Abstract
Productivity of Indian mustard (Brassica juncea L. Czern. and Coss.) is markedly reduced by salt stress. To develop salt tolerance in this important oilseed crop is a need of the hour. This study, based on analysis of growth parameters and antioxidant profile of fourteen Indian mustard genotypes treated with 50, 100, 150 and 200 mM of sodium chloride, was performed to identify the salt-sensitive and salt-tolerant genotypes. Salinity stress inhibited biomass accumulation and reduced the protein and chlorophyll contents in a dose-dependent manner. The reduction was the highest in genotype Pusa Agrani and lowest in CS-54, depicting their contrasting sensitivity to salt stress. Salt treatments triggered a concentration-dependent overproduction of reactive-oxygen species and a concurrent upregulation of the expression of different antioxidants. Genotype CS-54 showed the least damage and maintained a high antioxidant level with almost each salt treatment, exhibiting its competence to withstand the damage provoked by salinity stress. Genotype Pusa Agrani, on the contrary, depicted a salt-sensitive nature by way of its very high lipid peroxidation and low intensity of antioxidants. These two genotypes were further investigated through gel-based proteomic approach, which resulted in the identification and quantification of 42 salinity-responsive proteins related to different metabolic modifications. Molecular processes, including photosynthesis, redox homeostasis, nitrogen metabolism, ATP synthesis, protein synthesis and degradation, signal transduction and respiratory pathways, have exhibited significant changes. The identified stress-responsive proteins could pave the way to develop salt tolerance in Indian mustard plant, thus sustaining its productivity under salinity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil salinization has been causing shrinkage of limited and valuable agricultural land and decrease in crop productivity all over the globe. Increasing rate of secondary salinization (i.e. salinization due to anthropogenic activities) of arable land is expected to result in a loss of 30 % land within the next 25 years, and up to 50 % by the year 2050 (Wang et al. 2003). Accumulation of salts in the soil leads to water stress and nutrient deficiency in plants (Arshi et al. 2012). One of the primary strains induced by salinization is the alteration of redox homeostasis (Foyer and Noctor 2009). Salt stress causes divergence of electron flow from the main transport chains in organelles to the oxygen-reduction pathways leading to the overproduction of reactive oxygen species (ROS), which brings about oxidation of indispensable biomolecules such as lipids, proteins, nucleic acids and carbohydrates, thus altering their properties and functions, which ultimately lead to metabolic and physiological disorders (Anjum et al. 2014; Aref et al. 2016). Excessive production of ROS causes imbalance in the homeostasis at the cellular and sub-cellular levels and can eventually lead to cell death (Abogadallah 2010; Sharma et al. 2012). In order to scavenge the toxic ROS and combat the oxidative damage, plants possess an antioxidant system comprising of some enzymatic and non-enzymatic components that counteract the negative influence of ROS by converting them into less toxic forms (Munns and Tester 2008; Medeiros et al. 2012; Iqbal et al. 2015). The augmented levels of antioxidants are customarily indicative to enhanced plant tolerance (Iqbal et al. 2015; Lei et al. 2016).
The possible solutions to the salinization conundrum include either reclamation of waste lands, which is far from the economic boundaries of poor farmers, or development of salt-tolerant plant species (Miranda et al. 2016). Differences in salt sensitivity among genotypes of a species provide a basis for screening the salt-sensitive and salt-tolerant genotypes, which can be used for further experimentation. Salt stress induces changes in the cellular machinery by modifying expression of both specific as well as housekeeping genes (Sahi et al. 2006), which may affect the abundance of cellular proteins. Proteomics serves as the finest tool to work out environmental pressures, molecular manipulations, stress-interceded adaptations and genotypic variability (Hakeem et al. 2012a, b). Indian mustard (Brassica juncea L. Czern. and Coss), an important oil seed crop, is cultivated widely in India and ranks second in its production (Shah 2007). The main cultivation area is restricted to the north-west agro-climatic zone, which faces high salinity levels (Sharma et al. 2013). Proteomic data available on this oilseed crop are meagre and hence our awareness of the molecular mechanisms operative in this plant in response to salt stress is limited (Yousuf et al. 2016a). Given this, the present study was conducted to assess the sensitivity and acclimatization capacity of different genotypes of Indian mustard grown under salt stress by evaluating their growth parameters and antioxidative actions. Proteomic analysis of the identified salt-sensitive and salt-tolerant genotypes was also undertaken to figure out the regulatory mechanisms operative behind the salt-stress responses in Indian mustard.
Materials and methods
Plant growth and treatment conditions
Authenticated seeds of fourteen genotypes of Brassica juncea L. Czern and Coss, namely CS-52, P-Agr. (Pusa Agrani), P-Vij. (Pusa Vijay), P-Var. (Pusa Varuna), CS-54, P-Jai. (Pusa Jaikisan), P-Bah. (Pusa Bahar), P-Kran. (Pusa Kranti), P-Bold (Pusa Bold), Lax. (Laxmi), P-Bas. (Pusa Basant), ZEM-1, RC-781 and JM-1, were procured from the Genetics Division, IARI, New Delhi, India. Background information on the genotypes used is given in supplementary Table ST1. The seeds were washed thoroughly with water, surface-sterilized with 0.01 % mercuric chloride and washed again with distilled water prior to sowing in pots containing a mixture of sand and vermiculite (1:1). After germination, ten plants were maintained in each pot. The experiment was set in a random design. The plants were grown in the Hoagland’s growth solution of one-fourth strength for the first 10 days, in half-strength for the next 10 days and in full strength for the last 10 days, in a growth chamber under the controlled conditions of light (16 h photoperiods), temperature (27 °C) and humidity (60 %). Different treatments of sodium chloride, viz. 0 mM (T0), 50 mM (T1), 100 mM (T2), 150 mM (T3) and 200 mM (T4), were given to 20-day-old plants in a single frame. The leaves of 30-day-old plants were excised and used for experimental analysis. Images of plants obtained at the time of sampling are given in Supplementary Fig. S1. Three biological replicates were taken during the experimental procedure.
Biomass accumulation
At the time of harvest, plant weight was recorded before and after oven-drying the samples at 65 °C ± 2 °C for 72 h, when they attained a constant weight, in order to estimate the biomass accumulation.
