Abstract
Dendrobium is an epiphytic orchid, which is highly valued as a cut flowers. The assessment of genetic diversity is crucial to study the relationship between the species and also to develop the novel hybrids. In this study, 25 cross species simple sequence repeat (SSR) markers were screened for amplification in Dendrobium species. Eighteen polymorphic markers were used to estimate the genetic diversity in 15 Dendrobium species. The 18 SSR markers generated a total of 55 polymorphic bands, with an average of 3.05 bands per primer. The observed and expected heterozygosity within the species ranged from 0 to 0.62 and 0.30 to 0.86, respectively. Cluster analysis based on the unweighted pair group method with an average led to a dendrogram with three major groups that coincided for morphological characters mostly for flower colour, flower shape and nature of shoot, but did not coincide with their geographical locations. Results revealed that genetic variation exists amongst the 15 Dendrobium species, which will be helpful in selecting desirable species as a parent for crossing purpose to develop new interspecific hybrids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Orchid flowers are highly prized and admired by flower lovers around the world for their unique and diverse characteristics. They come in a wide range of forms, sizes, and colors, making them incredibly appealing and captivating. Orchids are indeed popular as both pot plants and cut flowers. Dendrobium is the second largest genera amongst the Orchidaceae family after Bulbophyllum (Basavaraj et al. 2020) and gained importance in cut flower, perfume and cosmetic industries. The use of Dendrobium orchids in the traditional medicine system have been known in many parts of the world since ancient times (Varma and Rajashekar 2022).
The northeast region of India is rich in genetic diversity and is one of the world’s eight mega biodiversity hotspots, hosting nearly 900 species of orchids out of 1350 species present in India; i.e., 69% of India’s orchid taxa (Ninawe and Swapna 2017). Keeping in view the enormity and diversity of indigenous ornamental plant species in India and orchids in particular, a networking approach is essentially required which could share the responsibility of germplasm collection, characterization, conservation, evaluation and maintenance of precious genetic resources.
Characterization of genetic relationship and diversity of orchids is very important for conservation and its utilization in crop improvement programmes (Roy et al. 2017). Molecular markers provide a very precise tool for evaluating genetic diversity of germplasm which helps to select more diverse parents in breeding programme which has led to an advancement in analysis of orchid genetic diversity (Wang et al. 2009). In spite of the evolutionary significance of the Dendrobium orchids, the available data of these species using reproducible and reliable markers is very less. With the rampant forest destruction, Dendrobium orchids is facing extinction and needs to be conserved through proper plant conservation strategies. At present the research reports on the genetic diversity of Dendrobium species in particular is scanty which is essential for genetic conservation and plant breeding programme. Keeping in view the need for conservation and future use in breeding programmes, the present study aimed to characterize and identify the genetic diversity of 15 important species of genus Dendrobium by using simple sequence repeat (SSR) markers which will be useful for further crop improvement programmes.
Materials and methods
Plant materials
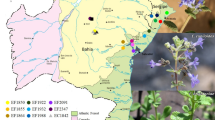
In the present study, fifteen wild Dendrobium species (Fig. 1) were characterized. These orchids were collected from different locations of East Siang District of Arunachal Pradesh (Table 1; Fig. 2) and maintained at College of Horticulture and Forestry, Central Agricultural University, Pasighat, Arunachal Pradesh, India. Collected genotypes were grown in pots having 1:1:1 ratio of brick pieces, coconut husk and cocopeat along with few charcoal pieces for purifying the media.
DNA Extraction
DNA was extracted from the young fresh leaves through CTAB method as performed by Doyle and Doyle (1987) with slight modifications. Tender young leaves of 0.5 g from each species were used to isolate genomic DNA. RNA was removed by treating DNA with RNase (Himedia Laboratories Pvt. Ltd. Thane, Maharashtra) at 65 °C for 20 min. The DNA quality was checked on 0.8% agarose gel stained with ethidium bromide (EtBr) and visualized in the gel documentation system.
