Abstract
Shoot branching is essential in ornamental chrysanthemum production and determines final plant shape and quality. Auxin is associated with apical dominance to indirectly inhibit bud outgrowth. Two non-mutually exclusive models exist for indirect auxin inhibition. Basipetal auxin transport inhibits axillary bud outgrowth by limiting auxin export from buds to stem (canalization model) or by increasing strigolactone levels (second messenger model). Here we analyzed bud outgrowth in treatments with auxin (IAA), strigolactone (GR24) and auxin transport inhibitor (NPA) using a split-plate bioassay with isolated chrysanthemum stem segments. Besides measuring bud length, dividing cell percentage was measured with flow cytometry and RT-qPCR was used to monitor expression levels of genes involved in auxin transport (CmPIN1) and signaling (CmAXR2), bud dormancy (CmBRC1, CmDRM1) and strigolactone biosynthesis (CmMAX1, CmMAX3). Treatments over a 5-day period showed bud outgrowth in the control and inhibition with IAA and IAA + GR24. Bud outgrowth in the control coincided with high dividing cell percentage, decreased expression of CmBRC1 and CmDRM1 and increased CmPIN1 expression. Inhibition by IAA and IAA + GR24 coincided with low dividing cell percentage and unchanged or increased expressions of CmBRC1, CmDRM1 and CmPIN1. Treatment with GR24 showed restricted bud outgrowth that was counteracted by NPA. This restricted bud outgrowth was still concomitant with a high dividing cell percentage and coincided with decreased expression of dormancy genes. These results indicate incomplete inhibition of bud outgrowth by GR24 treatment and suggest involvement of auxin transport in the mechanism of bud inhibition by strigolactones, supporting the canalization model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shoot architecture is a key aspect in the quality of ornamental plants. The garden mum and florist Chrysanthemum, Chrysanthemum morifolium (Ramat), is worldwide an economically important ornamental crop as cut flowers, garden plants and potted plants. A great number of varieties and shapes are produced by breeding and pruning to influence plant shape (Anderson 2007). Cut flowers require laborious pruning, while garden and potted varieties need to be compact and highly branched. Therefore investigating the physiological and genetic regulation of axillary bud outgrowth could offer potential treatments to control the branching process or genetic markers for breeding. Chrysanthemum has also been proposed as a suitable ornamental model plant for in-vitro research due to genotype-independent phenotypic plasticity (Shinoyama et al. 2006). The variety in plant habit is largely determined by the formation of secondary axes through axillary meristem development and shoot branching during post-embryonic growth, for review see McSteen and Leyser (2005). The regulation of axillary meristem and axillary bud outgrowth reflects the plasticity of the plant to the environment involving external cues (temperature, light, nutrients, decapitation) and endogenous control (plant hormones, sucrose). Domagalska and Leyser (2011) and Rameau et al. (2015) extensively reviewed these interactions. Endogenously, the main regulators that are ascribed to the control of bud outgrowth are sucrose (Mason et al. 2014; Barbier et al. 2015) and the cross-talk of plant hormones auxins, cytokinins and strigolactones (Ongaro and Leyser 2008; Ferguson and Beveridge 2009). Strigolactone is the most recent plant hormone that was proposed to play a part in shoot branching (Gomez-Roldan et al. 2008; Umehara et al. 2008); the exact control mechanism is still subject of debate. The two most important hypotheses about the role of strigolactone in shoot branching are the auxin canalization model (Bennett and Leyser 2006; Leyser 2009) and the second messenger model (Brewer et al. 2009). The canalization model states that auxin transport canalization from the axillary buds to the stem is necessary for bud outgrowth and that auxin in the main stem, along with strigolactone, influences auxin transport to inhibit bud outgrowth. The auxin transport happens from shoot to root (basipetal) and is canalized by the mobilization of PIN1 auxin efflux proteins in the basal membrane of xylem parenchyma cells. In this way a polar auxin transport (PAT) is established in the stem (Gälweiler et al. 1998; Friml et al. 2003). Strigolactone transport happens from root to shoot (acropetal) (Kohlen et al. 2011; Xie et al. 2015b) and involves PDR proteins as was shown in Petunia and Nicotiana tabacum (Kretzschmar et al. 2012; Xie et al. 2015a). According to the canalization model, the ability to transport auxin to the stem determines the outgrowth of axillary buds and these compete with the PAT in the stem and other axillary buds. The role of strigolactone in this model is to dampen the mobilization of PIN1 proteins at the axillary buds, and inhibits the auxin transport to the stem and consequent bud outgrowth (Ongaro and Leyser 2008). According to the second messenger model, auxin regulates the production of strigolactones, which are transported to the axillary buds to inhibit outgrowth through the involvement of a bud-specific transcription factor BRC1 (Domagalska and Leyser 2011). These models are not mutually exclusive as there is experimental evidence in support of both the canalization model (Prusinkiewicz et al. 2009; Crawford et al. 2010; Shinohara et al. 2013) and the second messenger model (Brewer et al. 2009, 2015; Dun et al. 2013). The main arguments in support of the auxin canalization model are the observation that auxin transport through the mobilization of PIN1 proteins is dampened by strigolactone (Crawford et al. 2010; Shinohara et al. 2013) and the fact that a competing auxin source is required to allow strigolactone inhibition of axillary bud outgrowth (Prusinkiewicz et al. 2009; Liang et al. 2010; Crawford et al. 2010). Conversely, in support of the second messenger model, strigolactone was reported to inhibit axillary bud outgrowth in Pisum sativum in the absence of an apical auxin source (Brewer et al. 2009, 2015; Dun et al. 2013). Furthermore, treatments with the auxin transport inhibitor naphtyl phthalamic acid (NPA) showed that strigolactone could inhibit axillary bud outgrowth regardless of auxin transport status (Brewer et al. 2015). In chrysanthemum, previous reports had shown bud outgrowth inhibition by strigolactone only in the presence of an apical auxin source (Liang et al. 2010) corroborating the auxin transport canalization model. In recent experiments we used the synthetic strigolactone GR24 in experiments with Chrysanthemum shoots. GR24 is a commercially available strigolactone analogue that is widely used in germination assays and is very active for repressing shoot branching (Zwanenburg and Pospísil 2013). We showed that in Chrysanthemum stem segments with two axillary buds it was possible to inhibit the outgrowth of both buds with strigolactone GR24 without an apical auxin source (Dierck et al. 2016b). Furthermore, we found that inhibition by GR24 treatment could be counteracted by auxin transport inhibition with NPA treatment. Both observations seem contradictory, with the former being in support of the second messenger model while the latter supports the auxin transport canalization model. To further investigate these observations, we set up a bioassay with Chrysanthemum stem segments treated with the auxin indole acetic acid (IAA), the synthetic strigolactone GR24 and the auxin transport inhibitor NPA. Aside from measuring bud outgrowth, the expression levels of several branching genes in samples of axillary buds and stems were used to indicate the status of bud development and auxin transport. Several Chrysanthemum genes that are involved in axillary bud outgrowth have been described, including the strigolactone biosynthesis gene MAX4 (Liang et al. 2010), strigolactone signaling genes MAX2 (Dong et al. 2013) and D14 (Wen et al. 2015) and the BRC1 gene (Chen et al. 2013). In previous work we isolated several genes in Chrysanthemum that are involved in bud dormancy and the pathways of auxin, cytokinin and strigolactone biosynthesis and regulation: CmDRM1, CmSTM, CmMAX1, CmMAX3, CmRR1, CmHK3a, CmHK3b, CmTIR3, CmPIN1, CmAXR1, CmTIR1, CmAXR6, CmIAA12, CmAXR2 and CmIAA16 (Dierck et al. 2016a).
