Abstract
With the increasing acidification of soil, aluminum (Al) toxicity has become one of the most important stress factors affecting seed germination quality and crop yield. To investigate the Al tolerance on seed germination, genome-wide association analysis (GWAS) of 19,949 SNPs with genome-wide coverage was used to identify the candidate genes, which were potentially related to germinate traits of rapeseed (Brassica napus L.) under Al stress. In the experiment, 169 rapeseed cultivars (lines) were treated with AlCl3 solution of 90 ppm, and distilled water was added to the control. At the 7th day, the phenotype data, including root length and dry weight, were measured and calculated. Using the TASSEL software, Al tolerance related traits were explored in rapeseed under germination with a 60 K Brassica Illumina® Infinium SNP array. Then, the structure of the population was analyzed with the STRUCTURE software, and the genetic relationship and LD attenuation were analyzed with the software TASSEL, respectively. The GWAS of relative root length (RRL) and relative dry weight (RDW) with SNP markers were carried out under the optimal model. Meanwhile, the candidate genes were predicted based on the LD interval sequence of the associated SNP locus. Subsequently, the homologous genes of rapeseed related to Al tolerance in the target genome region were screened in Arabidopsis thaliana (L.) Heynh. The results showed that 13 SNPs were significantly associated with these two traits. Among them, 8 SNPs were significantly associated with RRL and located on chromosomes A03, A07, A09, A10, C05, C06, and C09, respectively. Five SNPs were significantly associated with RDW and located on chromosomes A03, A04, A10, C05, and C07, respectively. Afterward, fifty-nine candidate genes related to Al tolerance were identified in the LD region of these SNP loci. Four of these genes were involved in the growth regulation about organic acid, ten were involved in growth-regulating substance, eleven were related to oxidative stress, and nineteen were involved in carbon and nitrogen metabolism. The results of this study provided a theoretical basis for Al tolerance in rapeseed and laid out a foundation for further functional verification of genes and cultivation of new Al tolerant rapeseed varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum (Al) is the most prevalent metal and the third most abundant element in the earth’s crust (Exley 2009). Al in solid or bound form does not cause toxicity to plants, environment, and human beings, but ionic (Al3+) or free Al [Al(OH)2+] can cause great toxicity (Kochian 1995). With the application of ammonium fertilizer and the increasing acid rain in recent years, the expanded area of acidic soil directly caused an increased dissociation of Al3+ ions (Meng et al. 2017). Studies are indicating that increasing dissociated Al3+ ions can damage the cell activity of root meristem, inhibit cell mitosis, root growth (Marciano et al. 2010; Doncheva et al. 2005), affect the uptake of nutrition (Li et al. 2015), and ultimately limit crop growth and yield (Furukawa et al. 2007). To alleviate the harm of Al toxicity, many efforts have been made, especially in the genetic mechanism and gene improvement. Variations in Al-tolerant genes in wheat (Hamel et al. 1998), barley (Cai et al. 2013), rice (Nguyen et al. 2001), oat (Castilhos et al. 2011), sorghum (Gourley et al. 1990), and maize (Mattiello et al. 2012) have been guessfully reported. Then high-yielding varieties with stronger tolerance to Al toxicity have been developed in breeding programs. Rapeseed (Brassica napus L.) is considered a plant with weak tolerance to Al (Qian et al. 2014), and it’s mainly planted in the Middle and Lower parts of Yangtze River, the regions with more acidic soil in China. Therefore, Al toxicity has become an important stress factor in its production (Ryan and Delhaize 2010). Seed germination is the initial stage of the plant life cycle, which directly affects the growth and development as well as the production benefits of rapeseed. Exploring the genetic mechanism of Al tolerance in rapeseed during germination is of great significance to the cultivation of high-yielding varieties of Al tolerant rapeseed.