SSR marker genotyping
The molecular diversity of the 15 Dendrobium species were characterized by using 25 SSR markers of Dendrobium officinale and Dendrobium loddigessi (Cai et al. 2012; Lu et al. 2012).
PCR amplification was carried out with 20 µl reaction mixture containing 2 µl of 50 ng template DNA, 2 µl of 10 × PCR buffer, 1.4 µl of 25 mM MgCl2, 0.3 µl of 10 mM dNTPs, 1 µl of 10 µM of each forward and reverse primer (AgriGenome Labs Pvt. Ltd. Kochi, Kerala) and 0.3 µl of Taq DNA polymerase (5U/µl). All the chemicals were procured from Himedia Laboratories Pvt. Ltd. Thane, Maharashtra. Thermal profiles were standardized for each SSR primer pair based on its melting temperature using a master cycler 5331 gradient –PCR machine (Eppendorf, Hamburg, Germany). The standard annealing temperatures of all the SSR primers are given in Table 2. The PCR fragments were resolved on a 3% (w/v) agarose gel with 2 µl ethidium bromide in 1 × TAE buffer at a constant voltage (75 V) for 120 min and visualized under UV light using a Fire reader gel documentation system (IGene Labserve Pvt. Ltd. New Delhi). The data were stored for further analysis.
Statistical data analysis
For the molecular analysis, amplification profiles of different primer combinations for all genotypes were analyzed by comparing DNA fragment sizes on agarose gel with 100 bp DNA ladder. The amplified fragments in each SSR marker were scored manually for their presence (1) and absence (0) among the 15 Dendrobium species. Number of alleles and values of polymorphic information content (PIC), observed heterozygosity and gene diversity (GD)/ expected heterozygosity (He) were calculated in GenAlEx 6.5 software (Peakall and Smouse 2006; Banks and Peakall 2012). The software NTSYS-pc Version 2.02i was used to generate the Jaccard’s similarity coefficient matrix (Rohlf 1987). The genetic distance was calculated by deducting the similarity index value from unity, which was used to produce the dendrogram by using Molecular Evolutionary Genetics Analysis v11 (MEGA11) software through Unweighted Pair Group Method with Average (UPGMA) (Tamura et al. 2021). Based on the SSR banding patterns, polymorphism percentages were calculated for different primers.
Results and discussion
Genetic diversity of species can be estimated by using molecular markers. Microsatellite markers have proved to be a powerful tool for cultivar identification, evaluation of the varietal purity and analysis of genetic diversity. SSR markers are useful tool for observing genetic diversity and these are PCR based, highly polymorphic, multi-allelic, co-dominant, easily reproducible and widely distributed across the genome (Wu et al. 2021; Li et al. 2022). Comparative genomic mining of SSR markers among closely related species, is one of the most efficient and cost-effective methods for discovering novel DNA markers. Sequence data from a variety of plant species revealed that there is enough homology between the genomes of two or more closely related genera or species. As a result, primer pairs designed for one species could be used to detect SSRs in related species and even other genera within the same family (Kalia et al. 2011).
SSR marker analysis
Twenty-five SSR primers were screened, out of which 18 polymorphic markers were used for molecular characterization and analysis of genetic diversity among the 15 Dendrobium species (Fig. 3). Fifty-six alleles with sizes ranging from 105 to 668 base pairs were detected with an average of 3.11 alleles per locus. A number of alleles produced by 18 primers ranged from 1 to 6 which is similar to the report in Dendrobium orchids by Kang et al. (2015); Lu et al. (2013b) and Lu et al. (2012) and in Cymbidium by Li et al. (2014). Similarly in another study conducted in Phalaenopsis orchids by Fattmah and Sukma (2011), 16 polymorphic SSR were identified with 2–5 alleles while, 14 SSR polymorphic loci were identified 1–12 alleles in Cymbidium by Moe et al. (2010). The obtained results indicated that the selected Dendrobium species have higher level of genetic diversity due to enormous phenotypic variation in flower colour, flower length, flower shape and nature of shoot (Table 3 and Table 4).
