osal50 mutant even higher than those in the wild type at the seven-leaf stage (Fig. 8b). These findings indicate that OsAL50 plays a vital role in the regulation of gene expression associated with chlorophyll biosynthesis, chloroplast development, and photosynthesis in rice.
Expression patterns of genes associated with photosynthetic pigment metabolism, photosynthesis, and chloroplast development in wild-type GP50 and osal50 mutant plants, determined by qRT-PCR. a, b Relative expression in wild-type and osal50 mutant leaves at the four- (a) and seven-leaf (b) stages. c, d Relative expression in wild-type and osal50 glumes (c) and branches (d) from young panicles at the heading stage. Relative gene expression was calculated using the comparative 2−△△CT method, OsActin served as a normalization control. Error bars indicate SD (n = 3). Student’s t-test was used for statistical analysis, *P < 0.05, **P < 0.01
Discussion
In addition to being a staple food crop throughout the world, rice also serves as a model monocotyledon plant in research settings. Variegated mutants with altered leaf or panicle coloration have long been the focus of intensive scientific interest in rice, particularly in the case of albino mutants (Peng et al. 2012). Efforts to characterize the genetic basis of these albino mutants have provided new insights into the molecular mechanisms governing leaf and panicle coloration and the roles that nuclear genes play in shaping chloroplast development and chlorophyll biosynthesis (Chen et al. 2009). Here, we identified and characterized a variegated rice mutant, osal50, which exhibits a relatively normal green phenotype through the L2 stage (Fig. 1a, b), during which the second leaf blade is well-expanded, but develops longitudinal white-striped leaves through the four-leaf stage (Fig. 1c, d); these gradually turn green such that plants exhibit normal green coloration at the seven-leaf stage (Fig. 1f). Additionally, osal50 plants exhibit mixed phenotypes at the tillering stage, with many white-striped leaves and fewer green leaves on a given plant (Fig. 1g), together with white panicles that are evident at the flowering stage and distinct from those exhibited by wild-type GP50 plants (Fig. 1i, j). The present findings imply that, in contrast to the majority of variegated mutants in rice that have been documented to date, the osal50 mutant phenotype is highly growth stage-dependent. Previous reports have described many mutants associated with albinism before the three-leaf stage that become more noticeable during the two-leaf stage, but the leaves return normal green coloration following the four-leaf stage, including gra(t) (Chen et al. 2009), hw-1(t) (Guo et al. 2012), graS (Du et al. 2020), albg (Jian et al. 2017), and ysa (Su et al. 2012). The osal50 mutant exhibits an albino leaf phenotype in a very different growth stage-dependent manner from these mutants under natural field conditions. Moreover, the albino phenotype exhibited by osal50 plants is unaffected by temperature, instead changing only over the course of development; this is in contrast to other previously reported green-revertible albino mutants, where leaf color is sensitive to temperature, including cisc (Lan et al. 2010), gra(t) (Chen et al. 2009), st1 (Yoo et al. 2009), v3 (Yoo et al. 2009), v2 (Sugimoto et al. 2007), and v1 (Kusumi et al. 2011). The phenotypic characteristics of osal50 mutant plants are also distinct from those of most other mutants with white panicles described to date, including wp1 (Wang et al. 2016), wp2 (Wang et al. 2021), wp3 (Li et al. 2018a), wsp1 (Zhang et al. 2017), wlp1 (Song et al. 2014), and wlp2 (Lv et al. 2017). Of these, the wp2 and wlp2 mutants exhibit more pronounced albinism under higher temperatures, while wsp1 and wlp1 mutant phenotypes are particularly sensitive to cold exposure. The wp3 mutant exhibits white panicles and white-striped leaves, and the generation of white-striped leaves in wp3 mutant plants is independent of changes in environmental conditions, such as temperature and light levels. Unusually, the white panicles in the wp3 mutant plants exhibit typical Mendelian inheritance, whereas the white-striped leaves instead show a pattern of maternal inheritance.