Genome-wide association study (GWAS) was first applied to human disease research, and then gradually applied to plant research. The continuous improvement of SNP marker mining technology covering the whole genome and GWAS based on Linkage Disequilibrium (LD) provided a new way of controlling complex traits in diverse natural populations (Gupta et al. 2014). Atwell et al. (2010) demonstrated the feasibility of GWAS application in Arabidopsis thaliana (L.) Heynh. for the first time and pointed out that this method can also be applied to other plants (Atwell et al. 2010). At present, GWAS has been widely used in studying crop stress resistance, such as salt tolerance (Shi et al. 2017), cold tolerance (Huang et al. 2013a), and drought-induced stress (Li et al. 2018). GWAS studies on Al tolerance in crops have also been reported. Tao et al. (2018) identified a total of 21 candidate genes for 7 important QTL regions associated with Al toxicity tolerance in rice. Cai et al. (2013) carried out GWAS on root length Al tolerance of Tibetan wild barley and cultivated some varieties based on SNP markers. With the publication of the whole genome sequence of rapeseed and the development of 60 K Illumina Infinium SNP chip, the application of GWAS in rapeseed has also made significant advances. Some loci associated with important agronomic and quality traits of rapeseed were identified (He et al. 2017; Liu et al. 2016; Huang et al. 2013b; Xue et al. 2018; Wei et al. 2016). Also, GWAS was already carried out on germination-related indexes such as germination vigor and rate in rapeseed (Hatzig et al. 2015; Tan et al. 2017). However, few GWAS studies were conducted on Al tolerance to rapeseed during germination. In this study, 169 rapeseed cultivars (lines) were treated with Al solution at seed germination stage. GWAS analysis of relative root length and relative dry weight was done to reveal the genetic and molecular mechanisms of Al tolerance in rapeseed, which will provide references for the cultivation of high-yield varieties with Al tolerance.
Materials and methods
Materials and reagents
One hundred and sixty-nine rapeseed germplasms, with different genetic backgrounds, and broad geographical origins, were collected as materials from universities and research institutes in China (Table 1). All varieties (lines) were provided by Chongqing Engineering and Technology Research Center for Rapeseed, China (106.40° E, 29.80° N).
The treatment agent was AlCl3·6H2O with 97% analytical purity, which was produced by Chengdu Chron Chemicals Co., Ltd., Sichuan province, China.
Design and phenotyping
In each variety (line), the uniform-size and full seeds were selected for the germination experiment. Then, 20 grains that were chosen from every variety (lines) and then washed with distilled water three times were evenly placed in Petri dishes on filter paper, respectively. 3 mL distilled water was added in every Petri dish for control and 3 mL Al solution (pH = 4.0) with a concentration of 90 ppm was added for stress (Gao et al. 2019). Each treatment was repeated three times, and the solutions were replaced every 2 days. All the materials were incubated at 25 °C under a photo-period of 16 h day/8 h night and 85% relative humidity.
On the 7th day of seed germination, 10 seedlings with similar growth were randomly selected from each dish to determine their root length and dry weight. Then the relative root length (RRL) and relative dry weight (RDW) of each variety (lines) were calculated as the ratio of treatment to control for GWAS analysis (Gao et al. 2019).
Data processing was carried out with Microsoft Excel 2016 software, while variance analysis and principal component analysis was performed with DPS 2005 software (Tang and Zhang 2013).
Genotype determination and analysis
60 k Illumina Infinium SNP chip was used to carry out the genotype analysis of 169 materials of rapeseed (Unterseer et al. 2014) using the Genome Studio version 2011.1 software (Illumina company, https://www.illumina.com.cn). SNP markers with more than 20% missing calls, and minor allele frequency lower than 0.05 or heterozygosity rate more than 20% across the panel were excluded. After preprocessing, 19,949 high-quality SNP markers with a unique physical location in the rapeseed genome were obtained for linkage disequilibrium (LD) and population structure analysis. The threshold in this study was set at 4.30 [− log (1/19,949)].
Genome-wide association analysis
According to 19,949 unlinked SNPs on 19 chromosomes of rapeseed, the population structure of the 169 rapeseed varieties (lines) was analyzed with STRUCTURE 2.3.4 software (Pritchard et al. 2000). Afterward, the results were added into the website of STRUCTURE HARVESTER (https://taylor0.biology.ucla.edu/structureHarvester/), and the appropriate ∆K value was chosen to determine the number of subgroups and obtaining Q matrix (Earl and Holdt 2012). The parameter settings for estimation of membership coefficients for varieties in each subpopulation were a burn-in of 10,000 generations followed by 100,000 iterations for each of the clusters (K) from 1 to 10, with each K running five times. The most likely K-value was determined by the log probability of the data [ln P(K)] and the value of ΔK, based on the rate of change of ln P(K) between runs using successive K-values as described by Earl et al. (2012). The maximum membership probability among sub-groups was applied to divide the accessions into different subgroups. The relationship among materials was analyzed in the kinship module of TASSEL 5.1.0 to obtain K matrix (Kinship) and PCA matrix (Principal Component Analysis) (Bradbury et al. 2007). Finally, combined with 19,949 high-quality SNP markers selected, GWAS built six models of the general linear model (GLM) and mixed linear model (MLM) with Q, PCA and K matrices as covariates.