Five of these genes were selected for this study: CmBRC1, CmDRM1, CmMAX1, CmMAX3, CmAXR2, and CmPIN1. BRC1 is a key integrator of branching signals that is expressed in inhibited axillary buds (Aguilar-Martínez et al. 2007). BRC1 transcription is also upregulated by strigolactone (Dun et al. 2012), making it an important strigolactone target in the second messenger model (Domagalska and Leyser 2011; Waldie et al. 2014; Brewer et al. 2015). In Chrysanthemum, CmBRC1 has shown high expression in dormant axillary buds and downregulation upon bud activation (Chen et al. 2013). In the same experiments, CmBRC1 expression was upregulated in treatments with auxin that inhibited bud outgrowth. DRM1 is a dormancy marker and its expression, like BRC1, is associated with bud activity (Rae et al. 2013), showing downregulation upon bud outgrowth (Aguilar-Martínez et al. 2007; Finlayson et al. 2010). The strigolactone biosynthesis genes MAX3 and MAX4 encode carotenoid cleavage dioxygenases (CCD7 and CCD8) involved in forming carlactone from carotenoid precursors. The MAX1 gene encodes a cytochrome P450 that converts carlactone to active strigolactones (reviewed by Waldie et al. 2014). These genes are also known to be expressed in the vascular tissue of the stem and their mutants display increased branching phenotypes (Booker et al. 2004). In Arabidopsis MAX3 expression decreased in response to decapitation (Hayward et al. 2009). More recent experiments with phosphate (Pi) starvation in Chrysanthemum have shown a link between strigolactone biosynthesis and the reduced branching under phosphate starvation (Xi et al. 2015). Plants under Pi starvation showed a significant decrease in bud outgrowth that coincided with increased levels of strigolactone and increased expression of the MAX3 and MAX4 genes. Furthermore, they showed that bud outgrowth inhibition by auxin application also caused increased expression of MAX3 and MAX4, as well as BRC1. AXR2 and PIN1 expression are both upregulated by auxin (Nagpal et al. 2000; Vieten et al. 2005) and could therefore be useful as markers for auxin accumulation. In a previous experiment CmPIN1 expression showed upregulation in outgrowing axillary buds (Dierck et al. 2016a).
In this work, we studied the axillary bud outgrowth upon treatments with GR24, IAA and NPA over a period of 5 days on stem segments bearing two nodes. At the start (D0), the first day (D1) and the second day (D2) of the 5-day experiment (D0–D4), samples of axillary buds and stems were taken for gene expression analysis to unravel underlying regulation of bud dormancy markers, strigolactone biosynthesis and auxin transport.
Materials and methods
Plant material
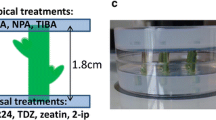
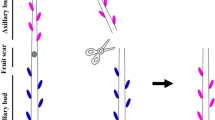
Chrysanthemum morifolium Ramat. cuttings of a cut flower genotype C17, were provided by Dekker Chrysanten BV., The Netherlands. Three batches of 825 cuttings were rooted during 3 weeks at standard greenhouse conditions: 20 °C, 16:8 LD (SON-T, 100 µmol m−2 s−1). Each batch was rooted separately, with 1 week in between, to provide plant material for three repetitions of the experiment in 3 consecutive weeks. Nodal stem segments of 1.8 cm were cut, containing two nodes with dormant axillary buds (Fig. 1 a, b). Leaves and stipules were removed from the nodal stem segments.
Chrysanthemum split-plate assay with stem segments containing two axillary buds. a Nodal positions under the apex that contained dormant axillary buds (example in upper right corner in contrast with grown out buds in lower right corner) were excised from defoliated chrysanthemum shoots. b Stem segments of 1.8 cm were cut from and placed in between two petri dishes to which a treatment was applied. c Samples of upper and lower node axillary buds and stems were taken separately. d Measurement of axillary bud length on stereomicroscopy pictures through ImageJ
Growth conditions, measurements and sampling
Split-plate experiments were set up as described in Dierck et al. (2016b). The experiment consisted of five treatments (1: Control, 2: 5 µM IAA, 3: 50 µM GR24, 4: 5 µM IAA + 50 µM GR24 and 5: 10 µM NPA + 50 µM GR24) that were applied over the course of 5 days with three repetitions in 3 consecutive weeks. Treatments were given to the apical or basal end of the stem segments by adding plant growth regulators from 1 mg/ml stock solutions to the solidifying agar (0.7%) in the top or bottom petri dish. IAA (5 µM) and NPA (10 µM) treatments were applied apically, in line with the basipetal auxin transport. GR24 (50 µM) was applied basally, in line with acropetal strigolactone transport (Fig. 1b). Plates containing stem segments were placed in a growth chamber (20 ± 1 °C, 19:5 LD) with fluorescent light (Philips TL-D Super 80 58W/840 cool white) with an intensity of 90 µmol m−2 s−1 at the height of the nodal stem segments. At the start of the experiment (D0) and on day 2 (D2) and day 4 (D4), axillary bud lengths were measured on stereomicroscopic (Leica M165FC) images of the stem segments using ImageJ software (Fig. 1d). Twenty nodal stem segments per treatment were measured in the 1st week and 25 stem segments per treatment were measured in the 2nd and 3rd week; as such data from a total of 70 stem segments were obtained over the 3 weeks. On D0, D1 and D2 for each treatment, in each of the 3 weeks, axillary bud and stem samples were taken for RT-qPCR on 150 stem segments. The axillary buds and stems of the lower and upper node on each stem segment were sampled separately (Fig. 1c) so that at each time-point for each treatment there were four samples (axillary buds and stems of both upper and lower node) with each sample a pool of 150 axillary buds or stems.