Estimation of soluble protein content
Bradford’s method (1976) was used for the quantification of soluble protein content. Fresh leaf material (0.5 g) was homogenized with the help of pre-cooled mortar and pestle in 0.1 M phosphate buffer with pH 6.8 at 4 °C. The homogenate was transferred to 2 mL tubes and centrifuged at 5000×g for 10 min at 4 °C. The supernatant taken was added with an equal amount of chilled 10 % TCA for protein precipitation and centrifuged at 3300×g for 10 min. The supernatant was then discarded, and the resulting pellet was washed with acetone and dissolved in 1 mL of 0.1 N NaOH. To 1 mL aliquot, 5 mL of Bradford’s reagent (50 mL of 90 % alcohol, 100 mL of o-phosphoric acid, 850 mL of double-distilled water, 0.1 g of Coomassie Brilliant Blue G-250) was added and vortexed. Tubes were kept in the dark for 10 min for optimal colour development, and the absorbance was noted at 595 nm. The soluble-protein content was estimated with the help of a standard curve, using bovine albumin serum (Sigma) as the standard, and expressed in mg g−1 FW.
Estimation of chlorophyll content
Chlorophyll content in fresh leaf samples was estimated by the method of Hiscox and Israelstam (1979). Briefly, 0.1 g leaves taken in moist filter paper in an icebox, were washed with cold double-distilled water and chopped. This leaf material was then transferred to vials, in triplicates, containing 5 mL of dimethyl sulfoxide (DMSO). The vials were kept in oven at 65 °C for 1 h for complete leaching of the pigments. DMSO was further added to make a final volume of 10 mL and optical density measured immediately. Absorbance of DMSO containing the pigments was noted at 663 nm and 645 nm, using a UV–Vis spectrophotometer (λ-Bio 20, Perkin Elmer). The contents of Chl a, and Chl b were estimated using the following formulae proposed by Arnon (1949).
Measurement of lipid peroxidation
Lipid peroxidation was estimated by the method of Heath and Packer (1968), using thiobarbituric acid (TBA) as the primary reagent, and measured from the levels of malondialdehyde (MDA), which is a principal constituent of the thiobarbituric-acid-reacting substance (TBARS). The absorbance was recorded at 532 and 600 nm, and the MDA content was calculated by subtracting the absorbance at 600 nm from that at 532 nm, using an extinction coefficient of 155 mM−1 cm−1. The MDA content was expressed as nmol g−1 fresh weight of the sample.
Analysis of the enzymatic antioxidants
SOD activity was estimated by its ability to inhibit nitroblue tetrazolium (NBT) formation from formazan, according to the method of Beyer and Fridovich (1987). The reduced NBT was measured at 560 nm using the absorbance coefficient of 100 mM−1 cm−1. The SOD activity was expressed in enzyme units per mg of protein.
APX activity was determined, following the method of Nakano and Asada (1981), in terms of its ability of catalyzing the reduction of hydrogen peroxide to water in the presence of 0.1 M phosphate buffer. The decrease in the absorbance was recorded at 240 nm, and the APX activity was calculated by using an extinction coefficient of 2.8 mM−1 cm−1.
Catalase activity was assayed using the method of Aebi (1984). Absorbance was recorded at 240 nm of the mixture of 0.1 ml enzyme extract and 0.1 M phosphate buffer both before and after adding 0.1 mL of hydrogen peroxide. The activity was calculated using an extinction coefficient of 0.036 mM−1 cm−1.
GR activity was determined by the method of Foyer and Halliwell (1976) modified by Rao (1992) and estimated by monitoring the glutathione-dependent oxidation of NADPH at its absorption maxima of 340 nm wavelength. The GR activity was calculated using an extinction coefficient of 6.2 mM−1 cm−1.
Analysis of non-enzymatic antioxidants
The total ascorbate content was estimated by the method of Law et al. (1983). It was determined by its ability to reduce Fe3+ to Fe2+ in acidic medium, which complexes with bipyridyl dye giving yellow colour with the absorption peak at 525 nm. The standard curve was prepared, and the resulting optical densities were compared with it to calculate the ascorbate content.
Glutathione content was determined by using the method of Anderson (1985) and estimated by its ability of getting oxidized by DTNB in the presence of NADPH to form TNB, which has the maximum absorbance at 412 nm.
Protein extraction
Leaf proteins of the selected genotypes were extracted by using the phenol method of Isaacson et al. (2006). Two grams of leaf material was pulverized to fine powder in liquid nitrogen and suspended in 10 mL of extraction buffer containing 50 mM HEPES, 2 % β-mercaptoethanol, 0.7 M sucrose, 1 mM PMSF, 50 mM EDTA and 100 mM KCl with pH adjusted to 7.5. Fifteen mL of phenol was added to dissolve the proteins. The solution was mixed up in a cold room rocker for half an hour. The resultant solution was centrifuged at the speed of 3000×g for 10 min at 4°C. The top phenolic phase containing the proteins was vigilantly retrieved in a separate tube. The proteins in phenolic phase were precipitated by adding 15 mL of ice-cold 0.1 M ammonium acetate solution and incubating at −20 °C for overnight. The proteins were centrifuged at 6000×g for 15 min at 4 °C. Methanol was added to the pellet for washing. The pellet in methanol was kept at −20 °C for 30 min and then centrifuged at 3000×g for 10 min. The pellet was again washed two times with acetone in the same manner. The protein was dried and solubilised in the solubilization buffer containing 2 M thiourea, 7 M urea, 4 % CHAPS and 50 mM DTT. Quantification of protein was carried out by the method of Bradford (1976), using BSA (Sigma) as a standard.
Two-dimensional gel electrophoresis
Two-dimensional electrophoresis was carried out in accordance with the method of O’Farrel (1975). 500 µg of protein dissolved in a total volume of 200 µL of solubilization buffer was carefully placed on an immobiline dry strip gel (11 cm, pH 4–7; Bio-rad, USA) and kept for rehydration at 20 °C for 14 h. After rehydration, the proteins were subjected to isoelectric focusing in a PROTEAN IEF apparatus (Bio-rad, USA). The programme of voltage set in IEF cell was a 250 V for 1 h, 500 V for 1 h, 1000 V for 2 h and 2000 V for 2 h, linear increase of 8000 V for 18 h and 500 V for 1 h. After the completion of IEF, the focused proteins were first reduced for 15 min by a solution containing 50 mM Tris (pH 8.8), 8 M urea, 20 % glycerol, 2 % SDS and 130 mM DTT and then alkylated for the same duration by alkylation buffer containing Tris (pH 8.8), 8 M urea, 20 % glycerol, 2 % SDS and 135 mM iodoacetamide. SDS-PAGE was performed in a PROTEAN plus Dodeca cell (Bio-rad, USA) for separation of focused proteins based on molecular weights, using 12 % acrylamide at a constant voltage of 250 V. The gels were stained with colloidal Coommassie brilliant blue dye and then destained by washing several times with Milli-Q water.