In the present study, map-based cloning confined the OsAL50 locus to a 160-kb physical interval on chromosome 1, flanked by the CAPS-08 and CAPS-37 SNP markers (Fig. 4). This region contained 26 annotated genes, of which 10 encoded transposons or retrotransposons, 13 encoded hypothetical or expressed proteins of unknown function, and 3 (LOC_Os01g19970, LOC_Os01g20140, and LOC_Os01g20160) encoded proteins with predicted functions (Online Resource 5). LOC_Os01g19970 is predicted to encode a member of the MYB transcription factor family, which is among the largest transcription factor families in plants, with members serving as key regulators of diverse processes related to development, secondary metabolism, hormone signaling, organ morphogenesis, cell differentiation, disease resistance, stress tolerance, and cell cycle progression (Katiyar et al. 2012). Several MYB transcription factors have recently been found to be regulators of anthocyanin and flavonoid biosynthesis in plants, thereby influencing the coloring of leaves, fruits, flowers, and other tissues (Li et al. 2019). LOC_Os01g20140 and LOC_Os01g20160 are respectively predicted to encode a latency-associated nuclear antigen (LANA) and an OsHKT1;5-Na+ transporter. LANAs are among the most widely expressed nuclear proteins during latency periods, serving as essential regulators of a range of nuclear processes including the recruitment of DNA replication machinery and proper chromatid segregation into daughter cells. The OsHKT1;5-Na+ transporter is found in the plasma membrane and the phloem of diffuse vascular bundles in basal nodes. It has crucial functions in preventing the transmission of Na+ to young leaf blades by excluding Na+ in the phloem. Furthermore, it performs a crucial role over different rice growth stages by safeguarding the subsequent generation of seeds and leaf blades against the detrimental effects of salt stress (Kobayashi et al. 2017). BLAST searches of available genomic and protein sequences indicated that multiple copies of LOC_Os01g20140 are present within the rice genome and that it does not exhibit any clear association with leaf coloration. As such, we focused on the two other functionally annotated genes, LOC_Os01g19970 and LOC_Os01g20160. No variations in allelic gene sequences between the wild type and the osal50 mutant were found when we amplified and sequenced the promoter and coding regions of these potential genes in both parental lines (data not shown). Sequence and expression analyses ultimately led to the identification of LOC_Os01g20110 as the OsAL50 candidate gene (Fig. 5), which was confirmed by knockout using the CRISPR/Cas9 system (Fig. 6). In total, 20 mutation sites were identified within the LOC_Os01g20110 promoter region in the osal50 mutant (Fig. 5d, e). Furthermore, a combination of phylogenetic analysis and multiple sequence alignment suggested that OsAL50 likely encodes an RNase E-like protein (Fig. 7). RNase functions by catalyzing the cleavage of phosphodiester bonds within substrate RNA molecules, with Endo-RNase generating RNA fragments by cleaving internal sequences. In contrast, Exo-RNase removes individual nucleotides from the 5’ or 3’ ends of RNA substrates (Ait-Bara and Carpousis 2015). RNase E is an Endo-RNase widely distributed throughout chloroplasts and bacterial species. In Escherichia coli, where it has been the most thoroughly studied. RNase E functions as a comprehensive regulator of gene expression, regulating various aspects of RNA metabolism. This includes the processing, maturation, and quality control of stable RNA molecules, the degradation of non-coding regulatory RNA sequences and RNA primers involved in DNA replication, as well as the turnover of mRNA molecules (Ait-Bara and Carpousis 2015; Schein et al. 2008). Higher plants include homologs of bacterial RNase E that are encoded in the nuclear genome. Their responsibility lies in the breakdown of RNA within the chloroplast, an organelle originating from a prokaryote resembling cyanobacteria, via a degradation pathway facilitated by polyadenylation (Schein et al. 2008). In the research they performed, Mudd et al. (2008) provided evidence that Arabidopsis contains a single nuclear gene, At2g04270, which produces a protein with a conserved catalytic domain similar to that of the RNase E/G-like protein found in chloroplasts. This gene, At2g04270, is essential for the appropriate development of chloroplasts and autotrophic growth. Additionally, it is expressed together with other nuclear genes involved in regulating RNA metabolism in plastids. The protein displays ribonucleolytic activity and influences the accumulation of plastid RNA. Plants with a homozygous lack of At2g04270 function display a pale green in color. The plastids in the leaves of these mutant plants are smaller, with fewer thylakoids and shorter grana stacks, compared to the chloroplasts of wild-type plants. The albino phenotype exhibited by the osal50 mutant, derived from a breeding population of the japonica cultivar GP50, is also associated with abnormal chloroplast morphology and development. While some chloroplast-like organelles are observed within cells from the white portions of osal50 plant tissues, they lack well-developed grana lamellar stacks and organized thylakoid structures, contributing to the white-striped leaves and white panicles that characterize these mutant plants (Fig. 3k–p). Given these findings, we speculate that impaired expression of OsAL50, which is predicted to encode an RNase E-like protein necessary for proper chloroplast development, leads to the albino phenotype observed in osal50 mutant rice plants. This impaired expression likely results from an abnormal promoter structure.
