Abstract
Although gibberellin (GA) has been reported to control branching, little is known about how GA mediates signals regulating the outgrowth of axillary buds (ABs). In the current study, the effect of the exogenous application of 5.0 mM GA3 on ABs outgrowth on 1-year-old ‘Nagafu No. 2’/T337/M. robusta Rehd. apple trees was investigated and compared to the bud-activating treatments, 5 mM BA or decapitation. Additionally, the expression of genes related to bud-regulating signals and sucrose levels in ABs was examined. Results indicated that GA3 did not promote ABs’ outgrowth, nor down-regulate the expression of branching repressors [MdTCP40, MdTCP33, and MdTCP16 (homologs of BRANCHED1 and BRC2)], which were significantly inhibited by the BA and decapitation treatments. MdSBP12 and MdSBP18, the putative transcriptional activators of these genes, which are expressed at lower levels in BA-treated and decapitated buds, were up-regulated in the GA3 treatment in comparison to the BA treatment. Additionally, GA3 did not up-regulate the expression of CK response- and auxin transport-related genes, which were immediately induced by the BA treatment. In addition, GA3 also up-regulated the expression of several Tre6P biosynthesis genes and reduced sucrose levels in ABs. Sucrose levels, however, were still higher than what was observed in BA-treated buds, indicating that sucrose may not be limiting in GA3-controlled AB outgrowth. Although GA3 promoted cell division, it was not sufficient to induce AB outgrowth. Conclusively, some branching-inhibiting genes and bud-regulating hormones are associated with the inability of GA3 to activate AB outgrowth.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pattern of shoot branching is a major determinant in the establishment of plant architecture. In many crops and fruit trees, branching has a profound effect on yield (Kebrom et al. 2013; Patrick and Colyvas 2014; Rameau et al. 2015). In modern apple orchards, fruit production needs to begin with a short juvenile stage as early as possible in order to ensure an early return on investment. In this regard, planting well-branched, rather than single-stem, apple trees on dwarfing rootstocks greatly contributes to early flowering and the onset of production (Kviklys 2006; Bostan 2010; Elfving 2010; Dun et al. 2012; Atay and Koyuncu 2013). Many nursery-grown apple trees, such as ‘Fuji’, exhibit poor levels of natural branching (Volz et al. 1994; Atay and Koyuncu 2013). Apple lateral branches are derived from the outgrowth of axillary buds (ABs), which is controlled by a complex interaction of phytohormones and environmental conditions (Domagalska and Leyser 2011; Rameau et al. 2015). Although plant branching is influenced by environmental factors, such as temperature (Labuschagné et al. 2003; Naor et al. 2003), light (Kebrom and Mullet 2016; Holalu and Finlayson 2017), nutrition (Yoneyama et al. 2007), and plant density (Aguilarmartínez et al. 2007), considerable research has focused on the role of phytohormones and their interaction in regulating AB outgrowth (Leyser 2009).
ABs can either undergo immediate outgrowth and turn into a lateral branch or they can remain dormant under natural environmental conditions. Auxin was the first hormone that was suggested to play a crucial role in the suppression of ABs by apical dominance (Thimann and Skoog 1934; Domagalska and Leyser 2011). There is a strong correlation between AB outgrowth and bud auxin export based on Sachs’ auxin transport model (Ho et al. 1981; Bennett et al. 2014); specifically, AB is activated when auxin is transported out of the bud. In contrast, cytokinins (CKs) have been found to directly promote AB outgrowth (Leyser 2009). Exogenous applications of CK, and the up-regulation of CK biosynthesis, induce AB outgrowth (Muller et al. 2015; Zeng and Zhao 2016). Cytokinin response-regulators (RRs), which are the key components in CK signal transduction pathways, were also reported to have an essential role in shoot branching (Muller et al. 2015). Additionally, strigolactones (SLs), which represent a new class of phytohormones (Brewer and Beveridge 2009; Agusti et al. 2011), have been reported to act antagonistically to CK in regulating AB outgrowth (Dun et al. 2012). However, the outgrowth of ABs on CK-treated shoots or decapitated shoots can be efficiently inhibited by the application of Germination Releaser 24 (GR24), a widely used analog of SL (Bergmann et al. 1993; Brewer and Beveridge 2009; Dun et al. 2012, 2013). Strigolactone deletion mutants, which exhibit a strong branching phenotype, have also been identified in diverse species, including A. thaliana (max), pea (rms), rice (dwarf), and petunia (dad) (Sorefan et al. 2003; Arite et al. 2007; Simons et al. 2007; Hao et al. 2009).
In addition to hormone signals, local control at the node and in the bud contributes to the regulation of bud activation. BRANCHED1 (BRC1), a member of class II TB1 CYCLOIDEA PCF (TCP) type transcription factors, is specifically expressed in non-growing ABs (Aguilarmartínez et al. 2007; Finlayson 2007; Martín-Trillo et al. 2011). As a repressor of branching, BRC1 is induced by SL and down-regulated by CK or decapitation in a variety of plant species (Aguilarmartínez et al. 2007; Minakuchi et al. 2010; Martín-Trillo et al. 2011; Braun et al. 2012; Muhr et al. 2016). BRC1 transcript abundance also decreases in activated, elongating ABs (Aguilarmartínez et al. 2007; Finlayson 2007). Additionally, BRC1 knock-out mutants also exhibit a highly branched phenotype, while overexpression results in an inhibition of AB outgrowth (Aguilarmartínez et al. 2007; Minakuchi et al. 2010; Martín-Trillo et al. 2011; Nicolas et al. 2015; Muhr et al. 2016).
Previous studies have demonstrated the role of sugar in the promotion of AB outgrowth (Nunes et al. 2013; Mason et al. 2014; François et al. 2015), including a significant increase in sugar metabolism and transport during the outgrowth of ABs (Henry et al. 2011; Rabot et al. 2012). In pea, more recent studies have found that an adequate supply of sugar is essential for the activation of AB outgrowth and down-regulation of BRC1 expression (Mason et al. 2014; François et al. 2015). Otori et al. (2017) demonstrated that improvements in the capacity of sucrose biosynthesis results in enhanced shoot branching. Additionally, trehalose 6-phosphate, which functions as a signal of sucrose availability in plants (Lunn et al. 2006; Yadav et al. 2014), has been suggested to be involved in the triggering AB outgrowth in garden pea (Fichtner et al. 2017). Additionally, defoliation, which reduces the availability of photoassimilates, suppressed the outgrowth of ABs following decapitation (Mason et al. 2014; Fichtner et al. 2017).
In comparison to the well-known role of gibberellins (GAs) in internode elongation (Daviere et al. 2014), the specific role of GA in shoot branching has not been characterized well. GA-deficient mutants in Arabidopsis (Silverstone et al. 1998), rice (Lo et al. 2008), and pea (Murfet et al. 1993) all exhibited greater levels of shoot branching in comparison to their wild-type counterparts. Additionally, the overexpression of GA catabolism genes to reduce GA1 level produces increased branching phenotypes (Agharkar et al. 2007; Lo et al. 2008; Mauriat et al. 2011; Zawaski and Busov 2014) and the application of GA3 suppressed tillering in rice (Lo et al. 2008). In LMP1::AtGA2ox2 hybrid aspen plants, a reduction in bioactive GAs was observed which resulted in a much higher percentage of lateral buds producing a branch as compared to wild-type (Mauriat et al. 2011). In GA2ox overexpressing plants, this was followed by a significant increase in lateral branch outgrowth as compared to WT plants (Zawaski and Busov 2014). Collectively, these observations suggest that GA acts as a negative regulator of branching. However, the promotion of AB outgrowth by GA3, GA4 or GA4+7 has been reported in some species, such as sweet cherry (Elfving et al. 2011), rose (Choubane et al. 2012), and the perennial woody plant Jatropha curcas (Ni et al. 2015), respectively. The loss of function mutants of DELLA, which are repressors of GA signaling (Peng et al. 1997), displayed reduced shoot branching (Cheng et al. 2004). These reports indicate that the effect of GAs on AB outgrowth varies in different species. Few studies have focused on identifying the potential pathways through which gibberellin modulates other branch-regulating signals in the control of ABs, and especially for apple ABs.