Flow cytometry
Two separate flow cytometrical analyses of cell division were performed in the second (week 2) and third (week 3) repetition of the experiment. Samples for flow cytometry consisted of excised shoot apices and axillary buds on D0 and axillary buds on D2 of the control and the treatments. Because of the small size of the axillary buds, ten buds were sampled (both lower and upper axillary buds on five stem segments) and pooled per analysis. For measurements on the shoot apices, 1 apex was sampled and analyzed. Sampling and measurements were repeated five times per treatment and per repeat. Sample preparation involved chopping of leaf tissue (Galbraith et al. 1983) and 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, Diegem, Belgium) staining according to Otto (1990). Flow cytometrical cell cycle analysis was performed on a CyFlow Space (Sysmex-Partec, Münster, Germany) flow cytometer equipped with a UV Light Emitting Diode and Flomax 2.9 software (Quantum Analysis, Münster, Germany). Based on the peak positions of cell count over fluorescence, the G1, S and G2 phases of the cell cycle were delimited in the acquired histograms. The number of nuclei analyzed in each phase was counted. The percentage of cells in mitosis (mitotic index) was represented by the sum of the percentages of cells in the S and G2 phase (S1 and S2).
Gene expression analysis
RNA extraction, using a modified CTAB protocol (Luypaert et al. 2017), was followed by cDNA synthesis including no reverse transcriptase (noRT) samples for all genes and gene expression analysis of branching genes (CmBRC1, CmDRM1, CmMAX1, CmMAX3, CmAXR2, and CmPIN1) as described previously by Dierck et al. (2016a). Due to some noRT problems (Cq differences < 5) using previously developed primers in the new dataset, a new RT-qPCR primerset was developed for CmBRC1 (JX870411; Chen et al. 2013) and CmMAX1 (KT124645; Dierck et al. 2016a) (Online Resource 1).
RNA concentration and quality was verified with a NanoDrop spectrophotometer (Isogen Life Sciences) (Online Resource 2) and Experion™ automated electrophoresis using the StdSense analysis kit (Bio-Rad) was performed on a subset of samples (Online Resource 3) as described in De Keyser et al. (2013). Gene expression data from the LightCycler® 480 (Roche) was analysed with qbase+ software (Biogazelle; Hellemans et al. 2007). NTCs (no template controls) were included for all genes; technical replicates were omitted but all analysis was done on three biological replicates (three repetitions). In case the difference between the Cq of noRT and samples was smaller than 5, samples were excluded from further analysis. Mean gene-specific amplification efficiencies (Online Resource 4) were calculated with LinRegPCR (Ramakers et al. 2003; Ruijter et al. 2009). Gene expression results are presented as fold changes, from D1 or D2 relative to D0, of calibrated normalized relative quantities (CNRQ), normalised using three reference genes for stem and apex/bud samples CmACT2, CmUBC and CmUBQ10 (Online Resource 5), based on the selection of optimal reference genes using Genorm (Vandesompele et al. 2002).
Statistical analysis
ANOVA, Mann Whitney U and Kruskal Wallis tests were performed with SPSS statistical software (SPSS 23, IBM Corp. Released 2015. IBM SPSS Statistics for Windows, Version 23.0. Armonk, NY: IBM Corp.).
Results
Bud outgrowth
To measure the effect of plant growth regulators on bud outgrowth, a modified split plate assay was used (Fig. 1). This method was established for Chrysanthemum cuttings in previous work to show bud outgrowth inhibition with strigolactone treatment and the influence of sucrose (Dierck et al. 2016b). With this approach it is possible to measure effects on bud outgrowth within a 5-day period for a large number of stem segments with similarly sized dormant axillary buds (ca. 1 mm). Besides bud length measurements, this allowed samples to be taken for analysis of cell division and gene expression. Furthermore, plant growth regulators can be applied basally or apically and the lower and upper node can be measured and sampled separately. In total five treatments were examined in this study: control (untreated), auxin (IAA), strigolactone (GR24), auxin combined with strigolactone (IAA + GR24) and an auxin transport inhibitor in combination with strigolactone (NPA + GR24). The initial bud length in all treatments was around 1 mm. On day 4 (D4) axillary buds in the control treatment showed the highest bud length (Online Resource 6). Apical treatment with auxin and apical auxin treatment in combination with a basal treatment with GR24 resulted in the lowest bud length. Treatment with only basal GR24 showed a reduced bud growth compared to the control treatment. The combined treatment with basal GR24 and apical NPA showed a more pronounced bud length compared to the treatment with only GR24. In general, the bud growth of the axillary buds at the upper node (UN) position was higher than at the lower node (LN), except for the apical treatment with IAA.
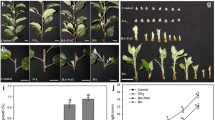
Additional to bud outgrowth measurements, for two repetitions of the experiment, samples of axillary buds were taken on D0 and D2 and analyzed with flow cytometry to show the amount of dividing cells (Fig. 2 and "Appendix" Table 1). As a reference for differentiated tissue and meristematic tissue, samples were included of leaf and shoot apex respectively ("Appendix" Fig. S7a, b). A high plateau at S and a high peak at G2 indicate the cell divisions in the shoot apex ("Appendix" Fig. 7b), and similarly in the growing axillary buds ("Appendix" Fig. 7d, f, h). In the leaf, lower amounts of mitotic cells are found ("Appendix" Fig. 7a), similar to dormant buds and IAA treated buds ("Appendix" Fig. 7c, e, g). In the first repetition of the experiment, the percentage of dividing cells did not differ in dormant buds on D0 compared to the inhibited axillary buds from the treatments with IAA and IAA + GR24 and was significantly lower than the percentage of dividing cells in the shoot apex and axillary buds of the control, GR24 and NPA + GR24 treatments on D2 (Fig. 2a).
Flow cytometrical analysis of dividing cells in the split-plate assay with control, IAA, GR24, GR24 + IAA and GR24 + NPA on chrysanthemum axillary buds. The flow cytometry analysis was repeated twice (a the first repetition in week 2, b the second repetition in week 3) (n = 5 pools of 10 buds or 1 apex) ± SD. Letters indicate homogenous subsets (different for the two repetitions) based on pairwise multiple comparisons with the Tukey post hoc test
The second repetition showed similar results except for the treatments with IAA + GR24 and NPA + GR24 (Fig. 2b). The percentage of dividing cells in the shoot apex and in the control and GR24 treatment on D2 was significantly higher than in the dormant axillary buds on D0 and the IAA treatment on D2. The treatment with IAA + GR24 showed a large standard deviation and was not significantly different from the shoot apex but did show a significantly lower percentage of dividing cells than the control and GR24 treatment on D2. Unlike in the first repetition, the percentage of dividing cells in the NPA + GR24 treatment was not significantly higher than in the dormant buds of control D0 or the inhibited buds of IAA and IAA + GR24 D2.