Gel analysis
The resolved gels were scanned with a densitometer (GS-800 Calibrated Densitometer Bio-rad) and examined with ImageMaster™ PD Quest software version 8.0 (BioRad, USA) for spot detection, background subtraction and intensity quantification. The gel with the highest number of spots was taken as the reference gel. Each spot value was normalised in terms of percentage of the total volume of all gel spots for the rectification of unevenness due to quantitative disproportion in spot intensities. The spots exhibiting more than two-fold change in their abundance during the treatment or a significant variation between the control and other treatments, as determined by the paired Student’s t test (p ≤ 0.05), were presumed as treatment-responsive proteins.
In-gel digestion and protein identification
The protein spots with more than two-fold change in their intensity with respect to reference gel were excised from gels and dehydrated with 50 µL of solution, containing 50 % acetonitrile (ACN) and 50 mM ammonium bicarbonate in 2:1 ratio, for 5 min. The protein spots were reduced with 15 mM DTT at 60 °C for 1 h and then subjected to alkylation by 100 mM IAA in dark for 15 min, rehydrated with 50 mM ammonium bicarbonate and then dried in a speed vac. Dried gel slices were rehydrated with 15 µL of working trypsin (Sequencing grade Modified Promega USA Ref V511A) at 37 °C for overnight. The supernatant was taken, and proteins in the remaining gel were further extracted by adding 20 % ACN and 1 % formic acid. The final supernatant was dried in speed vac until the volume was lowered to 25–50 µL. The proteins in the final volume were analysed with MALDI MS—AB Sciex 5800 TOF/TOF System with LC-MALDI. Peptide tolerance of 150 ppm, fragment mass tolerance of ±0.4 Da, and peptide charge of 1+ were selected. Only the significant hits, as defined by the MASCOT probability analysis (p < 0.05), were acknowledged. Peptides were searched with the following parameters: NCBInr database, taxonomy of green plants, trypsin of the digestion enzyme, one missed cleavage site, partial modification of cysteine carboamidomethylated and methionine oxidized.
Statistical analyses
Three biological replicates for both the treatments and control were used for application of statistical tests. Statistical analysis of the data, including data processing and variance analysis (ANOVA), was done using the SPSS software (16.0 version). A two-tailed Students t test with the significance of 95 % was performed on the normalised value of protein spots with the help of SPSS software. PCA analysis of the data of physiological and biochemical parameters was done by the statistical software R (R Core Team 2015) using the function princomp. Hierarchical clustering was done using the function hclust based on the Ward’s method on the squared Euclidean distance matrix of biomass obtained in all five treatments.
Results
Physiological growth evaluation
Biomass accumulation in Indian mustard was found to vary among genotypes and with the intensity of salt treatment. The biomass decreased progressively with increase in the salt concentration. The maximum reduction in biomass was observed in genotype Pusa Agrani with all the treatments, as compared with other genotypes. On the other hand, genotype CS-54 showed the least reduction in biomass (Table 1). Salt treatment reduced the soluble protein content of leaves, and this reduction was dose-dependent. The decrease was maximum in Pusa Agrani with all the treatments (Table 1). On the other side, CS-54 displayed the minimum decrease. The photosynthetic pigments of leaves, viz. chlorophylls a and b were found to vary both with treatments and genotypes. Chlorophyll a content displayed a decline of 0.33-1.33 mg g−1 fresh, the maximum decline occurring in Pusa Agrani and the minimum in CS-54 (Table 1). Chlorophyll b content also decreased with increase in NaCl concentration, as compared with the control. The decrease in chlorophyll content was maximum in Pusa Agrani and minimum in CS-54 (Table 1).
Oxidative stress and defence mechanism
MDA content was analysed to know the rate of lipid peroxidation among 14 genotypes of Indian mustard. MDA content varied significantly both with treatments and genotypes (Table 2). Compared to the control, MDA content increased in all genotypes with all treatments in a concentration-dependent manner. The increase was minimum in CS-54 and maximum in Pusa Agrani.
Superoxide dismutase (SOD) activity was augmented up to T3 treatment and then decreased in T4, in all genotypes except CS-54, which showed a progressive rise in all the treatments. This genotype showed the highest SOD activity, while the minimum was seen in Pusa Agrani (Table 2). Activity of ascorbate peroxidase (APX), the main peroxidase used for scavenging hydrogen peroxide in sub-cellular locations, including mitochondria, cytosol and chloroplasts, ranged from 1.08 to 2.87 EU (mg protein−1 min−1) with different salt treatments. Here also, activity was the highest in CS-54 and lowest in Pusa Agrani with all the treatments (Table 2).
Catalase (CAT) activity increased in Pusa Agrani, Pusa Varuna, Pusa Jai Kisan, Pusa Laxmi, Pusa Basant, ZEM-1 and RC-781 with T1 and T3, but decreased with T2 and T4 treatments. Genotypes CS-54 and Pusa Bold exhibited a linear upsurge in their CAT activities up to T3 and then a decline with T4. In genotype JM-1, the activity increased during the first two treatments and decreased thereafter. The lowest CAT activity was recorded in Pusa Agrani with T2 and T4 and in Pusa Basant and JM-1 with T1 and T3, respectively (Table 2). Glutathione reductase (GR) activity varied with genotypes under salt stress, with a range of 2.06–8.36 EU mg protein−1 min−1. With all the treatments, GR activity was the highest in CS-54 and lowest in Pusa Agrani (Table 3).
Of the non-enzymatic antioxidants studied, ascorbate (ASC) content varied markedly among genotypes under various salt treatments, ranging from 124.3 (Pusa Agrani) to 271.19 (CS-54) nmol gm−1 fresh wt (Table 3). Compared to the control, it increased significantly with T2, T3 and T4 treatments, attaining the maximum in CS-54 and minimum in Pusa Agrani. Likewise, a steep increase in glutathione (GSH) content of leaves was observed in all genotypes during different salt treatments, showing a dose-dependent progress. The highest value (118.19 nmol gm−1 fresh wt) was noted in CS-54 and the lowest (65.72 nmol gm−1 fresh wt) in Pusa Agrani with T4 treatment. The trend was similar for other treatments also (Table 3).
Proteomic modulations
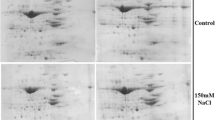
Leaf proteome of the different genotypes with contrasting salt tolerance efficiency was evaluated using two-dimensional gel electrophoresis at four different concentrations of sodium chloride. More than 420 reproducible spots were obtained by staining the gels with Colloidal Coomassie blue dye. Although numerous proteins showed differential expression, only 48 protein spots displayed more than two-fold change in their abundance during the course of experiment. Of these, 33 (69 %) increased in their intensity, while 15 (31 %) were down-regulated under salt stress. Position of these differentially expressed proteins on 2D profile is shown in Fig. 1.