All the markers have shown 100 percent polymorphism, except CXY03. Out of 56 SSR loci, 55 were polymorphic with 98.11% of polymorphism (Table 2). The possibility for high level of polymorphism in Dendrobium species is due to their diverse nature. Liu et al. (2014) reported that the overall level of polymorphism (92.50%) indicated the effectiveness of markers to investigate genetic diversity among the different species of Dendrobium germplasm. Similar results were obtained by Basavaraj et al. (2020); Kang et al. (2015) and Lu et al. (2013a) having 66.66%, 42% and 85.91% polymorphism in Dendrobium orchids, respectively. Huang et al. (2010) and Jantasuriyarat et al. (2012) also reported 90.80% and 64.80% polymorphism, respectively in different orchid genera.
The various genetic diversity parameters such as, polymorphic information content (PIC), observed heterozygosity and gene diversity (GD)/expected heterozygosity (He), were analyzed (Table 5). An average PIC value serves as the ideal index for measuring polymorphism. PIC values above 0.5 denote high polymorphism loci, between 0.25 and 0.5 which denotes intermediate polymorphism loci, and below 0.25 denoted low polymorphism loci (Ge et al. 2013; Bhargav et al. 2021). All the selected primers were polymorphic and the polymorphic information content ranged from 0.33 to 0.90 with an average of 0.74 which indicated that the loci displayed intermediate to high polymorphism and showed the occurrence of a broad gene pool amongst the species. The high levels of polymorphic markers can be used to track the introduction of genes into desirable genetic backgrounds that are advantageous to find species that are suitable for varietal development. The results are in concurrence with findings of Cai et al. (2012) where PIC value ranged from 0.36 to 0.84 with a mean of 0.64 in Dendrobium loddigesii. Similarly, in Cymbidium and Phalenopsis also reported PIC value ranging from 0.17 to 0.89 and 0.18 to 0.72 with a mean of 0.63 and 0.67, respectively (Moe et al. 2010; Fattmah and Sukma 2011).
The observed heterozygosity (Ho) ranged from 0.00 to 0.62 and found to be highest for the primers namely, CXY04 (0.62), followed by DOeSSR6 (0.50) and CXY09 (0.44) which reflects their ability to provide unique genetic profiles across the species. The expected heterozygosity (He) or gene diversity (GD) ranged from 0.30 to 0.86 with an average of 0.72. The values obtained are in agreement with the findings of Cai et al. (2012) in Dendrobium loddigesii where the observed heterozygosity ranged from 0.00 to 0.70 and the expected heterozygosity from 0.45 to 0.85. Estimates of genetic diversity helps in organizing germplasm, identifying cultivars, and choosing parents for hybridization. In plant breeding programmes, genotypes that exhibit a sufficient level of polymorphism based on genetic diversity are chosen (Joshi et al. 1997; Louarn et al. 2007; Ren et al. 2012).
Genetic relationship among Dendrobium species
Results of diversity analysis revealed that 15 genotypes have been grouped in 3 different clusters (Fig. 4), in which cluster-I comprised of 7 genotypes (Dendrobium densiflorum, D. devonianum, D. fimbriatum, D. nobile, D. nobile var. alba, D. thyrsiflorum and D. wardianum) Cluster-II comprised of 6 genotypes (D. aphyllum, D. chrysanthum, D. chrysotoxum, D. eriiflorum, D. lituiflorum and D. macraei) and Cluster-III comprises of 2 genotypes (D. jenkinsii and D. primulinum. Similar clustering pattern was observed by Lee et al. (2020), Kang et al. (2015), Li et al. (2014), Liu et al. (2014), Lu et al. (2013b) and Moe et al. (2010) using SSR markers in different orchid genera.