GA3 is a main form of biologically active gibberellins. In our previous study, we used GA3 for the exogenous application on apple trees to investigate the effect of GA3 on flowering (Zhang et al. 2016; Fan et al. 2017). Additionally, the exogenous application of GA3 was reported to trigger AB outgrowth in the perennial woody plant Jatropha curcas (Ni et al. 2015). As a result, we chose to examine whether the application of GA3 was effective for the outgrowth of ABs in apple.
In the present study, we examined the effect of GA3 on AB outgrowth in 1-year-old ‘Nagafu No. 2’ (a ‘Fuji’ cultivar) / T337 /M. robusta Rehd., a grafting combination widely produced in Chinese nurseries. The effect of an exogenous application of GA3 on AB outgrowth was compared with the effect of the bud-activating treatments, exogenous application of BA or decapitation. The expression pattern of several genes associated with the repression of branching, bud-regulating hormones, sugar metabolism, cell proliferation, and sucrose levels was also characterized to determine how GA3 modulates the control of branching. Our results provide basic information pertaining to the role of GA3 in regulating AB outgrowth via crosstalk with other branch-regulating signals.
Materials and methods
Plant material and treatments
One-year-old ‘Nagafu No. 2’ (a ‘Fuji’ cultivar) grafted onto a T337 interstem and M. robusta Rehd. rootstock were grown at the experimental nursery of the Northwest Agriculture and Forestry University in Yangling (34°52′N, 108°7′E), China. The plants were cultivated in the nursery using a spacing of 50 cm × 25 cm. The following steps were used in the propagation and cultivation of the apple trees: First, seeds of M. robusta Rehd. were sown during the Spring of 2014 and the T337 buds were subsequently grafted on the rootstocks at approximately 20 cm above the ground level during the Autumn of 2014. Second, the portions of rootstocks situated above the buds were cut off during the Spring of 2015; after pruning, only T337 buds were allowed to grow through the vegetation period. Third, the ‘Nagafu No. 2’ scion buds were grafted on the interstems at approximately 30 cm above the first grafting points during the Autumn of 2015; subsequently, the T337 part situated above the scion buds were cut off during the Spring of the following year (2016). Afterwards, we were able to obtain the 1-year-old ‘Nagafu No. 2’ (a ‘Fuji’ cultivar)/T337 /M. robusta Rehd. grafted combinations, which were used in our experiments during July (2016). The plant materials were subjected to standard management practices.
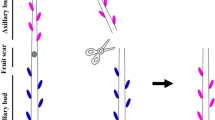
During July (2016), exogenous applications of GA3, cytokinin (6-benzylaminopruine, BA), and BA + PAC (paclobutrazol) were applied on the grafted apple trees when terminal shoots reached 70–80 cm in length, as measured from the point of the scion graft to the top of the main shoot. 5.0 mM working solutions of GA3 and BA amended with 0.05% dimethyl sulfoxide (DMSO) were applied with a sprayer. The 5.0 mM PAC solution amended with 0.05% DMSO was applied immediately after the BA treatment. Spraying treatments were applied by hand using a 1-L Solo Snazzy pressurized hand sprayer. The GA3, BA, and BA + PAC sprays were applied to the top sections (containing 8 ABs) of newly developing scion shoots. The control materials were sprayed with distilled water containing 0.05% DMSO. A single spray of each solution (approximately 5 mL) was applied to each tree. Each treatment consisted of three biological replicates where one replicate consisted of 100 plants. A schematic representing trees and replicates was prepared to help document our exogenous applications in the experiment blocks and this method was used to minimize the interaction of the applications (Fig. S1). To minimize variability between samples, only the ABs at node 3 from the apex were sampled in each of the treatments. ABs were sampled at 0, 4 and 8 h after the treatments were applied. For molecular analyses, 20 ABs were sampled from 10 plants at each sampled time-point; constituting one biological replicate. For the measurement of sucrose content, ABs in 70 plants were sampled and the 10 remaining plants were used for phenotyping. The sampled materials were immediately frozen in liquid nitrogen and stored at − 80 °C. Bud length at nodes 3 was measured using a vernier caliper every 2 days after the treatments were administered. As shown in Fig. 1a–e, ABs exhibiting expansion and elongation, with emergence of a ‘growing tip’, were defined as indicators of ABs outgrowth. The percentage of induced ABs was determined at 1 week after treatments.
Effect of the exogenous application of GA3, BA, or BA + PAC on the outgrowth of ABs in 1-year-old apple trees. a Inactive bud. b, c Induced outgrowth buds. d, e Lateral branches. f Branching phenotypes of treated shoots after the application of different hormonal treatments. Scale bars = 2 cm. g, h Representative ABs treated with GA3, BA, or BA + PAC. Scale bars = 2 cm. i The percentage of induced ABs (in treated section), which exhibited expansion and elongation with the ‘growing tip’ appearing was evaluated 1 week after treatments (n = 30). j ABs’ length at nodes 3 was measured using a vernier caliper every 2 days starting after treatments (n = 30). The data represent the mean ± standard error in i and j. Small letters (a, b, c) above the bars indicate significant differences according to Tukey test between the different treatments at each time-point. Red arrows indicate the induced outgrowth buds with the ‘growing tip’
Extraction and measurement of sucrose content in ABs
At each time point, approximately 30 frozen ABs obtained from the GA3 and BA treatments and the untreated trees were ground and subsequently weighted (0.3 g) for measurements of sucrose content. The extraction was performed as described by Rosa et al. (2009) and the identification was conducted using a high-performance liquid chromatography (HPLC) system with a model 1525 pump (Waters 1525, USA) and 2707 autosampler (Waters 2707, USA) with a thermostat. Solute elution was monitored using a Visible detector (Waters 2414, USA) and data acquisition and processing were performed by the Breeze software (Waters, USA). Twenty microliters of purified extract was injected by auto-sampler (Waters 2707). Analysis conditions were as follows: a Waters Sugar-Pak I column (Waters C18) was used; the column temperature was 90 °C; the mobile phase was 50 mg/L calcium ethylenediaminetetraacetic acid (EDTA-Ca; dissolved in redistilled water); the flow rate was 0.5 ml/min; the total run time was 20 min; and the Visible detector (Waters 2414) oven temperature was 40 °C.
RNA extraction and cDNA synthesis
At each time point, total RNA from ABs obtained from the GA3, BA and decapitation treatments and the untreated trees was extracted using a CTAB-based method (Gambino et al. 2008). Integrity of the total RNA, as indicated by three bright stripes, was verified by running the samples on 2% agarose gels and RNA concentrations were determined using a Nanodrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). All of the obtained OD260/280 values ranged between 1.8 and 2.0. Last, cDNA were synthesized using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa Bio, Shiga, Japan).
qRT-PCR analysis
The relative expression of genes involved in branching inhibition (MdTCP33, MdTCP40, MdTCP16, MdSBP12, and MdSBP18), cytokinin biosynthesis and signaling (MdIPT1, MdIPT3, MdRRB9, MdRRB11, MdRRA3, and MdRRA14), auxin biosynthesis and transport (MdYUCCA6a, MdYUCCA6b, MdYUCCA10a, MdAUX1-1, MdAUX1-2, and MdPIN1), DELLAs (MdRGL1a, MdRGL1b, MdRGL2a, MdRGL2b, MdRGL3a, and MdRGL3b), Tre6P biosynthesis (MdTPS5, MdTPS6, MdTPS7, MdTPS8, and MdTPS10) and cell proliferation (MdCYCA1;1, MdCYCB1;1, MdCYCB1;2, MdCDKB2;1, MdCDC20a, MdCDC20b, MdTCP13, MdTCP38, and MdTCP34) was analyzed by Quantitative real-time PCR (qRT-PCR). Related protein sequences that were reported in other species were used as queries in BLASTP searches with default parameters against the Malus domestica Genome database (Malus domestica Genome database v1.0, http://www.rosaceae.org/). The gene names (the abbreviation and full names) and the apple accession numbers, as well as the species and proteins of the homologue on which the apple protein is based, were listed in Table S1. The apple Actin gene served as a reference gene. Specific primers used in the qRT-PCR analyses were designed using Primer Premier 6.0 software. Primer sequences are listed in Table S2.