Protein identification and classification
Differentially regulated proteins exhibiting more than two-fold change in their intensities in at least one genotype or with any one salt treatment were excised from the stained gels and subjected to MALDI-TOF/TOF analysis. Protein identification was tried on the basis of combined peptide mass fingerprinting and MS/MS analysis. Among the identified proteins, 42 showed homology with already known proteins, whereas six were unknown. On the basis of their spatial distribution within the cell, the differentially-expressed known proteins were categorized into nine groups (Fig. 2). Most of them belonged to chloroplast, cytosol and nucleus, while others to such organelles as Golgi bodie, plasma membrane, ribosome, peroxisome, mitochondrion and vacuole. Based on their association with physiological processes, a functional cataloguing of proteins was done as illustrated in Fig. 3. The proteins were involved in carbohydrate metabolism (26 %), signal transduction (17 %), photosynthesis (16 %), oxidative stress (12 %), protein synthesis and degradation (6 %), amino-acid biosynthesis (6 %), energy metabolism (5 %) and nitrogen metabolism (5 %). Details of identified proteins, including their relative spot intensities, are given in Table 4.
Discussion
Physiological changes
Accumulation of biomass is one of the important markers for screening of salt tolerance in plants (Munns and James 2003). The decrease in biomass accumulation during the salt stress, as observed by us, was possibly due to nutrient imbalance and osmotically-induced water stress. The most affected genotype in terms of biomass accumulation was Pusa Agrani, implying its sensitivity towards salinity, while genotype CS-54 was least affected. Besides, salt treatments also induced considerable decrease in chlorophylls a and b, indicating the adverse impact of salinity on photosynthetic efficiency. Salt ions inhibit the uptake of other ions, like magnesium, which are essential for chlorophyll synthesis. The decline in chlorophyll synthesis and/or the degradation of chlorophyll molecules due to salt accumulation may be the reasons for the decreased chlorophyll content (Arshi et al. 2004; Santos 2004). In addition, abundance of proteins, an important determinant of physiological health of plants, also decreased due to salt stress, which often inhibits protein synthesis in a dose-dependent manner (Kong-Ngern et al. 2005). The decline in chlorophyll and protein contents was more pronounced in Pusa Agrani than in the other genotypes illustrating its higher vulnerability to physiological damage.
Clustering and PCA analysis based on physiological responses were carried out to classify the genotypes into salt-sensitive and salt-tolerant groups (Supplementary Figs S2 and S3). The changes in the antioxidant defense system of these genotypes were then evaluated.
Changes in the antioxidant defence system
This study of fourteen different genotypes of Indian mustard has revealed a salinity-induced overproduction of reactive oxygen species, as evident from the increase in abundance of MDA, an aldehyde produced as an outcome of lipid-peroxidation reaction. This confirms the earlier report of Ahmad et al. (2012). In our study, Genotype CS-54 grown under salt stress accumulated a relatively lesser amount of MDA than the other genotypes, signifying its higher efficiency of regulating the ROS concentration. On the contrary, the maximum MDA content was observed in Pusa Agrani, showing its greater susceptibility to lipid peroxidation. In response to the oxidative stress, activity of SOD and APX, the first line of defense in mitigating the oxidative damage, increased with increase in salt concentrations (Yousuf et al. 2016a). Our findings on SOD activity substantiate some earlier studies on Cicer arietinum (Kukreja et al. 2005), Morus alba (Harinasut et al. 2003), Carthamus tinctorius (Gengmao et al. 2015) and Solanum lycopersicum (Gapinska et al. 2008), while those on APX activity conform to earlier works on lentil and mungbean (Kulik et al. 2004; Nazar et al. 2011; Yasar et al. 2008). CAT detoxifies H2O2, overproduced mainly in peroxisomes during stressful conditions. Overexpression of CAT gene of Indian mustard introduced into tobacco cells enhanced tobacco tolerance to oxidative stress (Guan et al. 2009). The maximum CAT activity in CS-54 and the minimum in Pusa Agrani was possibly related to the rate of enzyme synthesis. Increase in the GR activity, which has a crucial role in providing the reducing potential for converting H2O2–H2O (Yousuf et al. 2012), develops stress tolerance in many plants including the Indian mustard (Wang et al. 2011). As to the non-enzymatic antioxidants. The level of ascorbate, a primary cellular redox buffer having a vital role in the elimination of H2O2 through glutathione-ascorbate pathway (Zhang et al. 2011; Venkatesh et al. 2012), was higher in genotype CS-54 than in Pusa Agrani, signifying its greater potential to maintain redox homeostasis under salt stress. Glutathione, a non-protein thiol acting as an important ROS scavenger by partaking in the regeneration of ascorbate, also showed a similar variation trend. The observed increase in glutathione content is in line with some earlier reports on crop plants (Sumithra et al. 2006; Anjum et al. 2012) grown under salinity stress.
In general, genotypes that showed a reduced physiological growth under salinity conditions exhibited a less efficient antioxidant system, whereas those with comparatively better growth showed an enhanced combating system. PCA analysis based on biochemical parameters (Fig. S3), and proteomic investigation of the salt-sensitive and salt-tolerant genotypes have duly strengthened our inferences.
Salt-induced proteomic changes
Comparative proteomics serves as a finest approach in monitoring the changes induced by abiotic stresses at protein level (Yousuf et al. 2016a, b). Leaves are the primary sites of photosynthesis, the main process being affected by salinity stress (Munns and Tester 2008), and hence provide the best material for proteomic studies in relation to salt stress (Manaa et al. 2013). The leaves of salt-sensitive and salt-tolerant genotypes analyzed to evaluate proteomic modulations and identify protein markers responsible for increased tolerance in CS-54.