Cluster I comprised of white, purple and yellow species of tall stature and big sized flowers. Cluster II represents the tall plants and small sized flowers, respectively. The genotypes in Cluster III are white and yellow flower coloured with bulbous shoot and less flower longevity (4 days) (Tables 3 and 4). Species located in Cluster I, II and III indicated more phenotypic variations. Similar findings were reported by Kang et al. (2015) and Lu et al. (2013b) in Dendrobium species. The results of the present findings indicated that the species from different regions genetically varied amongst the genus and conservation of the germplasm will be helpful for crop improvement. Since SSR markers produced sufficient information for classification of Dendrobium species into different groups, showing diversity for flower colour, flower shape and nature of shoot; the findings of this research can be efficiently used for genetic diversity assessments in Dendrobium genus.
Conclusion
The investigation revealed the successful utility of SSRs from closely related species in fingerprinting and estimation of genetic diversity amongst the Dendrobium species. Results revealed genetic variation that exists amongst the Dendrobium species, which will help in desirable species selection for crossing purpose to develop novel hybrids, with various flower forms and colours and to identify markers which are linked to desirable traits for marker-assisted selection (MAS).Thus the present study opens up new opportunities for targeted hybridization and advanced breeding techniques to enhance their aesthetic and economic value.
Data availability
Raw data are available upon request from the corresponding author.
References
Banks SC, Peakall ROD (2012) Genetic spatial autocorrelation can readily detect sex biased dispersal. Mol Ecol 21:2092–2105
Basavaraj B, Nagesha N, Jadeyegowda M (2020) Molecular characterization of Dendrobium orchid species from western ghat region of Karnataka using RAPD and SSR markers. Int J Curr Microbiol App Sci 9:2157–2169
Bhargav V, Kumar R, Sane A, Rao TM, Bharathi TU, Shankara KS, Reddy DL (2021) Molecular characterization of China aster (Callistephus chinensis (L.) Nees) genotypes using SSR markers. Isr J Plant Sci 68:287–296
Cai X, Feng Z, Hou B, Xing W, Ding X (2012) Development of microsatellite markers for genetic diversity analysis of Dendrobium loddigesii Rolfe, an endangered orchid in China. Biochem Syst Ecol 43:42–47
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Fattmah SD (2011) Development of sequence-based microsatellite marker for Phalaenopsis orchid. Hayati J Biosci 18:71–76
Ge H, Liu Y, Jiang M, Zhang J, Han H, Chen H (2013) Analysis of genetic diversity and structure of eggplant populations (Solanum melongena L.) in China using simple sequence repeat markers. Sci Hortic 162:71–75
Huang Y, Li F, Chen K (2010) Analysis of diversity and relationships among Chinese orchid cultivars using EST-SSR markers. Biochem Syst Ecol 38:93–102
Jantasuriyarat C, Ritchuay S, Pattarawat P, Huehne PS, Kate-Ngam S (2012) Development and transferability of EST-SSR and transferability of genomic SSR markers for genetic diversity assessment of Doritis. Biochem Syst Ecol 45:57–65
Joshi KD, Subedi M, Rana RB, Kadayat KB, Sthapit BR (1997) Enhancing on-farm varietal diversity through participatory varietal selection: a case study for chaite rice in Nepal. Exp Agric 33:335–344
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kang J, Lu J, Qiu S, Chen Z, Liu J, Wang H (2015) Dendrobium SSR markers play a good role in genetic diversity and phylogenetic analysis of Orchidaceae species. Sci Hortic 183:160–166
Lee HJ, Park HR, Lee AJ, Nam DE, Lee DG, Do Y, Chung KW (2020) Genetic authentication of cultivars with flower-variant types using SSR markers in spring orchid, Cymbidium goeringii. Hortic Environ Biotechnol 61:577–590