Plant chloroplasts retain their own genomic and translational systems, and chloroplast development is regulated by both nuclear and plastid genomes (Zheng et al. 2019). Moreover, chloroplasts harbor both nuclear- and plastid-encoded RNA polymerases (NEP and PEP, respectively), with coordination between the two being essential for normal plastid development and the biosynthesis of chlorophyll (Kusumi and Iba 2014). Loss of function of proteins associated with the PEP and NEP could lead to an albino phenotype (Zheng et al. 2019). Glutamyl-tRNA facilitates switching between NEP and PEP utilization during chloroplast development in Arabidopsis (Hanaoka et al. 2005). In the present study, we investigated expression levels of chlorophyll biosynthesis and chloroplast development-related genes in various tissues of wild type and osal50 during different stages of growth and development. The qRT-PCR analysis revealed down-regulation of genes associated with glutamyl-tRNA synthetase, involved in the chlorophyll biosynthetic pathway, in osal50 mutant leaves at the fourth-leaf stage and young panicles at the heading stage; these included genes such as HEMA1 and HEMD, involved in the processing of glutamyl-tRNA to aminolevulinic acid (ALA) and ALA to protoporphyrin IX, as well as genes involved in the processing of protoporphyrin to chlorophyll a and b (OsCHLI, OsCHLH, OsCHLM, OsCHLD, OsCAO1, and OsDVR) (Fig. 8a, c, d). In addition, nuclear-encoded genes (e.g., OsCAB1R, OsCAB2R, OsRPOTP, and RbcS), as well as plastid-encoded genes (e.g., OsPSAA, OsPSBA, OsPOLP, OsRPOB, and RbcL) related to photosynthesis and chloroplast development, exhibited comparable patterns of transcriptional suppression in osal50 mutant leaves and young panicles, the two albino stages (Fig. 8a, c, d); however, at the seven-leaf stage (when the leaves turn green), the transcript levels of these genes in osal50 mutant leaves were largely comparable to those in the wild type (Fig. 8b). These expression patterns closely matched the growth stage-dependent albino phenotype observed in osal50 mutant plants. Our findings demonstrate that OsAL50 is essential for regulating chlorophyll biosynthesis and chloroplast development in rice and that impaired OsAL50 expression disrupts the mRNA transcript levels of genes involved in these processes.
Conclusion
We identified and characterized a naturally occurring rice albino mutant designated osal50 (oryza sativa albino leaf 50), derived from a breeding population of the japonica cultivar GP50. Leaves and panicles of mutant plants displayed distinctive albino characteristics in a growth stage-dependent manner under natural field conditions. Moreover, the osal50 mutant exhibited decreased chlorophyll and carotenoid accumulation and abnormal chloroplast ultrastructure. Map-based cloning delimited OsAL50 to a region of 160-kb on chromosome 1 flanked by two SNP markers, CAPS-08 and CAPS-37; this region contained 26 putative open reading frames. Sequence and expression analyses revealed LOC_Os01g20110 as the candidate gene of OsAL50, which was confirmed by knockout using CRISPR/Cas9. In total, 20 mutation sites were detected in the LOC_Os01g20110 promoter region, resulting in severe suppression of mRNA expression. Subcellular localization and protein sequence analyses suggested that OsAL50 likely encodes a chloroplast-localized endoribonuclease E-like protein. Further investigation indicated that OsAL50 plays a vital role in the regulation of chlorophyll metabolism, chloroplast development, and photosynthesis. Together, these findings provide new molecular insights into the genetic basis for changes in leaf and panicle coloration in rice.