The reaction mixtures contained 10.0 µL of 2 × SYBR Premix Ex Taq II (TaKaRa, Beijing, China), 2.0 µL of cDNA template (150 ng/µl), 0.8 µL of the forward and reverse primers, and 6.4 µL of ddH2O, to make a total volume of 20 µL. The qRT-PCR assays were run on a CFX ConnectTM Optics Module (Bio-Rad, Singapore) with an initial denaturation of 95 °C for 180 s, followed by 39 cycles of 95 °C for 15 s, 58 °C for 20 s, and 72 °C for 20 s. The resulting fragments were subjected to a melting-curve analysis immediately after real-time PCR to verify the presence of gene-specific PCR products under the following conditions: 95 °C for 15 s, followed by a constant increase from 60 to 95 °C at a 2% ramp rate. Each reaction was performed in triplicate and the 2−DDCt method was used to calculate the relative expression level of the different genes (Livak and Schmittgen 2001).
Statistical analysis
Statistical analyses of the qRT-PCR data were performed in Excel 2003. Treatment effects were determined by ANOVA and the means were compared by the Tukey test using SPSS 18 (Chicago, IL, USA). Differences in values were considered statistically significant at P < 0.05. Figures were generated in Sigma Plot 10.0 (Systat Software, Inc.).
Results
Phenotypic changes in AB outgrowth in response to different exogenous applications
The impact of GA3 on the outgrowth of ABs was examined. In contrast to exogenous application of BA, GA3 was not effective for promoting apple ABs’ outgrowth (Fig. 1). ABs on GA3-treated shoots remained inactive (Fig. 1i, j). Additionally, a co-application of BA and paclobutrazol (an inhibitor of gibberellin biosynthesis, PAC) (Ghosh et al. 2011) did not affect the number of ABs that were activated compared to the BA treatment alone (Fig. 1i), but significantly reduced the length of lateral branches mediated by BA at node 3 (Fig. 1j). These data suggest that the inhibition of endogenous GA may limit the growth of lateral branches that are activated by BA in apple.
GA3 supply does not inhibit the expression of branching-inhibitor genes
BRANCHED1 (BRC1) and BRANCHED1(BRC2) act as inhibitors of AB outgrowth and are specifically expressed in ABs (Aguilarmartínez et al. 2007; Finlayson 2007). Transcript levels of MdTCP40, MdTCP16 and MdTCP33, which are homologs of AtBRC1 and AtBRC2 (Xu et al. 2014), were analyzed by quantitative real-time PCR (qRT-PCR) in different tissues in order to determine their role in the local control of AB outgrowth (Fig. S2 a–d). Results indicated that all of these genes were highly expressed in ABs. In addition, MdTCP40 and MdTCP16 also exhibited high levels of expression in flower buds, which could be considered as another axillary structure (Fig. S2 a, b). In contrast, the level of a class I TCP gene, MdTCP34, which is a homolog of AtTCP15 (Xu et al. 2014), was examined as a negative control (Fig. S2d). Results indicated that MdTCP34 was highly expressed in roots and fruits (Fig. S2 d), thus providing further support for the specific high expression of MdTCP40, MdTCP16, and MdTCP33 in ABs.
Since BRC1 and BRC2 transcript levels are often used as a marker for the inhibition of bud growth (Aguilarmartínez et al. 2007), their expression levels were analyzed in ABs obtained from GA3-, BA-, or decapitation-treated shoots (Fig. 2). MdTCP33, MdTCP40, and MdTCP16 were significantly down-regulated at 4 and 8 h after the BA and decapitation treatments, except for MdTCP16, which did not exhibit a difference at 8 h after decapitation (Fig. 2a–f). In contrast, these genes did not exhibit any difference in transcript levels in GA3-treated vs. non-treated bud samples at the 4 h sampling time. In fact, MdTCP40 and MdTCP16 expression levels were distinctly up-regulated at 8 h in GA3-treated ABs (Fig. 2b, c). Taken together, these data suggest that the inactivity of apple AB to activate in response to GA may be due to the inability of GA3 to suppress the putative branching-inhibitor genes. Transcript levels of MdSBP12 and MdSBP18, which are homologs of AtSPL2 and AtSPL9/15 (Li et al. 2013) that potentially activate TB1 (BRC1) expression (Lu et al. 2013; Liu et al. 2017), were also strongly reduced at 4 and 8 h after the BA and decapitation treatments (Fig. 2g–j). In contrast, no significant difference in MdSBP12 expression was observed at 4 h and 8 h in GA3-treated buds vs. untreated buds. In contrast, however, MdSBP18 was down-regulated at 4 and 8 h after the GA3 treatment.
Effect of GA3, BA and decapitation on the expression of branching regulators (MdTCP33, MdTCP40, MdTCP16, MdSBP12, and MdSBP18) in ABs located at node 3 of 1-year-old apple trees. Samples from non-treated and treated ABs were collected at 0, 4, and 8 h after the treatments. Decapitation was conducted on the 1-year-old nursery apple trees when the main stem reached to 90 cm by removing approximately 1 cm of the shoot tip. All transcript levels were normalized to their respective corresponding levels in non-treated controls (0 h). Apple actin was used as a reference gene. The data represent the mean ± standard error of three replicates. Small letters (a, b, c) above the bars indicate statistically significant differences (P < 0.05) according to Tukey test between non-treated and treated ABs at each time-point. Decap: decapitation
GA3 supply may not induce CK responses effectively
In order to examine the involvement of cytokinin-related responses in GA3-, BA + PAC-, and BA-treated buds, transcript levels of cytokinin synthesis-related genes (MdIPT1 and MdIPT3) and CK signaling-related genes (MdRRB9, MdRRB11, MdRRA3, and MdRRA14) were analyzed (Fig. 3). MdIPT1 was down-regulated by the GA3 treatment at 4 h, but significantly up-regulated at 8 h. In contrast, MdIPT1 transcript levels did not change in BA + PAC- or BA-treated buds at the 8-h sampling time point, relative to transcript levels in non-treated buds. MdIPT3, however, exhibited higher expression in buds treated with exogenous hormones than in untreated buds from 4 to 8 h, with an exception for in BA-treated buds at 8 h whose expression did not appreciably change. MdRRB9 and MdRRB11 (B-type MdRR genes) (Li et al. 2017) were significantly up-regulated during the first 8 h in the presence of BA, but not GA3 (Fig. 3c, d). MdRRA3 and MdRRA14 (A-type MdRRs genes) (Li et al. 2017) were markedly up-regulated by the BA treatment at all time points, while the co-application of BA + PAC did not effect on transcript levels; except for MdRRA14 which was up-regulated at 8 h in the BA + PAC treatment in comparison to the BA treatment (Fig. 3e, f). MdRRB9 and MdRRB11 did not exhibit any change in response to the GA3 treatment; however, their expression levels were lower than in the untreated buds at 8 h. MdRRA3 and MdRRA14 were up-regulated at 4 h in the GA3 treatment, but not as strongly as in the BA treatment and were down-regulated at 8 h. Collectively, these data indicate that GA3 does not have a major impact on cytokinin-signaling-related gene expression. DELLAs, which are repressors of GA signaling (Peng et al. 1997), have been reported to influence the transactivation ability of type-B ARR genes (Marínde et al. 2015). Transcript levels of six MdDELLA genes (MdRGL1a, MdRGL1b, MdRGL2a, MdRGL2b, MdRGL3a, and MdRGL3b), which were identified in the apple genome (Foster et al. 2006), were also investigated (Fig. S3). MdRGL1a and MdRGL3a were not up-regulated at 4 h, but exhibited significant up-regulation at 8 h after the GA3 treatment. MdRGL1b and MdRGL2a were significantly down-regulated at 4 h, whereas they were up-regulated at 8 h in GA3-treated buds. MdRGL2b and MdRGL3b exhibited higher expression in GA3-treated buds than in untreated buds at all sampling time points, except for MdRGL2b, which did not exhibit a significant increase in its transcript level at 8 h. MdRGL1a, MdRGL2b, MdRGL3a, and MdRGL3b were also strongly up-regulated at 4 or 8 h in the BA treatment. MdRGL1b and MdRGL2a, however, were down-regulated in the BA-treated buds at all of the sampled time points.