Li X, Jin F, Jin L, Jackson A, Huang C, Li K, Shu X (2014) Development of Cymbidium ensifolium genic-SSR markers and their utility in genetic diversity and population structure analysis in cymbidiums. BMC Genet 15(1):1–14
Li X, Qiao L, Chen B, Zheng Y, Zhi C, Zhang S, Cheng Z (2022) SSR markers development and their application in genetic diversity evaluation of garlic (Allium sativum) germplasm. Plant Divers 44:481–491
Liu X, Huang Y, Li F, Xu C, Chen K (2014) Genetic diversity of 129 spring orchid (Cymbidium goeringii) cultivars and its relationship to horticultural types as assessed by EST-SSR markers. Sci Hortic 174:178–184
Louarn S, Torp AM, Holme IB, Andersen SB, Jensen BD (2007) Database derived microsatellite markers (SSRs) for cultivar differentiation in Brassica oleracea. Genet Resour Crop Evol 54:1717–1725
Lu JJ, Suo NN, Hu X, Wang S, Liu JJ, Wang HZ (2012) Development and characterization of 110 novel EST-SSR markers for Dendrobium officinale (Orchidaceae). Am J Bot 99:e415–e420
Lu JJ, Gao L, Kang JY, Feng SG, He RF, Wang HZ (2013a) Thirteen novel polymorphic microsatellite markers for endangered Chinese endemic herb Dendrobium officinale. Conserv Genet Resour 5:359–361
Lu JJ, Kang JY, Feng SG, Zhao HY, Liu JJ, Wang HZ (2013b) Transferability of SSR markers derived from Dendrobium nobile expressed sequence tags (ESTs) and their utilization in Dendrobium phylogeny analysis. Sci Hortic 158:8–15
Moe KT, Zhao W, Song HS, Kim YH, Chung JW, Cho YI, Park YJ (2010) Development of SSR markers to study diversity in the genus Cymbidium. Biochem Syst Ecol 38:585–594
Ninawe AS, Swapna TS (2017) Orchid diversity of northeast India-Traditional Knowledge and strategic plan for conservation. J Orchid Soc India 31:41–56
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Ren R, Nagel BA, Kumpatla SP, Zheng P, Cutter G, Greene TW, Thompson SA (2012) Maize cytoplasmic male sterility (cms) c-type restorer rf4 gene, molecular markers and their use. U.S. Patent Application No.13/244,049
Rohlf FJ (1987) NTSYS-pc: Microcomputer programs for numerical taxonomy and multivariate analysis. Am Stat 41:330–330
Roy SC, Moitra K, De Sarker D (2017) Assessment of genetic diversity among four orchids based on ddRAD sequencing data for conservation purposes. Physiol Mol Biol Plants 23:169–183
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Varma AR, Rajshekhar I (2022) A review article on: an overview of medicinal importance of orchids. Acta Sci Med Sci 6:1–9
Wang HZ, Feng SG, Lu JJ, Shi NN, Liu JJ (2009) Phylogenetic study and molecular identification of 31 Dendrobium species using inter-simple sequence repeat (ISSR) markers. Sci Hortic 122:440–447
Wu F, Zhang S, Gao Q, Liu F, Wang J, Wang X (2021) Genetic diversity and population structure analysis in a large collection of Vicia amoena in China with newly developed SSR markers. BMC Plant Biol 21:1–12
Acknowledgements
The authors are thankful for the College of Horticulture and Forestry, Central Agricultural University, Pasighat, Arunachal Pradesh for extending the research facilities and Dr. Mahesh Kumar, College of Agriculture Pasighat for helping in editing the research article.
Funding
The research work was supported by College of Horticulture and Forestry, Central Agricultural University, Pasighat, Arunachal Pradesh.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that there are no conflicts of interest to disclose.
Consent to participate
All author’s have given consent to participate.
Consent for publication
All authors have given consent to publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Reddy, D.M., Momin, K.C., Singh, A.K. et al. Genetic diversity of Dendrobium species revealed by simple sequence repeat (SSR) markers. Genet Resour Crop Evol 71, 1453–1464 (2024). https://doi.org/10.1007/s10722-023-01701-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01701-w