References
Ait-Bara S, Carpousis AJ (2015) RNA degradosomes in bacteria and chloroplast: classification, distribution and evolution of RNase E homologs. Mol Microbiol 97:1021–1035
Bae CH, Abe T, Matsuyama T, Fukunishi N, Nagata N, Nakano T, Kaneko Y, Miyoshi K, Matsushima H, Yoshida S (2001) Regulation of chloroplast gene expression is affected in ali, a novel tobacco albino mutant. Ann Bot 88:545–553
Chen T, Zhang YD, Zhao L, Zhu Z, Lin J, Zhang SB, Wang CL (2009) Fine mapping and candidate gene analysis of a green-revertible albino gene gra(t) in rice. J Genet Genomics 36:117–123
Chen YP, Zhai Z, Yang WJ, Sun J, Shu XL, Wu DX (2015) Genetic analysis and fine mapping of St-wp gene in mutant rice with stripe white leaf and white panicle. Acta Agric Nucl Sin 29:1246–1252 (Chinese with English abstract)
Chen P, Hu HT, Zhang Y, Wang ZW, Dong GJ, Cui YT, Qian Q, Ren DY, Guo LB (2018) Genetic analysis and fine-mapping of a new rice mutant, white and lesion mimic leaf1. Plant Growth Regul 85:425–435
Deng XJ, Zhang HQ, Wang Y, HeF LJ, XiaoX S, Li W, Wang GH, Wang GL (2014) Mapped clone and functional analysis of lear-color gene Ygl7 in a rice hybrid (Oryza sativa L. spp. indica). PLoS ONE 9:e99564
Du ZX, Hao HY, He JP, Wang JP, Huang Z, Xu J, Fu HH, Fu JR, He HH (2020) GraS is critical for chloroplast development and affects yield in rice. J Integr Agr 19:2603–2615
Dunford R, Walden RM (1991) Plastid genome structure and plastid-related transcript levels in albino barley plants derived from another culture. Curr Genet 20:339–347
Gong XD, Su QQ, Lin DZ, Jiang Q, Xu JL, Zhang JH, Teng S, Dong YJ (2014) The rice OsV4 encoding a novel pentatricopeptide repeat protein is required for chloroplast development during the early leaf stage under cold stress. J Integr Plant Biol 56:400–410
Guo T, Huang X, Huang YX, Liu YZ, Zhang JG, Chen ZQ, Wang H (2012) Characterizations of a mutant gene hw-1(t) for green-revertible albino, high tillering and dwarf in rice (Oryza sativa L.). Acta Agron Sin 38:23–35 (Chinese with English abstract)
Han B, Xue YB, Li JY, Deng XW, Zhang QF (2007) Rice functional genomics research in China. Philos T R Soc B 362:1009–1021
Hanaoka M, Kanamaru K, Fujiwara M, Takahashi H, Tanaka K (2005) Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep 6:545–550
Hsieh MH, Googman HM (2005) The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138:641–653
Inada H, Kusumi K, Nishimura M, Iba K (1996) Specific expression of the chloroplast gene foe RNA polymerase (rpoB) at an early stage of leaf development in rice. Plant Cell Physiol 37:229–232
International Rice Genome Sequencing Project (2005) The mapbased sequence of the rice genome. Nature 436:793–800
Jian L, Wang ZK, Zeng DD, Qian R, Shi CH, Jin XL (2017) Characterization and fine mapping of a green-revertible albino (albg) mutant in rice. Acta Agric Nucl Sin 31:2289–2297 (Chinese with English abstract)
Kapoor S, Maheshwari SC, Tyagi AK (1994) Developmental and light-dependent cues interact to establish steady-state levels of transcripts for photosynthesis-related genes (pabA, psbD, psaA and rbcL) in rice (Oryza sativa L.). Curr Genet 25:362–366
Karlin-Neumann GA, Sun L, Tobin EM (1988) Expression of light-harvesting chlorophyll a/b-protein genes is phytochrome regulated in etiolated Arabidopsis thaliana seedlings. Plant Physiol 88:1323–1331
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544
Kobayashi NI, Yamaji N, Yamamoto H, Okubo K, Ueno H, Costa A, Tanoi K, Matsumura H, Fujii-Kashino M, Horiuchi T, Nayef MA, Shabala S, An G, Ma JF, Horie T (2017) OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J 91:657–670