The LD decline plot of each chromosome was plotted against the physical distance with TASSEL 5.1.0 using the full matrix and sliding window options for sub-genomes A and C (Bradbury et al. 2007). With the determinant coefficient R2, representing the attenuation threshold, 0.2, the LD decline distances of the chromosomes significantly associated with SNPs were calculated.
Annotation of candidate genes
According to the position of LD interval in rapeseed genome, the number of genes and the sequence of gene-coding proteins in LD interval were analyzed based on the genome annotation information of rapeseed “Darmor-Bzh,” published in France (https://www.genoscope.cns.fr/brassicanapus) (Li et al. 2016). The gene expression patterns, possible co-expression genes, and interacting proteins of Arabidopsis thaliana homologous genes were analyzed in the website of Arabidopsis Information Resource (https://www.arabidopsis.org/). With the E-value threshold at 1 × 10–10, functions of candidate gene were annotated according to Arabidopsis thaliana genes with the highest homology.
Results
Phenotypic data
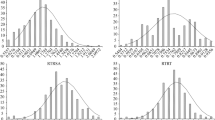
The RRL and RDW of the 196 rapeseed varieties (lines) exhibited extensive phenotypic variation under Al stress (Table 2). The RRL ranged from 0.271 to 1.874 with the mean as 0.705, while RDW ranged from 0.500 to 1.821 with the mean as 0.964. The variation coefficient of RRL and RDW was 49.656% and 15.366%, respectively. Frequency analysis of RRL and RDW showed that these two traits had a continuous distribution (Fig. 1), which was in accordance with quantitative traits and was suitable for GWAS analysis.
Population structure and genetic relationship
The highest ΔK was observed when K = 2 (Fig. 2A). Accordingly, the population could be divided into two subpopulations: P1 (46 accessions) and P2 (123 accessions) (Fig. 2B). The relative kinship analysis revealed that the population of rapeseed had a null or weak relationship, with 55.06% not related (Fig. 2C). The results indicated that GWAS analysis could be performed due to the distant genetic relationship among the tested population materials.
Linkage disequilibrium (LD)
The LD of sub-genome A and C of rapeseed was estimated according to the R2 among SNP markers. The attenuation distances of sub-genomes A and C were different, but their R2 decreased with the extension of genetic distance (Fig. 3). In sub-genome A, the attenuation distance of chromosome A08 was the longest, about 1250 Kb, A09 and A10 chromosomes were about 500 Kb and 330 Kb, respectively. The others followed, mostly around 150–200 Kb with little change. In sub-genome C, the attenuation distance of chromosome C02 was about 1490 Kb, obviously longer than other chromosomes, indicating that there were more genes affecting traits selected by artificial evolution in chromosome C02. Simultaneously, the attenuation speed of chromosome C05 was the slowest, with an attenuation distance around 260 Kb. The average attenuation distance of sub-genome A was 329 Kb with R2 = 0.2, and sub-genome C was 730 Kb (Table 3). The reason that the decline rate of sub-genome A was slower than the one of sub-genome C could be related to the large-scale recombination of sub-genome A in Chinese semi-winter rapeseed and the breakdown of linkage imbalance (Lu et al. 2016).
Genome-wide association analysis
General linear models (GLM) including GLM, Q, and PCA models as well as mixed linear models (MLM), including K, K + Q, and K + PCA models were made with GWAS. When comparing the Q-Q plots (Fig. 4) of distribution under these six models, the K + PCA models of RRL and RDW in MLM were found closer to the predicted line, so the K + PCA model was selected to find the related loci.
Eight SNP markers related to RRL were identified on chromosomes A03, A07, A09, A10, C05, C06, and C09, with phenotypic variation explained (PVE) ranging from 14.566% to 26.102% (Table 4, Fig. 5A). Five SNP markers related to RDW were identified on chromosomes A03, A04, A10, C05, and C07, which explained 14.679–17.83% of the phenotypic variation (Table 4, Fig. 5B).
Potential candidate genes
Through GWAS analysis, the genes significantly associated with RRL and RDW were identified in the LD interval of SNP loci. Then according to the gene function predicted with the Arabidopsis Information Resource (Xu et al. 2016), fifty-nine candidate genes related to Al tolerance were ultimately screened out (Table 5).