Effect of GA3, BA + PAC, or BA on the expression of cytokinin synthesis-related genes (MdIPT1 and MdIPT3) and signaling-related genes (MdRRB9, MdRRB11, MdRRA3, and MdRRA14) in apple ABs located at node 3. Samples from non-treated, GA3-, BA + PAC-, and BA-treated ABs were collected at 0, 4, and 8 h after the treatments. All transcript levels were normalized to their respective corresponding levels in non-treated controls (0 h). Apple actin was used as a reference gene. The data represent the mean ± standard error of three replicates. Small letters (a, b, c) above the bars indicate statistically significant differences (P < 0.05) according to Tukey test between non-treated, GA3-, BA + PAC-, and BA-treated ABs at each time-point
Effect of GA3 supply on auxin export out of ABs
The export of auxin out of ABs into subtending stems has been previously demonstrated in activated ABs (Li and Bangerth 1999; Balla et al. 2011). In order to investigate the involvement of bud auxin export in GA3-, BA + PAC-, and BA-treated ABs, the relative expression of auxin synthesis-related genes (MdYUCCA6a, MdYUCCA6b, and MdYUCCA10) and transport-related genes (MdAUX1-1, MdAUX1-2 and MdPIN1) was examined (Fig. 4). MdYUCCA6a and MdYUCCA6b were up-regulated, but MdYUCCA10 was strongly down-regulated by all hormone treatments at all of the sampled time points. The expression of MdAUX1-1, MdAUX1-2, and MdPIN1 did not exhibit a response to the GA3 treatment and their level of expression was similar to expression levels in untreated buds at 4 and 8 h. In contrast, transcripts of these genes were rapidly up-regulated in ABs with the presence of BA at the 8-h time point. The expression levels in BA-treated ABs were significantly higher than in the GA3 treatment. These data indicate that the export of auxin out of ABs may not be induced by the GA3 treatment.
Effect of GA3, BA + PAC, or BA on the expression of auxin synthesis-related genes (MdYUCCA6a, MdYUCCA6b, and MdYUCCA10a) and transport-related genes (MdAUX1-1, MdAUX1-2, and MdPIN1) in apple ABs located at node 3. Samples from non-treated, GA3-, BA + PAC-, and BA-treated ABs were collected at 0, 4, and 8 h after the treatments. All transcript levels were normalized to their respective corresponding levels in non-treated controls (0 h). Apple actin was used as a reference gene. The data represent the mean ± standard error of three replicates. Small letters (a, b, c) above the bars indicate statistically significant differences (P < 0.05) according to Tukey test between non-treated, GA3-, BA + PAC-, and BA-treated ABs at each time-point
Effect of GA3 supply on SL
To test the involvement of strigolactones in GA3-, BA + PAC-, and BA-treated buds, the expression of strigolactones synthesis-related genes (MdMAX1 and MdD14) and SL signaling-related genes (MdMAX2) were quantified (Fig. 5). MdMAX1 was significantly down-regulated by all hormone treatments at 4 and 8 h, respectively (Fig. 5a). MdD14 exhibited lower expression in GA3-treated buds than in untreated buds at 4 h, while its expression did not appreciably change in response to the BA treatment at all of the sampling time points (Fig. 5b). The expression of MdMAX2 was down-regulated in GA3-treated buds at 4 h, while no difference was observed in the presence of BA-treated vs. non-treated bud samples at 4 h (Fig. 5c). MdMAX2, however, was strongly up-regulated by all hormone applications at 8 h.
Effect of GA3, BA + PAC, or BA on the expression of strigolactone synthesis-related genes (MdMAX1 and MdD14) and signaling-related gene (MdMAX2) in apple ABs located at node 3. Samples from non-treated, GA3-, BA + PAC-, and BA-treated ABs were collected at 0, 4, and 8 h after the treatments. All transcript levels were normalized to their respective corresponding levels in non-treated controls (0 h). Apple actin was used as a reference gene. The data represent the mean ± standard error of three replicates. Small letters (a, b, c) above the bars indicate statistically significant differences (P < 0.05) according to Tukey test between non-treated, GA3-, BA + PAC-, and BA-treated ABs at each time-point
GA3 supply promotes Tre6P biosynthesis in ABs
In order to determine whether or not the activation of ABs in decapitated shoots is correlated with Tre6P levels, the expression of several MdTPS genes that are essential components of the Tre6P biosynthetic pathway (Du et al. 2017) were analyzed using qRT-PCR (Fig. 6a–e). As expected, all five MdTPS genes (MdTPS5, MdTPS6, MdTPS7, MdTPS8, and MdTPS10) were strongly up-regulated within 4 and 8 h after decapitation; especially MdTPS5 which exhibited more than a 120-fold increase in transcript levels at 8 h (Fig. 6a). MdTPS gene transcript accumulation was also investigated in response to the GA3 and BA treatments (Fig. 6f–j). MdTPS5 and MdTPS8 were strongly down-regulated by the GA3 treatment at 4 h, while MdTPS8 was significantly up-regulated at 8 h. MdTPS6, however, did not exhibit a significant difference in expression between GA3- treated and untreated buds at 4 and 8 h. MdTPS7 and MdTPS10 were up-regulated at all of the sampling time points of GA3 with respect to control, expect for MdTPS7, which did not exhibit a significant increase in its expression at 8 h. MdTPS5 was significantly down-regulated at 4 h, whereas, it was up-regulated at 8 h in the BA treatment. MdTPS7, however, was strongly up-regulated at 4 h. MdTPS6 and MdTPS8 did not exhibit a significant difference in expression between BA-treated and untreated buds at 4 and 8 h. The expression of MdTPS10, however, increased in response to the BA treatment. Collectively, the data indicate that there was no correlation between Tre6P biosynthesis and the inactivity of ABs in the GA3 treatment.
Effect of decapitation, GA3, and BA on the expression of Tre6P biosynthesis-related genes (MdTPS5, MdTPS6, MdTPS7, MdTPS8, and MdTPS10) in apple ABs located at node 3. Samples from non-treated and treated ABs were collected at 0, 4, and 8 h after the treatments. Decapitation was conducted on the 1-year-old nursery apple trees when the main stem reached to 90 cm by removing approximately 1 cm of the shoot tip. All transcript levels were normalized to their respective corresponding levels in non-treated controls (0 h). Apple actin was used as a reference gene. The data represent the mean ± standard error of three replicates. Small letters (a, b, c) above the bars indicate statistically significant differences (P < 0.05) according to Tukey test between non-treated and treated ABs at each time-point. Decap: decapitation
GA3 supply reduces endogenous sucrose content in ABs
To examine the effect of GA3 and BA on sucrose metabolism in ABs, endogenous sucrose levels were examined (Fig. 7). In general, sucrose content strongly decreased during the sampled time points in response to the GA3 and BA treatments. The levels of sucrose in GA3-treated ABs, however, were higher than in BA-treated ABs at all of the examined time points, except for 48 h. These data suggest that the effect of GA3 or CK on bud regulation may not be mediated by the supply of sucrose to ABs.
Changes in sucrose levels in ABs located at node 3 in response to the GA3 and BA treatments. Samples from non-treated, GA3, and BA-treated ABs were collected at 0, 4, 8, 24, and 48 h after the treatments. The data represent the mean ± standard error of three replicates. Small letters (a, b, c) above the bars indicate statistically significant differences (P < 0.05) according to Tukey test between non-treated, GA3-, and BA-treated ABs at each time-point
GA3 supply activates cell proliferation in ABs
The function of CK is primarily executed through the control of cell cycle activity (Roef and Onckelen 2010). Based on this fact and the lack of a stimulatory effect of GA3 on AB outgrowth, the expression of several cell division-related genes was analyzed in response to the GA3 and BA treatments (Fig. 8). Expression levels of three class I TCP genes (MdTCP13, MdTCP38, and MdTCP34), which are homologs of AtTCP14 and AtTCP15 (Xu et al. 2014) and are associated with the promotion of cell proliferation (Martin et al. 2011), were also examined. Results indicated that the expression of MdCYCA1;1, MdCYCB1;1, MdCYCB1;2, MdCDKB2;1, MdCDC20a, and MdCDC20b increased in BA-treated ABs at 4 or 8 h, relative to their expression in untreated ABs (Fig. 8a–f). MdCYCB1;1, MdCYCB1;2, and MdCDC20a, however, were also up-regulated by the GA3 treatment at 4 or 8 h (Fig. 8b, c, e). Specifically, the class I TCP genes, MdTCP13, MdTCP38, and MdTCP34, all exhibited higher expression at all of the sampled time points in both GA3- and BA-treated ABs, relative to their expression in untreated ABs (Fig. 8g–i).