Proteins related to photosynthesis
Salt stress affected the abundance of proteins involved in different steps of photosynthesis, such as photosystem regulation, electron transfer and carbon fixation. The intensity of protein related to light-harvesting complex (LHC), which harvests light energy and transfers it to the reaction centre of photosystems, and also protects photosystems against photodamage by dissipating the excess light energy in the form of heat (Murata et al. 2007; Takahashi and Badger 2011; Rowley and Mockler 2011), was found to increase due to salt stress (spot 15) in both the genotypes, the increase being greater in the salt-tolerant genotype. This could contribute to protection against photodamage. In addition, salt stress also affected the structural and functional aspects of PS II. The level of photosystem II protein 1 (spot 29), a structural component, declined due to salt stress. However, the abundance of the oxygen-evolving enhancer protein, which has a crucial role in stabilization of Mn cluster (in the oxygen-evolving complex) and functioning of the PSII (Suorsa and Aro 2007; Busheva et al. 2012), increased (spot 34). The stress conditions influenced the electron transport also. Concentration of ferredoxin-dependent NADP reductase (FNR), which catalyses the electron transfer from ferredoxin to NADP during light reactions, decreased in a dose-dependent manner. This decline in the enzyme level may be due to low electron flow resulting from tissue dehydration (Takahashi and Badger 2011). Salt stress also affected rubisco, an enzyme that catalyses the fixation of carbon dioxide. Two proteins pertaining to large (spot 33) and small (spot 14) subunits of rubisco, exhibited differential expression under salt stress. Rubisco abundance markedly decreased in a dose-dependent manner, possibly due to degradation (Galmes et al. 2013). Negative impacts of abiotic stresses on rubisco abundance have been reviewed recently (Feller et al. 2008; Bashir et al. 2015). The mode of rubisco activity, mainly associated with the regulation of activation state, is predominantly dependent on rubisco activase, a catalytic chaperone (Boex-Fontvieille et al. 2014). Rubisco activase (spot 36) increased in abundance with a higher expression level in tolerant genotype than in sensitive one. Possibly, this enabled the former to stabilize rubisco to carry out carbon fixation at low CO2 level due to closing of stomata under salinity stress. Our results go in line with earlier works of Kim et al. (2005), Parker et al. (2006) and Bandehagh et al. (2011).
Proteins related to carbohydrate metabolism
Two enzymes, fructose kinase (spot 8) and glyceraldehyde 3-phosphate dehydrogenase (spot 23), catalyzing two necessary steps of glycolysis during the production of primary as well as secondary metabolites (Jeong et al. 2001), were downregulated. Besides, four enzymes pertaining to TCA cycle, namely, malate dehydrogenase (spot 3), glycerate kinase (spot 17), NADP isocitrate dehydrogenase (spot 22) and isocitrate lyase (spot 30), showed a sharp decline in intensity in the treated samples, as noted earlier in arabidopsis (Jiang et al. 2007), rice (Ghaffari et al. 2014) and canola (Bandehagh et al. 2011). This low expression of glycolytic and TCA enzymes can be attributed to the restricted CO2 fixation induced by a limited stomatal conductance and downregulation of rubisco enzyme (Caruso et al. 2008). Sucrose synthase is a vital enzyme that regulates balance between metabolisms of starch and sucrose, catalyzing degradation as well as synthesis of sucrose, but preferring the former under energy-limiting conditions (Lu et al. 2010). Two enzymes related to sucrose metabolism, sucrose-phosphate synthase (spot 20) and sucrose synthase 2 (spot 37) were upregulated, possibly to meet the energy demands under salt stress.
Proteins involved in antioxidant defense
In order to scavenge the toxic ROS, plants regulate the level of their antioxidants. We identified three proteins involved in oxidative stress protection, namely (1) APX (spot 41), which forms an integral part of ascorbate–glutathione pathway reducing H2O2 to water, (2) heme oxygenase 3 (spot 40), which accelerates oxidative breakdown of heme to biliverdin releasing Fe2+ and CO, and also has a role in phytochrome biosynthesis (Shekhawat and Verma 2010), and (3) thioredoxin h like protein (spot 6) known to be involved in oxidative defense besides sustaining seed germination, early seedling growth, self-incompatibility, and C and N metabolisms (Yamamoto and Nasrallah 2009). All these enzymes showed a concentration-dependent upsurge during salt treatments to overcome the oxidative damage associated with overproduction of toxic oxidants. The observed increase in the intensity of APX substantiate these findings.
Proteins involved in nitrogen metabolism
GS is known to catalyze the ATP-dependent reaction of NH4 with glutamate to yield glutamine, besides being involved in the synthesis of precursors meant for the biosynthesis of proline, an osmoprotectant that overcomes the osmosis-induced water stress. GS (spot 27) was significantly increased in the salt-sensitive genotype. PII-like protein is an essential signal-transduction protein that regulates N assimilation and C metabolism in general (Uhrig et al. 2009; Huergo et al. 2013). In Indian mustard, PII-like protein (spot 32) accumulated more prominently in CS-54 than in Pusa Agrani, possibly to maintain nitrogen level within the plant, which is otherwise disturbed under stressful conditions (Rais et al. 2013; Kim et al. 2004; Ashraf and Harris 2004).
Proteins involved in energy and protein metabolisms
Two proteins having a role in ATP synthesis and identified as ATP synthase F0 subunit-β (spot 26) and ubiquinol: cytochrome c oxidoreductase 7 kDa subunit (spot 24), were differentially expressed during salt stress showed a dose-dependent decrease in their intensity, which was more prominent in the salt-sensitive genotype. Reduced abundance of ATP synthase subunits in salt-affected plants points to the role of this enzyme during acclimation phase, e.g. a down-regulation of ATP synthase subunit β-3 was noticed in cucumber (Du et al. 2010).
Five proteins related to protein metabolism showed more than two-fold change in their expression under salt stress with reference to the control. Two of these, viz. ribosomal protein S19 (spot 25) and ribosomal protein S4 (spot 35), which form the structural part of small subunit of ribosomes, were downregulated while the other three, ubiquitin (spot 4), ubiquitin-like specific protease (spot 42) and polyubiquitin (spot 16), which help in protein degradation, were upregulated. Ubiquitination regulates the transcriptional changes required for adaptation to various abiotic stresses by modulating the amount and activity of regulatory proteins (Lyzenga and Stone 2012).
Proteins related to signal-transduction
Salt stress brought about changes in abundance of 7 such proteins that function as transcriptional factors and/or signal transducers, regulating different phases of growth and defense. Two proteins, viz. Zn-finger protein (spot 2) and DC1 domain-containing protein (spot 9), belong to Zn finger family. DC1 domain binds to the secondary messenger diacylglycerol, which activates protein kinase C and transduces signal in a Ca2+-dependent way. DC1 domain-containing proteins play vital roles in modulating the plant response to abiotic factors (Li et al. 2010). Besides, bZIP transcription factor, belonging to the bZIP family of proteins and neighbouring to a leucine zipper dimerization domain, with a DNA-binding domain rich in basic amino acid residues, is involved in reactions with abiotic stresses, including drought and salinity (Hsieh et al. 2010). Both the DC1 domain-containing protein and bZIP transcription factor act in abscisic-acid-mediated-signal pathways. The intensity of spots corresponding to DC1 domain-containing protein (spot 9) and bZIP transcription factor (spot 12) significantly increased during the experiment. The other transcriptional factors, which exhibited more than two-fold change in their intensities over the control, were identified as the remorin-family protein (spot 18) and type 1 MADS-box transcriptional factor (spot 38); both are associated with plant response to environmental stress (Puig et al. 2013). With all the salt treatments, abundance of these proteins was greater in CS-54 than in Pusa Agrani. SNF1-related protein kinase 2 family is a relatively small plant-specific gene family, which regulates ABA-dependent stomatal closure, besides having other roles in sulphur metabolism and environmental strains such as osmotic stress and heavy-metal toxicity (Umezawa et al. 2004; Kulik et al. 2011). This protein is negatively regulated by protein phosphatase 2C (Meskiene et al. 2003). SNF1-related protein kinase (spot 39) was upregulated, while phosphatase 2C (spot 5) decreased in abundance possibly to regulate the activity of enzymes involved in salt tolerance.