Kuaumi K, Yara A, Mitsui N, Tozawa Y, Iba K (2004) Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol 45:1194–1201
Kusumi K, Iba K (2014) Establishment of the chloroplast genetic system in rice during early leaf development and at low temperatures. Front Plant Sci 5:386
Kusumi K, Sakata C, Nakamuta T, Kawasaki S, Yoshimura A, Iba K (2011) A plastid protein NUS1 is essential for buildup of the genetic system for early chloroplast development under cold stress conditions. Plant J 68:1039–1050
Kyozuka J, Mcelroy D, Hayakawa T, Xie Y, Wu R, Shimamoto K (1993) Light-regulated and cell-specific expression of tomato rbcS-gusA and rice rbcS-gusA fusion genes in transgenic rice. Plant Physiol 102:991–1000
Lan T, Wang B, Ling QP, Xu CH, Tong ZJ, Liang KJ, Duan YL, Wu WR (2010) Fing mapping of cisc(t), a gene for cold-induced seedling chlorosis, and identification of its candidate in rice. Chin Sci Bull 55:2183–2187 (Chinese with English abstract)
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li HC, Ji GB, Wang Y, Qian Q, Xu JC, Sodmergen LG, Zhao XF, Chen MS, Zhai WX, Li DY, Zhu LH (2018a) WHITE PANICLE 3, a novel nucleus-encoded mitochondrial protein, is essential for proper development and maintenance of chloroplasts and mitochondria in rice. Front Plant Sci 9:762
Li X, He Y, Yang J, Jia YH, Zeng HL (2018b) Gene mapping and transcriptome profiling of a practical photo-thermo-sensitive rice male sterile line with seedling-specific green-revertible albino leaf. Plant Sci 266:37–45
Li JL, Han GL, Sun CF, Sui N (2019) Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal Behav 14:e1613131
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Method Enzymol 148:350–382
Liu RH, Meng JL (2003) MapDraw: a Microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25:317–321
Long D, Martin M, Sundberg E, Swinburne J, Puangsomlee P, Coupland G (1993) The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Ds insertion. Proc Natl Acad Sci USA 90:10370–10374
Lv YS, Shao GN, Qiu JH, Jiao GA, Sheng ZH, Xie LH, Wu YW, Tang SQ, Wei XJ, Hu PS (2017) White leaf and Panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J Exp Bot 68:5147–5160
Lv J, Shang LG, Chen Y, Han Y, Yang XY, Xie SZ, Bai WQ, Hu MY, Wu H, Lei KR, Yang YN, Ge SZ, Trinh HP, Zhang Y, Guo LB, Wang ZW (2020) OsSLC1 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling survival. Rice 13:25
Matsuoka M (1990) Classification and characterization of cDNA that encodes the light-harvesting chlorophyll a/b binding protein of photosystem II from rice. Plant Cell Physiol 31:519–526
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Mudd EA, Sullivan S, Gisby MF, Mironov A, Kwon CS, Chung W, Day A (2008) A 125 kDa RNase E/G-like protein in present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J Exp Bot 59:2597–2610
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Peng Y, Zhang Y, Lv J, Zhang JH, Li P, Shi XL, Wang YF, Zhang HL, He ZH, Teng S (2012) Characterization and fine mapping of a novel rice albino mutant low temperature albino 1. J Genet Genomics 39:385–396
Rao YC, Yang YL, Xu J, Li XJ, Leng YJ, Dai LP, Huang LC, Shao GS, Ren DY, Hu J, Guo LB, Pan JW, Zeng DL (2015) EARLY SENESCENCE 1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol 169:1225–1239
Schein A, Sheffy-Levin S, Glaser F, Schuster G (2008) The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 14:1057–1068
Schmied J, Hedtke B, Grimm B (2011) Overexpression of HEMA1 encoding glutamyl-tRNA reductase. J Plant Physiol 168:1372–1379
Shang LN, Chen XL, Mi SN, Wei G, Wang L, Zhang YY, Lei T, Lin YX, Huang LJ, Zhu MD, Wang N (2019) Phenotypic identification and gene mapping of temperature-sensitive green-revertible albino mutant tsa2 in rice (Oryza sariva L.). Acta Agron Sin 45:662–675 (Chinese with English abstract)