These results show bud outgrowth inhibition in treatments with IAA, GR24 and IAA + GR24.
The inhibition by GR24 is weaker compared to treatments with IAA and IAA + GR24. This is also reflected by the low mitotic index of IAA treated segments compared to GR24 treatment.
Inhibition by GR24 treatment was reduced in combination with NPA treatment. With exception of the IAA treatment, all treatments showed a stronger bud outgrowth of the upper node compared to the lower node. To confirm the bud outgrowth inhibition of the treatments, expression of dormancy markers CmBRC1 and CmDRM1 was analyzed. The expression of the MAX genes was followed to show local effects on strigolactone biosynthesis regulation during bud outgrowth inhibition. CmAXR2 and CmPIN1 were chosen to show feedback to the auxin levels/transport.
Gene expression
In a previous study on shoot branching in Chrysanthemum, a set of candidate branching genes was developed for expression analysis by RT-qPCR (Dierck et al. 2016a). A subset of these genes was used in this study, including auxin signaling and transport genes CmAXR2 and CmPIN1, bud dormancy genes CmBRC1 and CmDRM1 and strigolactone biosynthesis genes CmMAX1 and CmMAX3 (Figs. 3, 4, 5).
RT-qPCR expression analysis of Chrysanthemum auxin signalling gene CmAXR2 and auxin transport gene CmPIN1. Expression of CmAXR2 and CmPIN1 is shown in the control, 5 µM IAA, 50 µM GR24, 5 µM IAA + 50 µM GR24 and 10 µM NPA + 50 µM GR24 treatments. Data are fold changes of mean CNRQ gene expression values (n = 3 pools of 150 axillary buds or 150 stems) on D1 and D2 relative to D0. Significant differences between CNRQ values on D1 and D2 against D0 are indicated with arterisk (Kruskal Wallis test, p < 0.05)
RT-qPCR gene expression analysis of Chrysanthemum bud dormancy genes CmBRC1 and CmDRM. Expression of CmBRC1 and CmDRM1 in the control, 5 µM IAA, 50 µM GR24, 5 µM IAA + 50 µM GR24 and 10 µM NPA + 50 µM GR24 treatments. Data are fold changes of mean CNRQ gene expression values (n = 3 pools of 150 axillary buds or 150 stems) on D1 and D2 relative to D0. Significant differences between CNRQ values on D1 and D2 against D0 are indicated with asterisk (Kruskal Wallis test, p < 0.05)
RT-qPCR gene expression analysis of Chrysanthemum strigolactone biosynthesis genes CmMAX1 and CmMAX3. Expression of CmMAX1 and CmMAX3 in the control, 5 µM IAA, 50 µM GR24, 5 µM IAA + 50 µM GR24 and 10 µM NPA + 50 µM GR24 treatments. Data are fold changes of mean CNRQ gene expression values (n = 3 pools of 150 axillary buds or 150 stems) on D1 and D2 relative to D0. Significant differences between CNRQ values on D1 and D2 against D0 are indicated with asterisk (Kruskal Wallis test, p < 0.05)
Expression of the auxin signaling gene CmAXR2 showed a different pattern in the axillary buds compared to stems (Fig. 3). In both stem parts, the expression of CmAXR2 decreased in the control, GR24 and NPA + GR24 treatments. A significantly increased expression was seen on D1 for the upper and lower node for the IAA treatment (Fig. 3c, d). No significant differences compared to D0 were seen in the axillary bud samples, although a noticeable increase was observed in the IAA treatment on D2 (Fig. 3a, b). For the expression of the auxin transport gene CmPIN1 the most pronounced change was seen in the axillary buds (Fig. 3e, f). Generally, the treatments with IAA had lower fold changes compared to the other treatments in both upper and lower nodes and the highest fold changes were seen in the treatment with NPA + GR24, especially in the upper node. Significantly increased expression was observed on D1 and D2 for both upper and lower nodes in the control, GR24 and NPA + GR24 treatments. In the IAA treatment, CmPIN1 expression did not change significantly from D0 in the upper node, but in the lower node this gene showed a significant difference on D1. The GR24 + IAA treatment showed a significant change in CmPIN1 expression on D2 in the upper node and on D1 and D2 in the lower node. In the stem samples a different CmPIN1 expression pattern was seen with generally low fold changes (Fig. 3g, h). In contrast with the axillary buds, the highest fold changes were seen in the IAA and IAA + GR24 treatments. Significant CmPIN1 downregulation was seen in the upper node stem at control D1 and GR24 D1 and in the lower node stem at GR24 D1 and NPA + GR24 D2. Significant CmPIN1 upregulation was seen in the upper node stem part at IAA D1 and NPA + GR24 D1 and D2 and in the lower node stem at control D2, and for IAA and GR24 + IAA at both time points.
The dormancy marker genes CmBRC1 and CmDRM1 showed similar expression patterns in the axillary buds of upper and lower nodes with decreased expression in the control and the treatments with GR24 (Fig. 4a, b, e, f). In the stem, CmBRC1 and CmDRM1 generally showed increased expression in the treatments with IAA (Fig. 4c, d, g, h).
Expression of CmBRC1 decreased significantly compared to D0 in the upper and lower node axillary buds on D1 and D2 in the control, GR24 and NPA + GR24 treatments. Increased expression was seen in the IAA and IAA + GR24 treatments, but was only significant on D2 for the IAA + GR24 treatment (Fig. 4a, b). In the stem significantly increased expressions of CmBRC1 were seen in the upper node on D1 and D2 for the IAA, IAA + GR24 and NPA + GR24 treatments. In the lower node, increased expression was significant on D1 for the IAA treatment and on D1 and D2 for the IAA + GR24 treatment (Fig. 4c, d). CmBRC1 expression in the upper node stem for the control and GR24 treatment, and in the lower node stem for the control, GR24 and NPA + GR24, remained virtually unchanged with only a slight decrease in the control on D1.
Significantly decreased expression of CmDRM1 in the axillary buds was seen in both upper and lower node buds for the control and NPA + GR24 treatment at both time points, and for the GR24 treatment only on D1 (Fig. 4e, f). In the stem CmDRM1 expression was only significantly increased in the upper node on D2 for the treatments with IAA (Fig. 4g, h).