Proteins involved in ion homeostasis
Salt stress impairs ion homeostasis in plants, which needs to be re-established for plant survival. The SOS pathway is an important mechanism present in plants to regulate ionic balance mainly by compartmentalising sodium ions in the vacuoles and extracellular spaces through transporters like SOS1 and NHX1 present on plasma membrane and tonoplast respectively (Ji et al. 2013). SOS2 protein, which regulates the activity of these two key transporters, was overexpressed under salt stress, and its abundance was significantly higher in the salt-tolerant genotype than in the salt-sensitive one, suggesting its high efficiency in maintaining ion homeostasis under salt stress.
Transporter proteins
Phosphate transporters help in the absorption and transport of phosphates within the plant. Although few reports point to the presence of PT4 in shoots (Paszkowski et al. 2002), we found for the first time a highly increased PT4 expression in the shoot (spot 19). Availability of P in the saline soil is greatly reduced due to its low solubility and a reduced PO4 3− activity via ionic strength effects (Parihar et al. 2015). Upregulation of this protein may help the plant maintain its potassium level, which is otherwise disturbed due to low uptake of phosphates under salt stress (Qadir and Schubert 2002).
Conclusion
In conclusion, the adaptive response of Indian mustard to salt stress is multifaceted, being dependent on genotype and salt dosage. Genotype CS-54 suffered the least in terms of biomass damage and the chlorophyll and protein contents. This genotype also showed the least MDA content and a high level of antioxidants, thus confirming its high salt-tolerance efficiency. Pusa Agrani, on the other hand, experienced the maximum reduction in growth parameters, high membrane damage and a low level of antioxidants, showing its salt-sensitive nature. The comparative proteomic analysis of Indian mustard leaves under salinity stress led to the identification of 42 differentially-expressed proteins, functionally involved in photosynthesis, antioxidant defense, energy metabolism, ion homeostasis, carbohydrate metabolism, signal transduction, and the nitrogen and protein metabolisms (schematically represented in Fig. 4). The salt-tolerant genotype appeared to have a higher capability of resisting the negative effect of salt stress on the carbohydrate and protein metabolisms, maintaining the osmotic homeostasis and possessing a better antioxidant defense. The recognition of several novel proteins such as PT4 transporter, SOS2, PII-like protein, oxygen-evolving enhancer protein 1 and rubisco activase (Fig. S4), along with other differentially-expressed proteins, must improve our understanding of the complex response-network associated with salt stress, and provide new openings for developing salt tolerance in Indian mustard. Our maiden report of overexpression of PT4 transporter in Indian mustard leaves under salinity stress may help in achieving phosphate regulation to enhance plant productivity.
Schematic representation of differentially-expressed salt-responsive leaf proteins in Indian mustard subjected to salt stress. Yellow coloured proteins were upregulated and orange coloured decreased in intensity. OEC oxygen evolving complex, FNR ferredoxin dependent NADP reductase; RA rubisco activase, GS glutamine synthetase, FK fructose kinase, GAP glyceraldehyde 3-phosphate dehydrogenase, IL isicitrate lyase; ID isocitrate dehydrogenase, MD malate dehydrogenase, GK glycerate kinase, ETC electron transport chain, ROS reactive oxygen species. (Color figure online)
References
Abogadallah GM (2010) Antioxidative defense under salt stress. Plant Signal Behav 5:369–374
Aebi H (1984) Catalase in vitro. Meth Enzymol 105:121–126
Ahmad P, Hakeem KR, Kumar A, Ashraf M, Akram NA (2012) Salt-induced changes in photosynthetic activity and oxidative defense system of three cultivars of mustard (Brassica juncea L.). Afr J Biotechnol 11(11):2694–2703
Anderson ME (1985) Determination of glutathione and glutathione disulfide in biological samples. Meth Enzymol 113:548–555
Anjum NA, Ahmad I, Mohmood I, Pacheco M, Duarte AC, Pereira E, Umar S, Ahmad A, Khan NA, Iqbal M, Prasad MNV (2012) Modulation of glutathione and its related enzymes in plants’ responses to toxic metals and metalloids: a review. Environ Exp Bot 75:307–324
Anjum NA, Sofo A, Scopam A, Roychoudhury A, Gill SS, Iqbal M, Lukatkin AS, Pereira E, Duarte AC, Ahmad I (2014) Lipids and proteins—major targets of oxidative modifications in abiotic stressed plants. Environ Sci Pollut Res 22(6):4099–4121. doi:10.1007/s11356-014-3917-1
Aref IM, Khan PR, Khan S, El-Atta H, Ahmed AI, Iqbal M (2016) Modulation of antioxidant enzymes in Juniperus procera needles in relation to habitat environment and dieback incidence. Trees Struct Funct. doi:10.1007/s00468-016-1399-0
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoxidase in Beta vulgaris. Plant Physiol 24:1–15
Arshi A, Abdin MZ, Iqbal M (2004) Changes in biochemical status and growth performance of Senna (Cassia angustifolia Vahl.) grown under salt stress. Phytomorphology 54:109–124
Arshi A, Ahmad A, Are IM, Iqbal M (2012) Comparative studies on antioxidant enzyme action and ion accumulation in soybean cultivars under salinity stress. J Environ Biol 33:9–20
Ashraf M, Harris PJC (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Sci 166:3–16
Bandehagh A, Salekdeh GH, Toorchi M, Mohammadi A, Komatsu S (2011) Comparative proteomic analysis of canola leaves under salinity stress. Proteomics 11:1965–1975
Bashir H, Qureshi MI, Ibrahim AM, Iqbal M (2015) Chloroplast and photosystems: impact of cadmium and iron deficiency. Photosynthetica 53:321–335
Beyer WF, Fridovich I (1987) Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal Biochem 161:559–566
Boex-Fontvieille E, Daventure M, Jossier M, Hodges M, Zivy M, Tcherkez G (2014) Phosphorylation pattern of Rubisco activase in Arabidopsis leaves. Plant Biol 16:550–557
Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Busheva M, Tzonova I, Stoitchkova K, Andreeva A (2012) Heat-induced reorganization of the structure of photosystem II membranes: role of oxygen evolving complex. J Photochem Photobiol, B 117:214–221