Song J, Wei XJ, Shao GN, Sheng ZH, Chen DB, Liu CL, Jiao GA, Xie LH, Tang SQ, Hu PS (2014) The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol Biol 84:301–314
Su N, Hu ML, Wu DX, Wu FQ, Fei GL, Lan Y, Chen XL, Shu XL, Zhang X, Wan JM (2012) Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol 159:227–238
Su Y, Hu SK, Zhang B, Ye WJ, Niu YF, Guo LB, Qian Q (2017) Characterization and fine mapping of a new early leaf senescence mutant es3(t) in rice. Plant Growth Regul 81:419–431
Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K (2007) The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J 52:512–527
Takeuchi R, Kimura S, Saotome A, Sakaguchi K (2007) Biochemical properties of a plastidial DNA Polymerase of rice. Plant Mol Biol 64:601–611
Tomiyama M, Inoue SI, Tsuzuki T, Soda M, Morimoto S, Okigaki Y, Ohishi T, Mochizuki N, Takahashi K, Kinoshita T (2014) Mg-chelatase I subunit 1 and Mg-protoporphyrin IX methyltransferase affect the stomatal aperture in Arabidopsis thaliana. J Plant Res 127:553–563
Walter M, Piepenburg K, Schottler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R (2010) Konckout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J 64:851–863
Wang PR, Gao JX, Wan CM, Zhang FT, Xu ZJ, Huang XQ, Sun XQ, Deng XJ (2010) Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiol 153:994–1003
Wang YL, Wang CM, Zheng M, Lyu J, Xu Y, Li XH, Niu M, Long WH, Wang D, Wang HY, Terzaghi W, Wang YH, Wang JM (2016) WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol 170:2110–2123
Wang ZW, Lv J, Xie SZ, Zhang Y, Qiu ZN, Chen P, Cui YT, Niu YF, Hu SK, Jiang HZ, Ge SZ, Trinh HP, Lei KR, Bai WQ, Zhang Y, Guo LB, Ren DY (2018) OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul 84:249–260
Wang YL, Wang YH, Ren YL, Duan EC, Zhu XP, Hao YY, Zhu JP, Chen RB, Lei J, Teng X, Zhang YY, Wang D, Zhang X, Guo XP, Jiang L, Liu SJ, Tian YL, Liu X, Chen LM, Wang HY, Wang JM (2021) White panicle2 encoding thioredoxin z, regulates plastid RNA editing by interacting with multiple organellar RNA editing factors in rice. New Phytol 229:2693–2706
Wei XY, Zeng YH, Zhang R, Huang JH, Yang WX, Zou WG, Xu XM (2019) Fine mapping and identification of the rice blast-resistance locus Pi-kf2(t) as a new member of the Pi2/Pi9 multigene family. Mol Breed 39:108
Wei XY, Zeng YH, Yang WX, Xiao CC, Hou XP, Huang JH, Zou WG, Xu XM (2023) Development of high-quality fragrant indica CMS line by editing Badh2 gene using CRISPR-Cas9 technology in rice (Oryza sativa L.). Acta Agron Sin 49:2144–2159 (Chinese with English abstract)
Wu LW, Ren DY, Hu SK, Li GM, Dong GJ, Jiang L, Hu XM, Ye WJ, Cui YT, Zhu L, Hu J, Zhang GH, Gao ZY, Zeng DL, Qian Q, Guo LB (2016) Mutation of OsNaPRT1 in the NAD salvage pathway leads to withered leaf tips in rice. Plant Physiol 171:1085–1098
Xu JM, Yang J, Wu ZC, Liu HL, Huang FL, Wu YR, Carrie C, Narsai R, Murcha M, Whelan J, Wu P (2013) Identification of a Dual-Targeted protein belonging to the mitochondrial carrier family that is required for early leaf development in rice. Plant Physiol 161:2036–2048
Yang YL, Xu J, Huang LC, Leng YJ, Dai LP, Rao YC, Chen L, Wang YQ, Tu ZJ, Hu J, Ren DY, Zhang GH, Zhu L, Guo LB, Qian Q, Zeng DL (2016) PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot 67:1279–1310
Yang F, Debatosh D, Song T, Zhang JH (2021) Light harvesting-like protein 3 interacts with phytoene synthase and is necessary for carotenoid and chlorophyll biosynthesis in rice. Rice 14:32
Yoo SC, Cho SH, Sugimoto H, Li JJ, Kusumi K, Koh HJ, Iba K, Paek NC (2009) Rice Virescent3 and Stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150:388–401