Expression of the strigolactone biosynthesis gene CmMAX3 gene showed a general decrease in all treatments in both stem parts (Fig. 5c, d). In the axillary buds an increased expression was generally seen in the treatments with IAA while a decreased expression was seen in the other treatments (Fig. 5a, b). In the upper node axillary buds, expression increased significantly in the IAA treatment on D1 and D2 and a significant decrease in expression was seen for the treatment with GR24 (Fig. 5a). In the lower node axillary buds, expression decreased significant on D1 and D2 for the control, GR24 and NPA + GR24 treatment and only on D1 for the IAA treatment (Fig. 5b). CmMAX1 expression in the axillary bud showed a similar response as CmMAX3 to IAA treatment, but only in the upper node differences compared to D0 were significant in the IAA and GR24 + IAA treatments on D2 (Fig. 5e). In the lower node there were no significant differences (Fig. 5f) and a general increased expression was measured in both stem parts (Fig. 5g, h).
Discussion
In their study on pea axillary buds, Devitt and Stafstrom (1995) used flow cytometry to measure an increase in dividing cells within 24 h after decapitation, which coincided with an increased expression of cell cycle genes. For chrysanthemum, previous reports have also shown a differential expression of several shoot architecture genes during the outgrowth of axillary buds (Chen et al. 2013; Dierck et al. 2016a). In this study a modified split plate assay (Dierck et al. 2016b) was used to induce a controlled axillary bud outgrowth under influence of hormone treatments. Bud outgrowth was followed using a combination of flow cytometrical analysis of dividing cells and the expression analysis of chrysanthemum genes as markers for bud dormancy and hormonal regulation.
The results of bud outgrowth that were presented here correspond with previous experiments described in Dierck et al. (2016b), where a basal application with GR24 inhibited axillary bud outgrowth in both buds of a two-nodal stem segment without an apical auxin source and where application with the auxin transport inhibitor NPA in combination with GR24 showed a reduced inhibition compared to the treatment with only basal GR24. However, it must be noted that this restricted outgrowth is not a complete inhibition as could be seen in the treatment with apical IAA. Furthermore, the lower node axillary bud was more inhibited than the upper node axillary bud. This was also the case in the treatment with IAA + GR24 (Online Resource 6).
The incomplete inhibition of axillary bud outgrowth by GR24 treatment was also reflected in the measurement of the mitotic index. Dormant chrysanthemum axillary buds and buds that were inhibited by IAA and IAA + GR24 treatment on D2 had a significantly lower percentage of dividing cells than growing axillary buds in the control on D2. Axillary buds in the treatment with GR24, however, had a higher mitotic index compared to the dormant and inhibited buds, which was also seen in the axillary buds in the treatment with NPA + GR24 and the control treatment. These results indicate an incomplete restriction of axillary bud outgrowth by strigolactone when compared to the inhibition by auxin treatment. It has been shown that strigolactone treatment can promote or inhibit axillary bud outgrowth depending on the auxin transport status (Shinohara et al. 2013) and this could be a possible explanation for the bud outgrowth that was still observed in the GR24 treatment. Since strigolactone inhibits the mobilization of PIN1 proteins it could be argued that auxin transport is hindered both in the stem (polar auxin transport stream), which would promote bud outgrowth, and in the buds, which would inhibit bud outgrowth, allowing an intermediate restricted bud outgrowth. Furthermore, in our study, the combination of GR24 with IAA or NPA showed variation between both repetitions for the percentage of mitotic cells, which could indicate a delicate balance in between strigolactone and auxin, although more repetitions in both upper and lower node axillary buds would be necessary to confirm this.
Axillary bud outgrowth or bud outgrowth inhibition in the first 2 days (D1, D2) after decapitation (D0) also coincided with altered expression of several chrysanthemum candidate branching genes: auxin response gene CmAXR2, auxin transport gene CmPIN1, bud dormancy genes CmBRC1 and CmDRM1 and strigolactone biosynthesis genes CmMAX1 and CmMAX3.
AXR2 is an AUX/IAA gene involved in the auxin response; its expression is induced by auxin and therefore useful as a marker for auxin (Abel and Theologis 1996; Nagpal et al. 2000). PIN1 is the main transport protein for basipetal auxin transport and its expression is upregulated in a positive feedback mechanism by auxin (Vieten et al. 2005). Expression of CmPIN1 therefore gives information about the auxin presence but also about the auxin transport canalization. Bud outgrowth on D1 and D2 in the control treatment was accompanied by a decreased expression of CmAXR2 in lower and upper node stem samples (Fig. 3c, d). This effect was also observed in the NPA + GR24 treatment (most notable in the lower node) and in the GR24 treatment (most notable in the upper node), where bud outgrowth was restricted. On the other hand, inhibition of axillary bud outgrowth in the treatment with IAA coincided with an increased expression of CmAXR2. The CmAXR2 expression pattern in the stem indicates that in the nodal stem segments, there is an increased auxin concentration following the IAA treatments and a reduced auxin level in the control, the GR24 and NPA + GR24 treatment. This coincided with either axillary bud growth (control, NPA + GR24) or with the restricted axillary bud outgrowth that occurred in the GR24 treatment. These observations reflect a reduced polar auxin transport in the decapitated stem segments that allows axillary bud outgrowth, while apical IAA application maintains polar auxin transport and inhibits bud outgrowth. This corresponds with the auxin transport canalization mechanism of bud outgrowth, in which auxin produced by activated buds is canalized to the stem by upregulating auxin transport proteins (Sauer et al. 2006). In the treatment with strigolactone GR24 it can be noted that there is also a reduced CmAXR2 expression that could indicate a reduced polar auxin transport. Strigolactone has been shown to reduce polar auxin transport by dampening PIN1 mobilization (Shinohara et al. 2013). In this case a reduced expression of CmAXR2 could reflect the reduced auxin transport as a consequence of strigolactone treatment.
The expression of CmPIN1 further reflects auxin levels and transport and showed a somewhat differential expression pattern in axillary buds and stem. In the axillary buds, high fold changes corresponded with the treatments that showed axillary bud outgrowth (control, GR24, NPA + GR24). Conversely, in the stem, increased expression was generally seen in the treatments that inhibited bud outgrowth (IAA, IAA + GR24). This observation corresponds with the function of PIN1 in the auxin transport canalization model, where axillary buds must export auxin in order to grow out (Prusinkiewicz et al. 2009; Crawford et al. 2010; Balla et al. 2011; Domagalska and Leyser 2011). In the stem, CmPIN1 expression increased in the IAA treatments, reflecting the upregulation by the increased auxin levels. These results also correspond with our earlier observations of gene expression in chrysanthemum, where CmPIN1 expression was increased in axillary buds during outgrowth and CmAXR2 expression increased in stem samples from plants with strong apical dominance but not in samples from plants during bud outgrowth (Dierck et al. 2016a).