Caruso G, Cavaliere C, Guarino C, Gubbiotti R, Foglia P, Lagana A (2008) Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal Bioanal Chem 391:381–390
Du CX, Fan HF, Guo SR, Tezuka T, Li J (2010) Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71:1450–1459
Feller U, Anders I, Demirevska K (2008) Degradation of rubisco and other chloroplast proteins under abiotic stress. Gen Appl Plant Physiol 34:5–18
Foyer CH, Halliwell B (1976) The presence of glutathione and glutathione reductase in chloroplasts, A proposed role in ascorbic acid metabolism. Planta 133:21–25
Foyer CH, Noctor G (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antiox Redox Sign 11:861–905
Galmes J, Aranjuelo I, Medrano H et al (2013) Variation in Rubisco content and activity under variable climatic factors. Photosynth Res 117:73–90
Gapinska M, Skodowska M, Gabara B (2008) Effect of short- and long-term salinity on the activities of antioxidative enzymes and lipid peroxidation in tomato roots. Acta Physiol Plant 30:11–18
Gengmao Z, Yu Z, Xing S et al (2015) Salinity stress increases secondary metabolites and enzyme activity in safflower. Indust Crops Prod 64(1):175–181. doi:10.1016/j.indcrop.2014.10.058
Ghaffari A, Gharechahi J, Nakhoda B et al (2014) Physiology and proteome responses of two contrasting rice mutants and their wild type parent under salt stress conditions at the vegetative stage. J Plant Physiol 171:31–44
Guan Z, Chai T, Zhang Y et al (2009) Enhancement of Cd tolerance in transgenic tobacco plants overexpressing a Cd-induced catalase cDNA. Chemosphere 76(5):623–630
Hakeem KR, Chandana R, Ahmad P, Iqbal M, Ozturk M (2012a) Relevance of proteomic investigations in plant abiotic stress physiology. OMICS 16(11):621–635
Hakeem KR, Khan F, Chandna R, Siddiqui TO, Iqbal M (2012b) Genotypic variability among soybean genotypes under NaCl stress and proteome analysis of salt-tolerant genotype. Appl Biochem Biotechnol 168(8):2309–2329
Harinasut P, Poonsopa D, Roengmongkol K et al (2003) Salinity effects on antioxidant enzymes in mulberry cultivar. Sci Asia 29:109–113
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophy 125:189–198
Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Hsieh TH, Li CH, Su RC et al (2010) A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231:1459–1473
Huergo LF, Chandra G, Merrick M (2013) PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol Rev 37:251–283
Iqbal M, Ahmad A, Ansari MKA et al (2015) Improving the phytoextraction capacity of plants to scavenge metal(loid)-contaminated sites. Environ Rev 23:44–65
Isaacson T, Damasceno CM, Saravanan RS et al (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1(2):769–774
Jeong MJ, Park SC, Byun MO (2001) Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer. Mol Cells 12:185–189
Ji H, Pardo JM, Batelli G et al (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6(2):275–286
Jiang Y, Yang B, Harris NS et al (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58:3591–3607
Kim Y, Arihara J, Nakayama T, Nakayama N (2004) Antioxidative responses and their relation to salt tolerance in Echinochloa oryzicola Vasing and Steraia virdis (L.) Beauv. Plant Growth Regul 44:87–92
Kim DW, Rakwal R, Agrawal GK et al (2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophor 26:4521–4539
Kong-Ngern K, Daduang S, Wongkham CH et al (2005) Protein profiles in response to salt stress in leaf sheaths of rice seedlings. Sci Asia 31:403–408
Kukreja S, Nandval AS, Kumar N et al (2005) Plant water status, H2O2 scavenging enzymes, ethylene evolution and membrane integrity of Cicer arietinum roots as affected by salinity. Biol Plant 49:305–308
Kulik A, Mazur AA, Bucholc M et al (2004) Antioxidant responses of shoots and roots of lentil to NaCl salinity stress. Plant Growth Regul 42:69–77
Kulik A, Wawer I, Krzywinska E et al (2011) SnRK2 protein kinases-key regulators of plant response to abiotic stresses. OMICS 15:859–872
Law MY, Charles SA, Halliwell B (1983) Glutathione and ascorbic acid in spinach chloroplasts. Biol Biochem J 210:899–903
Lei P, Xu Z, Liang J, Luo X, Zhang Y, Feng X, Xu H (2016) Poly (γ-glutamic acid) enhanced tolerance to salt stress by promoting proline accumulation in Brassica napus L. Plant Growth Regul 78(2):233–241
Li C, Lv J, Zhao X et al (2010) TaCHP: a wheat zinc finger protein gene down-regulated by abscisic acid and salinity stress plays a positive role in stress tolerance. Plant Physiol 154:211–221
Lu S, Li T, Jiang J (2010) Effects of salinity on sucrose metabolism during tomato fruit development. Afr J Biotech 9(6):842–849
Lyzenga WJ, Stone SL (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63(2):599–616
Manaa A, Mimouni H, Wasti S, Gharbi E, Aschi-Smiti S, Faurobert M, Ahmed HB (2013) Comparative proteomic analysis of tomato (Solanum lycopersicum) leaves under salinity stress. Plant Omics J 6(4):268–277
Medeiros CD, Oliveira MT, Rivas R et al (2012) Gas exchange, growth, and antioxidant activity in sugarcane under biological nitrogen fixation. Photosynthetica 50:519–528
Meskiene I, Baudouin E, Schweighofer A et al (2003) Stress-induced protein phosphatase 2C Is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278(21):18945–18952
Miranda RS, Gomes-Filho E, Prisco JT, Alvariz-Pizzaro JC (2016) Ammonium improves tolerance to salinity stress in Sorghum bicolor plants. Plant Growth Regul 78(1):121–131
Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253:201–218
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI (2007) Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta 1767:414–421
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22:867–880
Nazar R, Iqbal N, Syeed S et al (2011) Salicylic acid alleviates decreases in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidant metabolism differentially in two mungbean cultivars. J Plant Physiol 168:807–815
O’Farrel PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021
Parihar R, Singh S, Singh R et al (2015) Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res 22:4056–4075
Parker R, Flowers TJ, Moore AL et al (2006) An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf. J Exp Bot 57:1109–1118
Paszkowski U, Kroken S, Roux C, Briggs S (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99:13324–13329
Puig J, Meynard D, Khong GN et al (2013) Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Express Patter 13:160–170