Zhang HT, Li JJ, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC (2006) Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62:325–337
Zhang ZG, Cui XA, Wang YW, Wu JX, Gu XF, Lu TG (2017) The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol Plant 10:86–98
Zhang T, Feng P, Li YF, Yu P, Yu GL, Sang XC, Ling YH, Zeng XQ, Li YD, Huang JY, Zhang TQ, Zhao FM, Wang N, Zhang CW, Yang ZL, Wu RH, He GH (2018) VIRESCENT-ALBINO LEAF 1 regulates leaf color development and cell division in rice. J Exp Bot 69:4791–4804
Zhang LS, Mi SN, Wang L, Wei G, Zheng YJ, Zhou K, Shang LN, Zhu MD, Wang N (2019) Physiological and biochemical analysis and gene mapping of a novel short radicle and albino mutant sra1 in rice. Acta Agron Sin 45:556–567 (Chinese with English abstract)
Zheng H, Wang ZR, Tian YL, Liu LL, Lv F, Kong WY, Bai WT, Wang PR, Wang CL, Yu XW, Liu X, Jiang L, Zhao ZG, Wang JM (2019) Rice albino 1, encoding a glycyl-tRNA synthetase, is involved in chloroplast development and establishment of the plastidic ribosome system in rice. Plant Physiol Biochem 139:495–503
Zhou KN, Xia JF, Ma TC, Wang YL, Li ZF (2018) Mapping and mutation analysis of stripe leaf and white panicle gene SLWP in rice. Chin J Rice Sci 32:325–334 (Chinese with English abstract)
Acknowledgements
This study was supported by the China Agriculture Research System of MOF and MARA (CARS-01), the Fujian Provincial Natural Science Foundation (2021J01535, 2021J01536), and the Sanming Municipal Science and Technology Project (2022-N-7).
Funding
Funding was supported by Agriculture Research System of China,CARS-01 ,Natural Science Foundation of Fujian Province,2021J01536, 2021J01535, Sanming Municipal Science and Technology Project,2022-N-7.
Author information
Authors and Affiliations
Contributions
YZ and XW contributed equally to this work. YZ carried out molecular-marker development, genetic analysis, molecular mapping, candidate gene analysis, physical-map and phylogenetic-tree construction, and wrote the manuscript. XW carried out molecular-marker development, genetic analysis, molecular mapping and expression analysis. CX and RZ participated in genetic analysis, molecular mapping, and physical-map construction. JH carried out mapping-population construction and participated in genetic analysis and molecular mapping. XX designed the research and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Dawei Xue.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10725_2023_1116_MOESM4_ESM.pdf
Supplementary file4 (PDF 554 KB)—Primers used for expression analysis of genes associated with chloroplast development, chlorophyll biosynthesis, and photosynthesis
10725_2023_1116_MOESM5_ESM.pdf
Supplementary file5 (PDF 554 KB)—Functional annotation of 26 candidate genes for OsAL50 based on the RGAP http://rice.plantbiology.msu.edu/
10725_2023_1116_MOESM6_ESM.pdf
Supplementary file6 (PDF 457 KB)—Mutation analysis within the promoter region of LOC_Os01g20110 in 20 rice varieties, where “--”indicates base deletion
10725_2023_1116_MOESM8_ESM.pdf
Supplementary file8 (PDF 404 KB)—Subcellular localization of OsAL50 in rice protoplasts. a Observation of free green fluorescent protein (GFP) signals in rice protoplasts. b Observation of OsAL50-GFP fluorescent signals in rice protoplasts. Bars: 10 μm
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeng, Y., Wei, X., Xiao, C. et al. Fine mapping and identification of a novel albino gene OsAL50 that is required for chlorophyll biosynthesis and chloroplast development in rice (Oryza sativa L.). Plant Growth Regul 103, 389–407 (2024). https://doi.org/10.1007/s10725-023-01116-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-023-01116-8