In the treatment with GR24 it is remarkable that CmPIN1 expression in the buds was also increased, as it was hypothesized in the auxin canalization model that strigolactone inhibits axillary bud outgrowth by restricting the polar auxin transport and increasing competition between buds for the auxin sink (Crawford et al. 2010; Domagalska and Leyser 2011; Shinohara et al. 2013). When considering the incomplete inhibition of axillary bud outgrowth in the GR24 treatment, which was reflected by the high percentage of dividing cells, it is possible to consider the axillary buds as outgrowing buds with a restricted growth. This could be a possible explanation for the increased CmPIN1 expression. Furthermore, the action of strigolactone on the mobilization of PIN1 proteins is known to be independent of new protein synthesis (Shinohara et al. 2013), indicating that CmPIN1 expression more likely reflects auxin levels and not the effect of GR24 on the PIN1 mobilization that is associated with restricted bud outgrowth. PIN1 expression levels also do not correspond reliably to the protein accumulation (Bennett et al. 2006, 2016).
In previous reports on the action of the strigolactone GR24 on shoot branching, basal application with GR24 could only inhibit bud outgrowth in the presence of an auxin source (Liang et al. 2010; Crawford et al. 2010). In these cases, there was also a competition between axillary buds with one bud growing out and dominating the dormant lower bud. In our treatment with GR24 there was similarly a slight difference between the two axillary buds, with the upper node showing higher bud length growth. Therefore, as was previously mentioned, it can be argued that strigolactone treatment could result in an intermediate, restricted bud growth by restricting auxin transport in both axillary bud and stem.
Expression of the dormancy markers CmBRC1 and CmDRM1 was generally decreased in the axillary buds of the control and the treatments with GR24 and NPA + GR24 (Fig. 4a, b, e, f), whereas increased expression was found in the stem in the treatments with IAA (Fig. 4c, d, g, h). The decreased expression during axillary bud outgrowth is consistent with earlier reports of expression and bud activity (Rae et al. 2013; Chen et al. 2013). These genes also showed a general downregulation during axillary bud outgrowth in chrysanthemum plants (Dierck et al. 2016a). For the treatment with GR24 it can be noted that the trend of decreased expression is similar to the control treatment. This seems contradictory to the role of strigolactone to inhibit axillary bud outgrowth and one would expect an increase or no change in expression compared to dormant buds on D0. Furthermore BRC1 is also regarded to be downstream of strigolactones in inhibiting bud outgrowth (Aguilar-Martínez et al. 2007; Minakuchi et al. 2010) and therefore it could be expected that treatments with strigolactone would induce CmBRC1 expression. However, this relation between BRC1 and strigolactones can be different, as was observed in rice, where strigolactone treatments do not alter BRC1 expression (Arite et al. 2007; Minakuchi et al. 2010). As was mentioned earlier, some, albeit restricted, bud outgrowth did occur in the treatment with GR24, which was also reflected in the percentage of dividing cells. This furthers explains the increased expression of CmDRM1 and CmBRC1 as these dormancy markers have been shown to be involved in blocking G1/S phase transition in dormant axillary buds (Wood et al. 2013; González-Grandío et al. 2013). Another possible explanation could be that the decreased expression of CmBRC1 is induced by decapitation and dominates GR24 impingement.
CmMAX3 expression in the axillary buds was generally increased in the treatments with IAA and decreased in the control, GR24 and NPA + GR24 treatment (Fig. 5a, b). In the stem a general decreased expression was seen (Fig. 5c, d). The upregulation of CmMAX3 in the treatments with IAA and the downregulation that coincided with bud outgrowth, corresponds to the MAX3 expression seen in Arabidopsis during bud outgrowth or inhibition by IAA (Hayward et al. 2009). Reduction of MAX3 expression by strigolactone treatment has also been reported as a feedback mechanism in Arabidopsis (Mashiguchi et al. 2009) but it is more likely that the decreased CmMAX3 expression, similarly to the CmBRC1 and CmDRM1 expression, reflects the restricted bud outgrowth in the GR24 treatment. Likewise, for CmMAX1 expression in the upper node axillary buds, lower fold changes coincided with the bud outgrowth of control, GR24 and NPA + GR24 treatments. It was further noted that in the IAA treatment, the CmMAX3 expression in the upper axillary bud was higher than in the lower bud and that this expression increased from D1 to D2. This was similarly observed in the expression of the strigolactone biosynthesis gene CmMAX1 (Fig. 5e). A possible explanation for this observation could be that the auxin treatment causes the upregulation of strigolactone biosynthesis genes, but this is established first in the upper axillary bud that is closest to the apical auxin source and only later in the lower axillary bud.
Conclusions
Our results have demonstrated a split plate bioassay in chrysanthemum where treatment with IAA and IAA + GR24 inhibited bud outgrowth, application with strigolactone GR24 restricted bud outgrowth and NPA application reduced the restricted outgrowth by GR24 (Fig. 6). These results were in line with the auxin transport canalization model. The bud outgrowth status was reflected by flow cytometrical measurements of cell divisions in the axillary buds and the expression of dormancy markers genes CmBRC1 and CmDRM1 as well as the strigolactone biosynthesis genes CmMAX3 and CmMAX1. The auxin transport status during bud outgrowth was reflected by CmAXR2 and CmPIN1 expression with an increased CmPIN1 expression in the axillary buds coinciding with bud outgrowth.