Through GWAS analysis of RRL, 31 candidate genes related to Al tolerance were the most reliable, distributed on chromosomes A03 (3), A07 (1), A09 (3), A10 (4), C05 (1), C06 (7), and C09 (12), respectively. Among them, two MATE family proteins located on chromosomes A03 and C09 were found, namely BnaA03g51020D and BnaC09g40230D. MATE transporter ABS4 (BnaA09g14730D) located in chromosome A09 was found, which could alleviate Al toxicity by regulating the citric acid synthesis pathway. Besides, BnaA10g20060D (JAZ10) on chromosome A10 and BnaC09g40420D (RGL3) on chromosome C09 were found to participate in jasmonic acid (JA) signal regulation. Also, BnaC09g40420D (RGL3) took part in salicylic acid (SA) signal transduction together with BnaC09g41020D (OPR1) on chromosome C09. A gene called BnaA09g13940D (GA20OX4) located on chromosome A09 was found to encode gibberellin oxidase. A gene called AT3G53280 (CYP71B5) was screened out, which belonged to the cytochrome P450 family. Some genes that participated in carbohydrate metabolism were screened out, including BnaA03g51300D (O-Glycosyl hydrolases family 17 protein) on chromosome A03, BnaA10g19920D (TRA2) on chromosome A10, as well as BnaC09g39650D (UTP), BnaC09g40390D (MEX1), and BnaC09g40960D (CSLD2) on chromosome C09. They mostly involved glucose, galactose and maltose metabolism, cellulose polysaccharide biosynthesis, and starch catabolism. When it came to nitrogen compound metabolism, genes were found in BnaC06g14440D (NODGS) on chromosome C06 and BnaC09g39680D (GDH1) on chromosome C09.
Through GWAS analysis of RDW, 28 candidate genes related to Al tolerance were screened out, distributed on chromosomes A03 (5), A04 (4), A10 (6), C05 (4), and C07 (9), respectively. Among them, BnaC07g27050D on chromosome C07 was found to belong to the MATE family gene. Two genes of BnaA03g15910D (JAZ7) on chromosome A03 and BnaA04g10280D (MKK3) on chromosome A04 involved in JA signal regulation were found. BnaA03g15880D (ATGA2OX3) and BnaA10g23640D (GA20OX3) on chromosome A10 encoded gibberellin oxidase and participated in oxidative stress response. BnaC07g26120D (ERD5) on chromosome C07 took part in oxidation and was also related to proline decomposition. BnaA03g16180D on chromosome A03 belonged to copper amine oxidase family protein and participated in nitrogen metabolism. Besides, 11 genes located on chromosomes A03 (1), A04 (3), A10 (1), C05 (1), and C07 (5) were found and involved in carbon metabolism, all of which involved starch and sucrose biosynthetic metabolism, and glycogen decomposition.
Discussion
SNP loci
The development of SNP chip provides a new channel for exploring genes and loci related to crop stress tolerance. It has been widely used in GWAS analysis, such as cold tolerance of maize (Huang et al. 2013a), salt and drought tolerance of sesame (Li et al. 2018), salt tolerance of rapeseed (He et al. 2017), as well as Al tolerance of rice (Tao et al. 2018) and barley (Cai et al. 2013). Many related SNP loci were successfully detected, and candidate genes were screened in these studies. To investigate the Al tolerance on seed germination, genome-wide association analysis (GWAS) of 19,949 SNPs with genome-wide coverage was used to identify the candidate genes related to germinate traits of rapeseed under Al stress. After doing a GWAS analysis of 169 rapeseed cultivars (lines), eight SNP loci were found to be associated with RRL and five SNP loci were correlated with RDW, respectively. These SNP markers, which were significantly associated with Al tolerance traits, could be selected as markers with higher contribution rate in breeding experiments. They can be used for the improvement of Al tolerance in rapeseed after further validation.