Effect of GA3 and BA on the expression of cell proliferation-related genes (MdCYCA1;1, MdCYCB1;1, MdCYCB1;2, MdCDKB2;1, MdCDC20a, MdCDC20b, MdTCP13, MdTCP38, and MdTCP34) in apple ABs located at node 3. Samples from non-treated, GA3-, and BA-treated ABs were collected at 0, 4, and 8 h after the treatments. All transcript levels were normalized to their respective corresponding levels in non-treated controls (0 h). Apple actin was used as a reference gene. The data represent the mean ± standard error of three replicates. Small letters (a, b, c) above the bars indicate statistically significant differences (P < 0.05) according to Tukey test between non-treated, GA3-, and BA-treated ABs at each time-point
Discussion
Although a positive regulatory role of GA3 in branching has been observed in some species such as sweet cherry (Elfving et al. 2011), the perennial woody plant J. curcas (Ni et al. 2015; Rinne et al. 2016) and hybrid aspen (Rinne et al. 2016), the outgrowth of ABs in nursery apple trees was not found to be stimulated by the exogenous application of GA3 in our study. Additionally, we also investigated how the ABs respond to different concentrations of GA3 and BA. Treatment with higher concentrations of GA3 did not lead to promotion in AB outgrowth, while the application of higher concentrations of BA (7 mM) led to a similar increased number of stimulated lateral buds (data was not shown). These data indicated that the inactive ABs in GA3-treated shoots may not be caused by the selected concentration of GA3. The co-application of BA and PAC results in a reduced length of developing lateral branches (Fig. 1j) indicating that bud dormancy may inhibit the role of GA3 in internodal elongation.
To further characterize the observation that gibberellin is unable to activate AB outgrowth, we examined the expression of the bud-specific gene, BRC1 (BRANCHED1) and BRC2 (Aguilarmartínez et al. 2007), both of which are important regulators of shoot branching. BRC1 and BRC2 are class II TCP transcription factors that possess a conserved function in negatively regulating AB outgrowth in various plant species, such as Arabidopsis (Aguilarmartínez et al. 2007), sorghum (Kebrom and Finlayson 2006), rice (Minakuchi et al. 2010), pea (Braun et al. 2012), tomato (Martín-Trillo et al. 2011), potato (Nicolas et al. 2015), and poplar (Muhr et al. 2016). In the present study, expression analyses were used to identify likely apple BRC orthologs based on their typical expression patterns including high levels of expression in non-growing ABs (Aguilarmartínez et al. 2007; Finlayson 2007; Martín-Trillo et al. 2011) and down-regulation under conditions that promote AB outgrowth (Aguilarmartínez et al. 2007; Minakuchi et al. 2010; Braun et al. 2012; Ni et al. 2015). The identified apple BRC orthologs were highly expressed in ABs; however, MdTCP40 and MdTCP16 were also highly expressed in flower buds (Fig. S2a–c). This pattern of expression is similar to the reported pattern of expression of AtBRC1 and AtBRC2, which also exhibited considerable expression in floral organs (Aguilarmartínez et al. 2007; Finlayson 2007). Moreover, BRC1 has also been reported to inhibit the floral transition of the axillary meristems in Arabidopsis by interacting with FLOWERING LOCUS T (FT) (Niwa et al. 2013). In contrast, the expression of BRC1 and BRC2 was down-regulated during the outgrowth of ABs (Aguilarmartínez et al. 2007). Similar patterns of expression were observed in ABs in decapitation or CK treatments in Arabidopsis (Aguilarmartínez et al. 2007), tomato (Martín-Trillo et al. 2011), rice (Minakuchi et al. 2010), pea (Braun et al. 2012), and poplar (Muhr et al. 2016). Therefore, it was reasonable to hypothesize that BRC1 and BRC2 homologues in apple ABs may exhibit a similar reduction of expression in response to the BA and decapitation treatments. Indeed, the expression of MdTCP33, MdTCP40, and MdTCP16 was significantly down-regulated in BA- and decapitation-treated ABs relative to their expression in untreated ABs (Fig. 2a–f). These results support the conserved role of MdTCP33, MdTCP40, and MdTCP16 as functional BRC1 and BRC2 orthologs. Based on these results, we also investigated their expression in ABs following the application of GA3. Transcript levels for all of these genes remained unchanged at 4 h by the GA3 treatment, and levels of MdTCP40 and MdTCP16 expression were highly up-regulated at 8 h (Fig. 2a–c). These data suggest that GA3 may limit the activation of ABs. Additionally, the expression of TB1 (BRC1) can be activated by OsSPL14 (homologs of Arabidopsis AtSPL9) in rice (Lu et al. 2013); as well as in bread wheat (Lu et al. 2013; Liu et al. 2017). In our study, the expression levels of apple BRC genes were correlated with related MdSPL genes (MdSBP12 and MdSBP18) in GA3-, BA- and decapitation-treated ABs, respectively (Fig. 2g–j). These results raise the possibility that MdSBP12 or MdSBP18 activates the expression of apple BRC-homologues.
Exogenous treatments with hormones have a significant effect on the balance of branch-related endogenous hormones that are present in ABs. CKs are involved in the activation of AB outgrowth in various species, including apple. CK-responsive type-B RR genes are believed to act as DNA-binding transcription factors that mediate CK signaling in the regulation of plant development (Sakai et al. 2001; Moubayidin et al. 2010). Many CK-regulated genes require the activation of type-B ARRs (Taniguchi et al. 2007; Argyros et al. 2008). The strongest evidence for the function of type-B ARRs in regulating the CK signaling has been obtained from the functional analysis of ARR1. Specifically, the over-expression of ARR1 in Arabidopsis results in the production of ectopic shoots and causes increased sensitivity to CK, while arr1-1 mutants exhibit diminished sensitivity to CK (Sakai et al. 2001). Our previous study indicated that the increase in apple floral induction by the application of exogenous CK might be due to the transcriptional activation of apple ARR1 on the flowering integrator, MdSOC1a/b (Li et al. 2017). It is also possible that ARR1 may target the expression of branching-related genes to promote AB outgrowth. In this scenario, the maintenance of AB dormancy after treatment with GA3 may be due to the lower expression of two homologues of ARR1 (MdRRB9 and MdRRB11) relative to their expression in BA-treated buds. In Arabidopsis, GA3 can suppress cytokinin responses through SPY-independent pathways, in which SPY suppresses GA signaling and promotes cytokinin responses (Greenboim-Wainberg and Weiss 2005; Maymon et al. 2009; Steiner and Weiss 2012). Additionally, DELLAs, which are considered as repressors of GA signaling (Peng et al. 1997; Cheng et al. 2004), have been reported to be recruited by type-B ARRs to enhance their transcription ability (Marínde et al. 2015). As a result, della mutants consistently exhibit reduced shoot branching (Cheng et al. 2004). Indeed, four (MdRGL1a, MdRGL2b, MdRGL3a, and MdRGL3b) out of six apple MdDELLA genes exhibited high levels of expression in response to the BA treatment (Fig. S3). GAs trigger DELLA degradation and thereby promote growth (Davière and Achard 2013). Transcript levels of apple MdDELLA genes, however, increased in response to the GA3 treatment (Fig. S3). Similar results have been previously reported (Itoh et al. 2002; Richter et al. 2010), indicating that the enhancement of DELLA gene expression may be due to a feedback mechanism. CK-responsive type-A RRs act as negative inhibitors of the CK signaling. CK-response in multiple type-A RR mutants has been examined and results indicate that the ABs in wild-type plants were activated after CK treatment, whereas the ABs in arr3,4,5,6,7,15 multiple mutants were not (Muller et al. 2015). In our study, the transcript abundance of MdRRA3 and MdRRA14 increased in BA-treated ABs at 4 and 8 h (Fig. 3e, f). Activation of the expression of these genes is required for the CK-mediated activation of ABs (Muller et al. 2015). Conversely, GA3 suppressed the induction of the CK primary-response gene, A-ARR5 (Greenboim-Wainberg and Weiss 2005), and similar results were also found in the present study. The expression of MdRRA3 and MdRRA14, which cluster with ARR5 (Li et al. 2017), was not significantly induced by exogenous application of GA3. Muller et al. (Muller et al. 2015) reported, however, that IPT-mediated CK synthesis in bud initiation did not require the ARRs involved in bud activation. We speculate that the up-regulation of CK biosynthesis observed in GA3-treated buds at 4 or 8 h (Fig. 3) may play a role in bud development, but not the initial activation of ABs.