Qadir M, Schubert S (2002) Degradation processes and nutrient constraints in sodic soils. Land Degrad Develop 13(4):275–294
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org/
Rais L, Masood A, Inam A, Khan N (2013) Sulfur and nitrogen co-ordinately improve photosynthetic efficiency, growth and proline accumulation in two cultivars of mustard under salt stress. J Plant Biochem Physiol 1:101
Rao MV (1992) Cellular detoxifying mechanisms determine age dependent injury in tropical plants exposed to SO2. J Plant Physiol 140:733–740
Rowley ER, Mockler TC (2011) Plant abiotic stress: insights from the genomics era. In: Shankar A, Venkateswarlu B (eds) Abiotic stress response in plants—physiological, biochemical and genetic perspectives. InTech Open, Rijeka, Croatia, pp 221–268
Sahi C, Singh A, Blumwald E, Grover A (2006) Beyond osmolytes and transporters: novel plant salt stress tolerance-related genes from transcriptional profiling data. Physiol Plant 127:1–9
Santos CV (2004) Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Sci Hort 103:93–99
Shah SH (2007) Effects of salt stress on mustard as affected by gibberellic acid application. Gen App Plant Physiol 33(1–2):9–106
Sharma P, Jha AB, Dubey RS et al (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot 2012:1–26
Sharma P, Sardana V, Banga SS (2013) Salt tolerance of Indian mustard (Brassica juncea) at germination and early seedling growth. Environ Exp Biol 11:39–46
Shekhawat GS, Verma K (2010) Heme oxygenase (HO): an overlooked enzyme of plant metabolism and defence. J Exp Bot 61(9):2255–2270
Sumithra K, Jutur PP, Carmel BD, Reddy AR (2006) Salinity-induced changes in two cultivars of Vigna radiata: responses of antioxidative and proline metabolism. Plant Growth Regul 50:11–22
Suorsa M, Aro EM (2007) Expression, assembly and auxiliary functions of photosystem II oxygen-evolving proteins in higher plants. Photosynth Res 93:89–100
Takahashi S, Badger MR (2011) Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci 16(1):53–60
Uhrig GR, Kenneth KS, Greg BG et al (2009) PII in higher plants: a modern role for an ancient protein. Trend Plant Sci 14(9):505–511
Umezawa T, Yoshida R, Maruyama K et al (2004) SRK2C a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. PNAS 101(49):17306–17311
Venkatesh J, Upadhyaya CP, Yu JW, Hemavathi A, Kim DH, Strasser RJ, Park SW (2012) Chlorophyll a fluorescence transient analysis of transgenic potato overexpressing d-galacturonic acid reductase gene for salinity stress tolerance. Hort Environ Biotechnol 53:320–328
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang X, Wei Z, Liu D et al (2011) Effects of NaCl and silicon on activities of antioxidative enzymes in roots, shoots and leaves of alfalfa. Afr J Biotechnol 10(4):545–549
Yamamoto M, Nasrallah JB (2009) In planta assessment of the role of thioredoxin h proteins in the regulation of S-locus receptor kinase signaling in transgenic Arabidopsis. Plant Physiol 163:1387–1395
Yasar F, Ellialtioglu S, Yildiz K (2008) Effect of salt stress on antioxidant defense systems, lipid peroxidation and chlorophyll content in green bean. Russ J Plant Physiol 55(6):782–786
Yousuf PY, Hakeem KR, Chandna R, Ahmad P (2012) Role of glutathione reductase in plant abiotic stress. In: Ahmad P, Prasad MNV (eds) Abiotic stress responses in plants. Springer, New York, pp 149–158
Yousuf PY, Ahmad A, Ganie AH, Iqbal M (2016a) Salt stress-induced modulations in the shoot proteome of Brassica juncea genotypes. Environ Sci Pollut Res 23(3):2391–2401. doi:10.1007/s11356-015-5441-3
Yousuf PY, Ahmad A, Aref IM, Ozturk M, Hemant Ganie AH, Iqbal M (2016b) Salt-stress-responsive chloroplast proteins in Brassica juncea genotypes with contrasting salt tolerance and their quantitative PCR analysis. Protoplasma. doi:10.1007/s00709-015-0917-z
Zhang C, Liu J, Zhang Y, Cai X, Gong P, Zhang J, Wang T, Li H, Ye Z (2011) Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep 30:389–398
Acknowledgments
The first author is grateful to Hamdard National Foundation (HNF) New Delhi, India for granting a fellowship. The second author (A Ahmad) is now working at the Department of Botany, Aligarh Muslim University, Aligarh 202002, India.
Author contributions
P. Y. Yousuf conducted experimental research with the help of A. Ahmad and A. H. Ganie. Statistical analysis was done by O Sareer and V Krishnapriya. Data interpretation and MS preparation were done by M. Iqbal, A. Ahmad and I. M. Aref.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors have no conflict of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2016_182_MOESM1_ESM.jpg
Fig. S1: Plants of salt-tolerant genotype (CS-54) grown under (a) control and (b) 200 mM NaCl treatment and salt-sensitive genotype Pusa Agrani grown under (c) control and (d) 200 mM NaCl treatment, at the time of sampling (JPEG 244 kb)
10725_2016_182_MOESM2_ESM.jpg
Fig. S2: (a) Clustering and (b) PCA analysis based on biomass of 14 different genotypes of Indian mustard grown under salt stress. Correlation matrix was used for PCA. Clustering was done using the Ward’s method of Squared Euclidean distance matrix. PCA and clustering of biomass clearly differentiate V2 and V5. Arrows for all traits are directed towards V5 indicating that it is indeed a tolerant variety, whereas V2 presents an opposite case indicating its salt-sensitive nature. Cluster dendrogram also reflects similar results (JPEG 1077 kb)
10725_2016_182_MOESM3_ESM.jpg
Fig. S3: PCA analysis of 14 different genotypes of Indian mustard under different salt treatments (a) T0 (b) T1 (c) T2 (d) T3 (e) T4, based on physiological and biochemical parameters. Percentage values given in brackets in the axis labels denote R2 values (JPEG 1586 kb)
10725_2016_182_MOESM4_ESM.jpg
Fig. S4: Spot images of five representative proteins differentially-expressed during different salt treatments in salt-sensitive (Pusa Agrani) and salt-tolerant (CS-54) genotypes of Indian mustard (JPEG 189 kb)
Rights and permissions
About this article
Cite this article
Yousuf, P.Y., Ahmad, A., Ganie, A.H. et al. Antioxidant response and proteomic modulations in Indian mustard grown under salt stress. Plant Growth Regul 81, 31–50 (2017). https://doi.org/10.1007/s10725-016-0182-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0182-y