Phenotypic data and Al toxicity
Aluminum toxicity is a considerable hindrance of crop production in acidic soils, which constitute about 50% of the world's potentially arable lands (Kochian et al. 2005). At pH values beneath 5, Al tends to be dissolved as Al3+ ions, which are quite toxic to plant roots and can further limit crop production (Kochian et al. 2015). Germination and root growth tests have been used to evaluate the effect of Al tolerance in crops (Marciano et al. 2010). In this study, the RRL and RDW were used to evaluate the tolerance of 169 rapeseed germplasm resources at the germination stage under Al stress. The results showed that the average value of RRL was 0.727 and that of RDW was 0.964 in 169 rape germplasm resources, respectively, which indicated root length were seriously inhibited than dry weight though both of them were decreased under aluminum stress. Generally, Al toxicity inhibits root elongation, further leading to roots stunting accompanied by reduced water and nutrients uptake (Kochian et al. 2004), and result in the reduction of dry weight by altering carbohydrate synthesis and metabolism (Yuriko et al. 2007). Recently, the root apex has also recommended to role in Al tolerance, which is related to serious changes in the root system, including cell differentiation in root tips and lateral roots, interfering with several enzymes, increasing cell wall inflexibility, modifying the structure and capacity of plasma membranes, and disrupting signal transduction pathways (Sade et al. 2016; Min et al. 2019). The phenomenon was described shoots might be delayed relative to the root damages, or even be an indirect response to Al toxicity since they are generally a consequence of the root inhibition (Kochian et al. 2005).
Potential candidate genes
As a very complex process, seed germination depends both on the external environmental factors and the inner biochemical mechanisms controlled by a complex network of diverse but functionally interrelated phytohormones (Shu et al. 2016). Different cations [e.g., Al3+, Al(OH)2+] exhibit different degrees of toxicity to root, and the morphology of aluminum depends on the pH degree around rhizosphere (Kihara et al. 2003). Two genes consisting of BnaC05g05800D and BnaC05g05810D were found on chromosome C05 in this study, they could play a role in alleviating Al toxicity by regulating the pH level. Similar result was reported, which identified a mutant, ALR-104, proving that the pH gradient on the root surface can produce tolerance in Arabidopsis thaliana (Degenhardt et al. 1998).
On the other hand, plant hormones play an important role in the defense system of plant stress. Of them, phytohormones including cytokinin, auxin, delay senescence while salicylic acid (SA), abscisic acid (ABA), ethylene (ETH), and jasmonic acid (JA) promote senescence. GA can promote plant growth and development. In this study, three genes encoding gibberellin oxidase were detected, which named BnaA09g13940D (GA20OX4), BnaA10g23640D (GA20OX3) and BnaA03g15880D (GA2OX3), respectively. Amongst these, GA20-oxidase (GA20OX) is involved in the synthesis of GA, which can catalyze GA12 and GA53 to synthesize GA1 and GA4 with activity (Giacomelli et al. 2013), and GA20OX deficiency reduces the amount of synthesis of bioactive GA. While GA2-oxidase (GA2OX) is involved in the catabolism of GA, which can inactivate the biologically active GAs, its precursor as well as other intermediates, and maintain the balance between the biologically active GAs and intermediates in plants (Wuddineh et al. 2015). Besides, the plant hormone auxin (IAA) regulated many aspects of plant growth and development, including stem elongation, lateral branching of roots and shoots, establishment of embryonic polarity (Chapman and Estelle 2009). These processes are controlled by auxin-mediated changes in cell division, expansion, and differentiation (Nemhauser et al. 2006). We detected BnaC09g39410D (SAUR21) and BnaC09g39770D (SAUR20), which associated with cell expansion of plants. Likewise, Arabidopsis seedlings, which expressed an artificial microRNA targeting multiple members of the SAUR19–24 subfamily, exhibited short hypocotyls and reduced leaf size, and led to root waving, increased hypocotyl elongation, larger leaf size, reduced phototropism, and impaired apical hook maintenance (Spartz et al. 2012).
During defense, phytohormone-mediated signaling [e.g., abscisic acid (ABA), ethylene (ETH), jasmonic acid (JA), salicylic acid (SA)] is critically important for increasing resistance to stress. BnaA04g10280D (MKK3) and BnaC09g40420D (RGL3) were identified to activate ABA and ETH signaling pathway, positively regulate the synthesis of ABA and ETH, and alleviate Al toxicity damage through the transduction pathway of JA and SA, as well as negatively regulating the signal of GA and seed germination. In Arabidopsis thaliana, MKK3 is involved in defense against pathogens, drought tolerance, JA signal transduction, and ABA and auxin responses (Li et al. 2017; Danquah et al. 2015; Enders et al. 2017). Similarly, it was suggested that the over-expression of OsMKK3 (oe-MKK3) increased levels of jasmonic acid (JA), and abscisic acid (ABA), and decreased SA levels in rice after biotic stress (Zhou et al. 2019). Our results and these studies confirmed that MKK3 played a pivotal role in the signaling pathway and defense responses under abiotic and biotic stresses. It is proposed that MKK3 mediated positive regulation of rapeseed resistance to aluminum toxicity by inducing phytohormone dynamics. Furthermore, BnaA03g15910D and BnaA10g20060D belonged to ZIM domain proteins of JA and participated in the plant endogenous growth regulation. Previous studies had shown that JAZ proteins inhibit JA response, and likely to induce systemic resistance by regulating and affecting the signal transduction pathway of JA (Melotto 2008).