Auxin acts antagonistically to cytokinins, which inhibits the outgrowth of AB mediated by apical dominance. There is a strong correlation, however, between AB outgrowth and the export of auxin out of the bud that AB outgrowth occurs along with bud auxin export (Li and Bangerth 1999; Balla et al. 2011; Domagalska and Leyser 2011). As reviewed by Muller and Leyser (2011), CK-mediated activation of ABs has been reported to be dependent on the change of endogenous auxin levels in buds. In our results, CK was found to up-regulate the biosynthesis of auxin in ABs. Indeed, the application of BA increased the expression of two auxin biosynthesis genes (YUCCA6a and YUCCA6b) (Fig. 4a, b). The effect of cytokinin on auxin biosynthesis has been previously reported in associated stem tissues (Li and Bangerth 2003). In our study, the auxin transcript genes, MdAUX1-1, MdAUX1-2, and MdPIN1, were up-regulated in response to exogenous application of BA (Fig. 4d–f). We also cannot determine, however, if the promotion of auxin biosynthesis in ABs is caused by CK directly or by the increased export of auxin from AB tissues; which would also stimulate endogenous auxin synthesis. ABs treated with GA3 exhibited increased auxin synthesis, but no significant difference in the expression of auxin transport genes (MdAUX1-1, MdAUX1-2 and MdPIN1) was observed relative to untreated buds. It should be noted that GA also enhances apical dominance (Willige et al. 2011). Therefore, the application of GA3 may increase the auxin levels in the main stem, which intensifies the competitiveness of the main shoot apex for access to a common polar auxin transport stream (PATS) (Bennett et al. 2006; Crawford et al. 2010); hence preventing auxin export from ABs.
Recent studies have shown that AB outgrowth can be regulated by strigolactones. In buds, the expression of the strigolactone synthesis-related genes, MdMAX1, was down-regulated by GA3 and BA (Fig. 5a). MdMAX2, a key regulatory gene in the signal transduction of strigolactones, was inhibited at 4 h, while its expression was up-regulated at 8 h by GA3 treatment (Fig. 5c). These effects may result from the independence of GAs and SLs in repressing AB outgrowth (De et al. 2013). However, in Rosa hybrida plants, RwMAX2 was repressed early by sucrose; which could trigger the outgrowth of ABs (François et al. 2015). It was considered that the SL response through RwMAX2 may be involved in bud outgrowth that is promoted by sucrose. In contrast, the expression of MdMAX2 was strongly induced in BA-treated buds. It is plausible that this contrasting result may indicate that the outgrowth of ABs promoted by CK was not caused by the inhibition of SL, which was reported to act antagonistically with CK in bud outgrowth control (Dun et al. 2012). Additionally, SLs and CK can act directly in buds to control their outgrowth (Dun et al. 2012). Our previous physiological study demonstrated that the co-application of GA3 with BA did not inhibit the BA-triggered bud outgrowth, but promoted an increase in the length of developing shoots derived from the outgrowth of ABs (data was not shown). However, the outgrowth of ABs induced by CK can be efficiently inhibited by the application of GR24 (Brewer and Beveridge 2009; Dun et al. 2012, 2013). We hypothesize that GA3 may play a role in internode elongation in the presence of a promoter of branching rather than the inhibition of AB outgrowth, similar to what SL does. However, in rice, there is evidence that DELLA SLR1 can interact with DWARF14 (D14; the putative SL receptor) in an SL-dependent manner (Nakamura et al. 2013). As a result, further investigations are warranted and necessary to elucidate the probable mechanism underlying the interplay of GA and SL signaling in controlling AB outgrowth.
In addition to the role of branch-related hormones, sucrose and Tre6P have been recently demonstrated to have a strong influence on shoot branching (Mason et al. 2014; François et al. 2015; Fichtner et al. 2017). Tre6P, a signal of sucrose availability in plants (Lunn et al. 2006; Yadav et al. 2014), is synthesized by Tre6P synthase (TPS) and dephosphorylated to trehalose by Tre6P phosphatase (TPP). Consistent with the results obtained by Fichtner et al. (Fichtner et al. 2017), all of the examined MdTPS genes (MdTPS5, MdTPS6, MdTPS7, MdTPS8, and MdTPS10) were rapidly induced and maintained high levels of expression in response to decapitation in the present study (Fig. 6a–e). In sorghum, the expression of several TPS and TPP genes changed during the outgrowth of sorghum buds (Kebrom and Mullet 2016). Constitutive over-expression of TPS raises Tre6P levels, thus generating a ‘high-sucrose’ signal which also results in enhanced branching (Yadav et al. 2014). As a bud-activating treatment, the application of BA, however, did not lead to changes in the expression of MdTPS genes that were similar to those observed in response to decapitation (Fig. 6). However, contrary to the effect of GA3 on AB activation (Fig. 1), an increased abundance of MdTPSs transcripts was observed in GA3-treated buds (Fig. 6g–j). We speculate that the supply of sucrose to ABs may act directly in buds’ outgrowth after removal of the apex. On the other hand, the application of exogenous hormones may not affect the import of sucrose into ABs, but it may impact the synthesis in ABs, which may not be the dominant factor in the control of AB outgrowth. Additionally, examination of endogenous sugar levels in ABs indicated that sucrose levels were lower in GA3-treated buds than in untreated buds (Fig. 7). The lowest sucrose level at each time point, however, was observed in BA-treated buds, suggesting that the effect of GA3 or CK on bud regulation may not be mediated by the level of endogenous sucrose to ABs. Interestingly, CKs were not reported to be necessarily involved in sucrose-triggered AB outgrowth in Rosa hybrida plants (François et al. 2015). These data support that supposition that the function of CK and sucrose in bud activation may involve independent signaling pathways. Considering the positive correlation between Tre6P biosynthesis and AB outgrowth in decapitated shoots, Tre6P or sucrose, at least, do not appear to be repressors of AB outgrowth in apple. In our previous study, the exogenous application of sucrose on apple trees significantly enhanced floral induction (Du et al. 2017); whereas a positive effect on AB outgrowth was not observed (data not shown) as found in previous studies (Mason et al. 2014; François et al. 2015). We suggest that perhaps the timing or location, concentration, or the sucrose type (such as sucrose analogues) of the sucrose-treatment are crucial factors for eliciting the response. Mason et al. (2014) reported that a relatively low sucrose supply to ABs may limit bud release. Therefore, further studies are required to determine whether sucrose application has a positive effect on apple AB outgrowth.
Since the exogenous application of CK can activate buds, and due to their known roles in promoting cell division in the shoot apical meristem (SAM), an intuitively obvious mechanism for CK-mediated bud activation is their ability to promote cell proliferation. GAs and CKs are known to exert antagonistic regulation of SAM activity, which is enhanced by CKs and restricted by GAs (Yanai et al. 2005). Therefore, it seems logical to presume that the activation of AB outgrowth may be related to the activation of cell proliferation in buds. Indeed, there is evidence that the proliferating cell nuclear antigen (PCNA/DNA polymerase auxiliary protein) gene is induced in response to decapitation and repressed if buds reentered a dormant state (Shimizu and Mori 1998). Some TCP genes were also found to be involved in the control of growth and cell cycling, such as axillary meristem activity (Kosugi and Ohashi 1997; Aguilarmartínez et al. 2007; Finlayson 2007). In our study, the expression of selected cell proliferation-related genes increased in response to the BA treatment after 4 or 8 h (Fig. 8). Cell proliferation can also be activated by GA3; however, GA3 did not result in the activation of ABs. These data indicate that although the bud remains inactive, the expression of cell division-related genes in ABs can also be induced by GA. In transgenic tobacco plants, the overexpression of AtcyclinD2 and AtcyclinD3 was not reflected by an increase in shoot branching (Boucheron et al. 2005). Considering the role of GA in the control of stem elongation (Daviere et al. 2014), which occurs from early stages along with cell proliferation (Martin et al. 2011), we hypothesize that the induction of cell proliferation by the supply of GA3 may be related to the preparation of internode elongation rather than the activation of AB outgrowth. Thus, if a bud is not activated, simply inducing cell proliferation is not sufficient to trigger AB outgrowth. In contrast, the regulation of CK synthesis and response can have a direct effect on AB outgrowth (Muller et al. 2015). These findings suggest that CK must do more than simply up-regulate cell proliferation to promote AB outgrowth. In Fig. 9, we present a working model for some of the factors that are putatively involved in the regulation of AB outgrowth by GA3.