Schematic overview of experimental results obtained from bud outgrowth measurements, flow-cytometry and gene expression analysis on axillary buds and stem segments. a Scheme of a stem segment under the control treatment in the split-plate assay. Predicted auxin flow, according to the canalization model, is represented by blue arrows. Decapitation in the stem segments reduces auxin flow in the stem (indicated by a dashed arrow) and facilitates auxin export from the axillary buds (blue arrows), resulting in axillary bud outgrowth (green arrows). The upper node (UN) auxin flow creates competition in the stem and reduces the lower node (LN) auxin transport (dashed arrow), resulting in stronger bud outgrowth in the UN compared to the LN (dashed green arrow). b Scheme of a stem segment under the IAA treatment. Apical IAA treatment induces a strong auxin flow in the stem (blue arrow) and inhibits auxin export from the axillary buds (indicated by red lines on the blue arrows), resulting in dormant axillary buds. c Scheme of a stem segment under the GR24 treatment in the split-plate assay. Basal GR24 is transported upwards (orange arrow) and reduces auxin flow from axillary buds to the stem (dashed red lines on blue arrows), resulting in restricted outgrowth of axillary buds (small, dashed, green arrows). d Scheme of a stem segment under the IAA + GR24 treatment. IAA treatment induces a strong auxin flow in the stem (blue arrow) and together with GR24 (orange arrow) inhibits auxin export from the axillary buds (indicated by red lines on the blue arrows), resulting in dormant axillary buds. e Scheme of a stem segment under the NPA + GR24 treatment. Decapitation and apical NPA strongly reduces auxin flow in the stem (dotted blue arrow). The weaker auxin flow reduces competition for auxin transport of the buds to the stem (blue arrows) and allows a less restricted bud outgrowth by GR24 (big dashed green arrows). f Change in the mitotic index of axillary buds corresponding to the different treatments on D2 compared to D0. An increase is indicated by a green plus sign, while equivalent levels are indicated by an equals sign. g Fold changes (p < 0.05) in gene expression (D1 and D2 compared to D0) in buds (oval) and stem segments (square) corresponding to the different treatments. Positive fold changes are indicated by an upward arrow and negative fold changes are indicated by a downward arrow. (Color figure online)
The buds in the GR24 treatment were not completely inhibited as in the IAA treatment and the amount of dividing cells as well as the downregulation of CmBRC1 and CmDRM1 dormancy markers suggested bud outgrowth, although restricted when compared to bud lengths in the control. This could be seen as an intermediate bud outgrowth state, considering strigolactone to inhibit or promote axillary bud outgrowth depending on the auxin transport status according to the auxin transport canalization model. Due to the large amount of samples needed for gene expression analysis, this study was limited to five treatments and could not include hormone measurements. Further investigations could include a combination of auxin measurements with PIN1 localization together with bud outgrowth measurements, flow cytometry and gene expression analysis. Additional treatments, such as the use of strigolactone inhibitors, like TIS108, would also be interesting to elucidate the exact GR24 inhibition mechanism and the role of auxin transport.
References
Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111:9–17. https://doi.org/10.1104/pp.111.1.9
Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19:458–472. https://doi.org/10.1105/tpc.106.048934
Anderson NO (2007) Flower breeding and genetics issues, challenges and opportunities for the 21st century. Springer, Dordrecht
Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51:1019–1029. https://doi.org/10.1111/j.1365-313X.2007.03210.x
Balla J, Kalousek P, Reinohl V, Friml J, Prochazka S (2011) Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J 65:571–577. https://doi.org/10.1111/j.1365-313X.2010.04443.x
Barbier F, Peron T, Lecerf M, Perez-Garcia M-D, Barriere Q, Rolcik J, Boutet-Mercey S, Citerne S, Lemoine R, Porcheron B, Roman H, Leduc N, Le Gourrierec J, Bertheloot J, Sakr S (2015) Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J Exp Bot 66:2569–2582. https://doi.org/10.1093/jxb/erv047
Bennett T, Leyser O (2006) Something on the side: axillary meristems and plant development. Plant Mol Biol 60:843–854. https://doi.org/10.1007/s11103-005-2763-4
Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16:553–563. https://doi.org/10.1016/j.cub.2006.01.058
Bennett T, Hines G, van Rongen M, Waldie T, Sawchuk MG, Scarpella E, Ljung K, Leyser O (2016) Connective auxin transport in the shoot facilitates communication between shoot apices. PLoS Biol 14:e1002446. https://doi.org/10.1371/journal.pbio.1002446
Booker J, Auldridge M, Willis S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14:1232–1238. https://doi.org/10.1016/j.cub.2004.06.061
Brewer PB, Dun E, Ferguson BJ, Rameau C, Beveridge C (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150:482–493. https://doi.org/10.1104/pp.108.134783
Brewer PB, Dun EA, Gui R, Mason M, Beveridge CA (2015) Strigolactone inhibition of branching independent of polar auxin transport. Plant Physiol. https://doi.org/10.1104/pp.15.00014
Chen X, Zhou X, Xi L, Li J, Zhao R, Ma N, Zhao L (2013) Roles of DgBRC1 in regulation of lateral branching in chrysanthemum (Dendranthema × grandiflora cv. Jinba). PLoS ONE 8:e61717. https://doi.org/10.1371/journal.pone.0061717
Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska M, Leyser O (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137:2905–2913. https://doi.org/10.1242/dev.051987
De Keyser E, Desmet L, Van Bockstaele E, De Riek J (2013) How to perform RT-qPCR accurately in plant species? A case study on flower colour gene expression in an azalea (Rhododendron simsii hybrids) mapping population. BMC Mol Biol 14:13. https://doi.org/10.1186/1471-2199-14-13
Devitt ML, Stafstrom JP (1995) Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Mol Biol 29:255–265. https://doi.org/10.1007/BF00043650
Dierck R, De Keyser E, De Riek J, Dhooghe E, Van Huylenbroeck J, Prinsen E, Van Der Straeten D (2016a) Change in auxin and cytokinin levels coincides with altered expression of branching genes during axillary bud outgrowth in chrysanthemum. PLoS ONE 11:e0161732. https://doi.org/10.1371/journal.pone.0161732
Dierck R, Dhooghe E, Van Huylenbroeck J, De Riek J, De Keyser E, Van Der Straeten D (2016b) Response to strigolactone treatment in chrysanthemum axillary buds is influenced by auxin transport inhibition and sucrose availability. Acta Physiol Plant 38:271. https://doi.org/10.1007/s11738-016-2292-6
Domagalska M, Leyser O (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12:211–221. https://doi.org/10.1038/nrm3088
Dong L, Ishak A, Yu J, Zhao R, Zhao L (2013) Identification and functional analysis of three MAX2 orthologs in chrysanthemum. J Integr Plant Biol 55:434–442. https://doi.org/10.1111/jipb.12028
Dun E, de Saint Germain A, Rameau C, Beveridge C (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158:487–498. https://doi.org/10.1104/pp.111.186783
Dun E, de Saint Germain A, Rameau C, Beveridge CA (2013) Dynamics of strigolactone function and shoot branching responses in Pisum sativum. Mol Plant 6:128–140. https://doi.org/10.1093/mp/sss131
Ferguson BJ, Beveridge C (2009) Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol 149:1929–1944. https://doi.org/10.1104/pp.109.135475