Hypothetical model of the regulation of AB outgrowth by GA3 and other bud-regulating factors in apple. Branching inhibitors and MdSBP12/18 are up-regulated by GA3 compared with BA, inhibiting ABs from being activated. Auxin transport-related genes (MdPIN1 and MdAUX1-1/1-2) are down-regulated by GA3, which may limit auxin export out of bud, thus preventing bud outgrowth. The CK response, required for bud activation, is inhibited by GA3 through the down-regulation of the CK response-regulators, MdRRB9/11 and MdRRA3/14, thus preventing the ABs from being activated
In summary, the present study demonstrated that exogenous applications of GA3 do not activate the outgrowth of ABs in apple, but they effectively increase lateral branch length. It is plausible that the inability of ABs to activate in response to GA3 may be partly due to the up-regulation of branching inhibitors and the expression of the MdSBP1 and MdSBP18 genes. Additionally, GA3 induced a decrease in the transcription of CK response-regulator genes and auxin transport-related genes in ABs, which resulted in the inability of GA3 to induce CK response and the export of auxin out of the bud and subsequent AB outgrowth. It is also possible that changes in Tre6P biosynthesis and sucrose levels may not be a limiting factor in the GA3- and CK-mediated regulation of AB outgrowth. Additionally, the activation of cell proliferation in response to the exogenous application of GA3 is not sufficient for inducing AB outgrowth. Collectively, our results provide basic information pertaining to the role of GA3 in bud regulation via crosstalk with other branching-regulating signals.
References
Agharkar M, Lomba P, Altpeter F, Zhang H, Kenworthy K, Lange T (2007) Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnol J 5:791–801
Aguilarmartínez JA, Pozacarrión C, Cubas P (2007) Arabidopsis BRANCHED1 Acts as an integrator of branching signals within axillary buds. Plant Cell 19:458
Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA 108:20242–20247
Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20:2102–2116
Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51:1019–1029
Atay E, Koyuncu F (2013) A new approach for augmenting branching of nursery trees and its comparison with other methods. Sci Hortic 160:345–350
Balla J, Kalousek P, Reinöhl V, Friml J, Procházka S (2011) Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J Cell Mol Biol 65:571–577
Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16:553–563
Bennett T, Hines G, Leyser O (2014) Canalization: what the flux? Trends Genet 30:41–48
Bergmann C, Wegmann K, Frischmuth K, Samson E, Kranz A, Weigelt D, Koll P, Welzel P (1993) Stimulation of Orobanche crenata seed germination by (+)-strigol and structural analogues dependence on constitution and configuration of the germinatio stimulants. J Plant Physiol 142:338–342
Bostan M (2010) Influence of crown formation method on development of the apple trees in the nursery. J Am Soc Hortic Sci 7:1193–1198
Boucheron E, Healy JC, Sauvanet A, Rembur J, Noin M, Sekine M, Riou KC, Murray JA, Van OH, Chriqui D (2005) Ectopic expression of Arabidopsis CYCD2 and CYCD3 in tobacco has distinct effects on the structural organization of the shoot apical meristem. J Exp Bot 56:123–134
Braun N, Germain ADS, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Xin L, Maia-Grondard A, Signor CL, Bouteiller N (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158:225–238
Brewer PB, Beveridge CA (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150:482–493
Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131:1055
Choubane D, Rabot A, Mortreau E, Legourrierec J, Péron T, Foucher F, Ahcène Y, Pelleschi-Travier S, Leduc N, Hamama L (2012) Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp. J Plant Physiol 169:1271–1280
Crawford S, Shinohara NT, Williamson L, George G, Hepworth J, Muller D, Domagalska MA, Leyser O (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137:2905
Daviere JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24:1923–1928
Davière JM, Achard P (2013) Gibberellin signaling in plants. Development 140:1147–1151
De SGA, Ligerot Y, Dun EA, Pillot JP, Ross JJ, Beveridge CA, Rameau C (2013) Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol 163:1012–1025
Domagalska MA, Leyser O (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12:211
Du L, Qi S, Ma J, Xing L, Fan S, Zhang S, Li Y, Shen Y, Zhang D, Han M (2017) Identification of TPS family members in apple (Malus × domestica Borkh.) and the effect of sucrose sprays on TPS expression and floral induction. Plant Physiol Biochem 120:10–23
Dun EA, Germain ADS, Rameau C, Beveridge CA (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158:487
Dun EA, Germain ADS, Rameau C, Beveridge CA (2013) Dynamics of strigolactone function and shoot branching responses in Pisum sativum. Mol Plant 6:128
Elfving DC (2010) Plant bioregulators in the deciduous fruit tree nursery. XI Int Symp Plant Bioregul Fruit Prod 884:159–166
Elfving D, Visser D, Henry J (2011) Gibberellins stimulate lateral branch development in young sweet cherry trees in the orchard. Int J Fruit Sci 11:41–54
Fan S, Zhang D, Gao C, Zhao M, Wu H, Li Y, Shen Y, Han M (2017) Identification, classification, and expression analysis of GRAS gene family in Malus domestica. Front Physiol 8:253
Fichtner F, Barbier FF, Feil R, Watanabe M, Annunziata MG, Chabikwa TG, Hofgen R, Stitt M, Beveridge CA, Lunn JE (2017) Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J 92:611–623
Finlayson SA (2007) Arabidopsis teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot teosinte Branched1. Plant Cell Physiol 48:667
Foster T, Kirk C, Jones WT, Allan AC, Espley R, Karunairetnam S, Rakonjac J (2006) Characterisation of the DELLA subfamily in apple (Malus × domestica Borkh.). Tree Genet Genomes 3:187–197
François B, Thomas P, Marion L, Maria-Dolores PG, Quentin B, Jakub R, Stéphanie BM, Sylvie C, Remi L, Benoît P (2015) Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J Exp Bot 66:2569
Gambino G, Perrone I, Gribaudo I (2008) A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem Anal 19:520–525
Ghosh A, Chikara J, Chaudhary DR (2011) Diminution of economic yield as affected by pruning and chemical manipulation of Jatropha curcas L. Biomass Bioenergy 35:1021–1029
Greenboim-Wainberg Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17:92
Hao L, Wang RX, Qian Q, Yan MX, Meng XB, Fu ZM, Yan CY, Jiang B, Zhen S, Li JY (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21:1512
Henry C, Rabot A, Laloi M, Mortreau E, Sigogne M, Leduc N, Lemoine R, Sakr S, Vian A, Pelleschitravier S (2011) Regulation of RhSUC2, a sucrose transporter, is correlated with the light control of bud burst in Rosa sp. Plant Cell Environ 34:1776–1789
Ho KM, English S, Bell J (1981) The control of the patterned differentiation of vascular tissues. Adv Bot Res 9:151–262
Holalu SV, Finlayson SA (2017) The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J Exp Bot 68:943–952
Itoh H, Ueguchitanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14:57
Kebrom TH, Finlayson SA (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140:1109
Kebrom TH, Mullet JE (2016) Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in Sorghum. Plant Physiol 170:2232–2250
Kebrom TH, Spielmeyer W, Finnegan EJ (2013) Grasses provide new insights into regulation of shoot branching. Trends Plant Sci 18:41–48
Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9:1607
Kviklys D (2006) Induction of feathering of apple planting material. Agron Vestis 9:58–63
Labuschagné IF, Louw JH, Schmidt K, Sadie A (2003) Selection for increased budbreak in apple. J Am Soc Hortic Sci 128:363–373
Leyser O (2009) The control of shoot branching: an example of plant information processing. Plant Cell Environ 32:694–703
Li C-J, Bangerth F (1999) Autoinhibition of indoleacetic acid transport in the shoots of two-branched pea (Pisum sativum) plants and its relationship to correlative dominance. Physiol Plant 106:415–420
Li C, Bangerth F (2003) Stimulatory effect of cytokinins and interaction with IAA on the release of lateral buds of pea plants from apical dominance. J Plant Physiol 160:1059–1063
Li J, Hou H, Li X, Jiang X, Yin X, Gao H, Zheng Y, Bassett CL, Wang X (2013) Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant Physiol Biochem 70:100
Li Y, Zhang D, Zhang L, Zuo X, Fan S, Zhang X, Shalmani A, Han M (2017) Identification and expression analysis of cytokinin response-regulator genes during floral induction in apple (Malus domestica Borkh). Plant Growth Regul:1–10
Liu J, Cheng X, Liu P, Sun J (2017) miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol 174:1931