Finlayson S, Krishnareddy SR, Kebrom TH, Casal JJ (2010) Phytochrome regulation of branching in Arabidopsis. Plant Physiol 152:1914–1927. https://doi.org/10.1104/pp.109.148833
Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426:147–153. https://doi.org/10.1038/nature02085
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220:1049–1051. https://doi.org/10.1126/science.220.4601.1049
Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282:2226–2230. https://doi.org/10.1126/science.282.5397.2226
Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun E, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, Bouwmeester H, Bécard G, Beveridge C, Rameau C, Rochange SF (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194. https://doi.org/10.1038/nature07271
González-Grandío E, Poza-Carrión C, Sorzano COS, Cubas P (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25:834–850. https://doi.org/10.1105/tpc.112.108480
Hayward A, Stirnberg P, Beveridge C, Leyser O (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151:400–412. https://doi.org/10.1104/pp.109.137646
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. https://doi.org/10.1186/gb-2007-8-2-r19
Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska M, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol 155:974–987. https://doi.org/10.1104/pp.110.164640
Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483:341–344. https://doi.org/10.1038/nature10873
Leyser O (2009) The control of shoot branching: an example of plant information processing. Plant Cell Environ 32:694–703. https://doi.org/10.1111/j.1365-3040.2009.01930.x
Liang J, Zhao L, Challis R, Leyser O (2010) Strigolactone regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum). J Exp Bot 61:3069–3078. https://doi.org/10.1093/jxb/erq133
Luypaert G, Witters J, Van Huylenbroeck J, De Clercq P, De Riek J, De Keyser E (2017) Induced expression of selected plant defence related genes in pot azalea, Rhododendron simsii hybrid. Euphytica 213:227. https://doi.org/10.1007/s10681-017-2010-5
Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T (2009) Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci Biotechnol Biochem 73:2460–2465. https://doi.org/10.1271/bbb.90443
Mason MG, Ross JJ, Babst B, Wienclaw BN, Beveridge C (2014) Sugar demand, not auxin, is the initial regulator of apical dominance. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.1322045111
McSteen P, Leyser O (2005) Shoot branching. Annu Rev Plant Biol 56:353–374. https://doi.org/10.1146/annurev.arplant.56.032604.144122
Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, Kyozuka J (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51:1127–1135. https://doi.org/10.1093/pcp/pcq083
Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123:563–574. https://doi.org/10.1104/pp.123.2.563
Ongaro V, Leyser O (2008) Hormonal control of shoot branching. J Exp Bot 59:67–74. https://doi.org/10.1093/jxb/erm134
Otto F (1990) DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods Cell Biol 33:105–110
Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O (2009) Control of bud activation by an auxin transport switch. Proc Natl Acad Sci USA 106:17431–17436. https://doi.org/10.1073/pnas.0906696106
Rae GM, David K, Wood M (2013) The dormancy marker DRM1/ARP associated with dormancy but a broader role in planta. Dev Biol J 2013:12. https://doi.org/10.1155/2013/632524
Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. https://doi.org/10.1016/S0304-3940(02)01423-4
Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher F, Sakr S (2015) Multiple pathways regulate shoot branching. Front Plant Sci 5:1–15. https://doi.org/10.3389/fpls.2014.00741
Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. https://doi.org/10.1093/nar/gkp045
Sauer M, Balla J, Luschnig C, Wis J, Reinöhl V, Benková E (2006) Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20:2902–2911. https://doi.org/10.1101/gad.390806
Shinohara N, Taylor C, Leyser O (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11:e1001474. https://doi.org/10.1371/journal.pbio.1001474
Shinoyama H, Anderson N, Furuta H, Mochizuki A, Nomura Y, Singh R, Datta SK, Wang B, da Silva J (2006) Chrysanthemum biotechnology. In: Teixeira da Silva JA (ed) Floriculture, ornamental and plant biotechnology: advances and topical issues, vol II. Global Science Books
Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200. https://doi.org/10.1038/nature07272
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132:4521–4531. https://doi.org/10.1242/dev.02027
Waldie T, McCulloch H, Leyser O (2014) Strigolactones and the control of plant development: lessons from shoot branching. Plant J. https://doi.org/10.1111/tpj.12488
Wen C, Xi L, Gao B, Wang K, Lv S, Kou Y, Ma N, Zhao L (2015) Roles of DgD14 in regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum “Jinba”). Plant Physiol Biochem 96:241–253. https://doi.org/10.1016/j.plaphy.2015.07.030
Wood M, Rae GM, Wu RM, Walton EF, Xue B, Hellens RP, Uversky VN (2013) Actinidia DRM1—an intrinsically disordered protein whose mRNA expression is inversely correlated with spring budbreak in kiwifruit. PLoS ONE. https://doi.org/10.1371/journal.pone.0057354
Xi L, Wen C, Fang S, Chen X, Nie J, Chu J, Yuan C, Yan C, Ma N, Zhao L (2015) Impacts of strigolactone on shoot branching under phosphate starvation in chrysanthemum (Dendranthema grandiflorum cv. Jinba). Front Plant Sci 6:1–15. https://doi.org/10.3389/fpls.2015.00694
Xie X, Wang G, Yang L, Cheng T, Gao J, Wu Y, Xia Q (2015a) Cloning and characterization of a novel Nicotiana tabacum ABC transporter involved in shoot branching. Physiol Plant 153:299–306. https://doi.org/10.1111/ppl.12267
Xie X, Yoneyama K, Kisugi T, Nomura T, Akiyama K, Asami T, Yoneyama K (2015b) Strigolactones are transported from roots to shoots, although not through the xylem. J Pestic Sci 40:214–216. https://doi.org/10.1584/jpestics.D15-045
Zwanenburg B, Pospísil T (2013) Structure and activity of strigolactones: new plant hormones with a rich future. Mol Plant 6:38–62. https://doi.org/10.1093/mp/sss141
Acknowledgements
This research was funded by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen Grant No. 110771). The authors would like to thank Magali Losschaert for technical assistance with gene expression analysis and Laurence Desmet, Jorien Oomen, Veerle Buysens, Katrijn Van Laere and Evelien Calsyn for their help in preparing the split-plate assays and sampling of plant tissue. Frederik Delbeke, Roger Dobbelaere and Jo De Groote are acknowledged for maintenance of the plant material in the greenhouse.
Author information
Authors and Affiliations
Contributions
RD conceptualized and performed the experiments and wrote the manuscript. LL performed flow cytometry analysis. EDK, LL, DVDS, ED, JVH and JDR assisted in conceptualizing the experiment and proofread and revised the manuscript.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
The mitotic index (percentage of dividing cells) is represented by the sum of the amount of cells in the S and G2 phase divided by the total number of nuclei analyzed (G1 + S + G2), as was delineated on a flow-cytometrical histogram. a leaf; b shoot apex; c, d control axillary buds at day 0 (c) and day 2 (d); e–h hormone treated (e IAA, f GR24, g GR24+IAA, h GR24+NPA) axillary buds at D2
Rights and permissions
About this article
Cite this article
Dierck, R., Leus, L., Dhooghe, E. et al. Branching gene expression during chrysanthemum axillary bud outgrowth regulated by strigolactone and auxin transport. Plant Growth Regul 86, 23–36 (2018). https://doi.org/10.1007/s10725-018-0408-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0408-2