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408
Lo SF, Yang SY, Chen KT, Hsing YL, Zeevaart JAD, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20:2603–2618
Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, Jing Y, Meng X, Hu X, Qian Q, Fu X, Wang Y, Li J (2013) Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25:3743–3759
Lunn EJ, Feil R, Hendriks Janneke HM, Gibon Y, Morcuende R, Osuna D, Scheible W, Carillo P, Hajirezaei MR, Stitt M (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADP glucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397:139
Marínde lRN, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Grønlund AL, Wanchookohli A, Thomas SG, Bennett MJ (2015) Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. Plos Genet 11:e1005337
Martin K, Vera M, Richard W, Brendan D (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J Cell Mol Biol 68:147–158
Martín-Trillo M, Grandío EG, Serra F, Marcel F, Rodríguez-Buey ML, Schmitz G, Theres K, Bendahmane A, Dopazo H, Cubas P (2011) Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J Cell Mol Biol 67:701
Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA (2014) Sugar demand, not auxin, is the initial regulator of apical dominance. Proc Natl Acad Sci USA 111:6092–6097
Mauriat M, Sandberg LG, Moritz T (2011) Proper gibberellin localization in vascular tissue is required to control auxin-dependent leaf development and bud outgrowth in hybrid aspen. Plant J Cell Mol Biol 67:805
Maymon I, Greenboim-Wainberg Y, Sagiv S, Kieber JJ, Moshelion M, Olszewski N, Weiss D (2009) Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J Cell Mol Biol 58:979
Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51:1127
Moubayidin L, Perilli S, Ioio RD, Mambro RD, Costantino P, Sabatini S (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol Cb 20:1138
Muhr M, Prüfer N, Paulat M, Teichmann T (2016) Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytol 212:613–626
Muller D, Leyser O (2011) Auxin, cytokinin and the control of shoot branching. Ann Bot 107:1203–1212
Muller D, Waldie T, Miyawaki K, To JP, Melnyk CW, Kieber JJ, Kakimoto T, Leyser O (2015) Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J 82:874–886
Murfet IC, Reid JB, Casey R, Davies DR (1993) Developmentalmutants. In: Casey R, Davies DR (eds) Peas genetics molecular biology and biotechnology Developmental mutants. Peas genetics molecular biology and biotechnology, (Wallingford: CAB), pp 165–216
Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K, Shi X, Ito E, Ito S, Park SH (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 4:2613
Naor A, Flaishman M, Stern R, Moshe A, Erez A (2003) Temperature effects on dormancy completion of vegetative buds in apple. J Am Soc Hortic Sci Am Soc Hortic Sci 128:636–641
Ni J, Gao CC, Chen MS, Pan BZ, Ye KQ, Xu ZF (2015) Gibberellin promotes shoot branching in the perennial woody plant Jatropha curcas. Plant Cell Physiol 56:1655–1666
Nicolas M, Rodríguez-Buey María L, Franco-Zorrilla José M, Cubas P (2015) A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr Biol 25:1799–1809
Niwa M, Daimon Y, Kurotani K, Higo A, Pruneda-Paz JL, Breton G, Mitsuda N, Kay SA, Ohme-Takagi M, Endo M (2013) BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25:1228–1242
Nunes C, O’Hara LE, Primavesi LF, Delatte TL, Schluepmann H, Somsen GW, Silva AB, Fevereiro PS, Wingler A, Paul MJ (2013) The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol 162:1720–1732
Otori K, Tamoi M, Tanabe N, Shigeoka S (2017) Enhancements in sucrose biosynthesis capacity affect shoot branching in Arabidopsis. Biosci Biotechnol Biochem 81:1
Patrick JW, Colyvas K (2014) Crop yield components—photoassimilate supply- or utilisation limited-organ development? Funct Plant Biol 41:893
Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194
Rabot A, Henry C, Ben BK, Mortreau E, Azri W, Lothier J, Hamama L, Boummaza R, Leduc N, Pelleschi-Travier S (2012) Insight into the role of sugars in bud burst under light in the rose. Plant Cell Physiol 53:1068
Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher F, Sakr S (2015) Multiple pathways regulate shoot branching. Front Plant Sci 5:741
Richter R, Behringer C, Muller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and phytochrome-interacting factors. Genes Dev 24:2093–2104
Rinne PL, Paul LK, Vahala J, Kangasjarvi J, van der Schoot C (2016) Axillary buds are dwarfed shoots that tightly regulate GA pathway and GA-inducible 1,3-beta-glucanase genes during branching in hybrid aspen. J Exp Bot 67:5975–5991
Roef L, Onckelen HV (2010) Cytokinin Regulation of the Cell Division Cycle. In: Davies PJ (ed) Plant Hormones. Springer, Dordrecht
Rosa M, Hilal M, González JA, Prado FE (2009) Low-temperature effect on enzyme activities involved in sucrose-starch partitioning in salt-stressed and salt-acclimated cotyledons of quinoa (Chenopodium quinoa Willd.) seedlings. Plant Physiol Biochem 47:300–307
Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294:1519–1521
Shimizu S, Mori H (1998) Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant Cell Physiol 39:255–262
Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10:155
Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC (2007) Analysis of the decreased apical dominance genes of petunia in the control of axillary branching. Plant Physiol 143:697
Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17:1469
Steiner E, Weiss D (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24:96–108
Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48:263–277
Thimann KV, Skoog F (1934) On the inhibition of bud development and other functions of growth substance in Vicia faba. In: Proceedings of the Royal Society of London series B-containing papers of a biological character, vol 114, pp 317–339
Volz RK, Gibbs HM, Popenoe J (1994) Branch induction on apple nursery trees: effects of growth regulators and defoliation. New Zealand J Crop Hortic Sci 22:277–283
Willige BC, Isono E, Richter R, Zourelidou M, Schwechheimer C (2011) Gibberellin regulates pin-formed abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23:2184–2195
Xu R, Sun P, Jia F, Lu L, Li Y, Zhang S, Huang J (2014) Genomewide analysis of TCP transcription factor gene family in Malus domestica. J Genet 93:733–746
Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) Arabidopsis KNOXI Proteins Activate Cytokinin Biosynthesis. Current Biology 15(17):1566–1571
Yadav UP, Ivakov A, Feil R, Duan GY, Walther D, Giavalisco P, Piques M, Carillo P, Hubberten HM, Stitt M (2014) The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J Exp Bot 65:1051
Yoneyama K, Xie X, Dai K, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K (2007) Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227:125–132
Zawaski C, Busov VB (2014) Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees. Plos One 9:e86217–e86217
Zeng XF, Zhao DG (2016) Expression of IPT in Asakura-sanshoo (Zanthoxylum piperitum (L.) DC. f. inerme Makino) alters tree architecture, delays leaf senescence, and changes leaf essential oil composition. Plant Mol Biol Report 34:649–658
Zhang S, Zhang D, Fan S, Du L, Shen Y, Xing L, Li Y, Ma J, Han M (2016) Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ apple (Malus domestica Borkh.). Plant Physiol Biochem 107:178–186
Funding
This work was supported by the National Apple Industry Technology System of Agriculture Ministry of China (CARS-28); Yangling Subsidiary Center Project of National Apple Improvement Center and Collaborative Innovation of Center Shaanxi Fruit Industry Development (C000088); Chinese postdoctoral project (2015M582713); Innovation project of science and technology plan projects of Shaanxi province (2016TZC-N-11-6).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest. Ming Tan declares that she has no conflict of interest. Guofang Li declares that he has no conflict of interest. Xiaojie Liu declares that he has no conflict of interest. Fang Cheng declares that she has no conflict of interest. Juanjuan Ma declares that she has no conflict of interest. Caiping Zhao declares that she has no conflict of interest. Dong Zhang declares that he has no conflict of interest. Mingyu Han declares that he has no conflict of interest.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tan, M., Li, G., Liu, X. et al. Exogenous application of GA3 inactively regulates axillary bud outgrowth by influencing of branching-inhibitors and bud-regulating hormones in apple (Malus domestica Borkh.). Mol Genet Genomics 293, 1547–1563 (2018). https://doi.org/10.1007/s00438-018-1481-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1481-y