Plants have evolved different kinds of strategies to deal with Al toxicity in acid soils. In summary, two major elementary sorts of Al resistance mechanisms are characterized: Al exclusion mechanisms, which means to prevent Al particles from entering the roots, and Al tolerance mechanisms, which prefer to detoxify internal Al in the symplast (Sade et al. 2016). One of the most well-documented mechanisms is secretion of Al-induced root organic acid anions, malate, citrate, and oxalate, to chelate Al apoplastically (Lou et al. 2020; Ryan 2001; Yang et al. 2019), thus protecting cell wall from Al binding. Multi drug and toxin compounds extrusion (MATE) and aluminium-activated malate transporter (ALMT) are two transporter families responsible for Al-activated malate and citrate secretion, respectively (Kochian et al. 2015). In this study, the MATE efflux family proteins were identified on chromosomes A03, C07, and C09, which were BnaA03g51020D, BnaC07g27050D, and BnaC09g40230D, respectively. Some studies were showing that the MATE family genes can promote citrate excretion into the rhizosphere to protect roots from Al toxicity (Magalhaes et al. 2007; Ryan et al. 2009). Meanwhile, there was another gene found called BnaA09g14730D (ABS4) belonging to MATE transporter. Wang et al. (2015) reported that ABS4 encodes a second putative MATE family transporter, and Arabidopsis MATE family of transporter genes including ABS4 can regulate cell elongation, further supporting the notion of a close functional relationship between the plant endomembrane system and cell elongation. In addition, ABS4 was able to transport citric acid, and participate in some physiological processes (Liu et al. 2009), and contributed to multidrug resistance and play major determinants of aluminum (Al) tolerance in plants (Min et al. 2019). However, there were no gene mutations related to malic acid transport in this study though the first Al tolerance gene about organic acids found in previous studies was malic acid transporter 1 activated by Al (ALMT1) (Sasaki et al. 2004). It is inferred that there might be two reasons for this: (1) The GWAS results are related to the effect value of traits caused by gene expression. It is difficult to detect them in GWAS analysis if the genes express little in RRL and RDW at the germination stage. (2) The ideal test for GWAS is a single factor test, which means that the interference of other factors within the study could lead to false-negative results. Moreover, two ABC transporters including ABCG37/PDR9 and ABCG38/PDR10 were identified, which belonged to the gene family encoding pleiotropic drug resistance (Fourcroy et al. 2016; Smart 2002). Some ABC transporters were associated with Al resistance (Xu et al. 2019). Among them, ABCG37/PDR9 contributed primarily highly oxygenated coumarins to root exudation in Arabidopsis (Strader and Bartel 2009), while ABCG38/PDR10 was a plasma membrane protein that plays a role in maintaining the proper distribution and function of a subset of other membrane proteins (Rockwell et al. 2009).
Apart from the genes above mentioned, we also found some genes related to growth and antioxidant stress, including GDH, RBOHD, P4H5, NODGS, CYP71B5, OPR1, PDR9 and PDR10, and so on. Specifically, Glutamate dehydrogenase (GDH) is an important enzyme in nitrogen (N) metabolism. And it served as a link between C and N metabolism, in its role of assimilating ammonia into glutamine or deaminating glutamate into 2-oxoglutarate and ammonia (Magadlela et al. 2019). Furthermore, P4H5 and NODGS were related to root growth. Previous study certified that P4H5 was pivotal for root hair tip growth (Velasquez et al. 2015) and downregulation of NODGS resulted in plants with a short main root, reduced meristematic activity and disrupted development of the root cap (Doskocilova et al. 2011). In addition, a cytochrome P450 family gene CYP71B5 on chromosome C06, which function was involved in oxidation reduction and alleviate oxidative damage via monooxygenase activity, iron ion binding, oxygen binding (Murgia et al. 2011). And its catalytic metabolic pathways can produce some important secondary metabolites, which strengthen plants resistant to pests, diseases, and other stresses (Sappl et al. 2009). So, it can be inferred that the gene is involved in the biological detoxification after aluminum stress. Also, RBOHD is responsible for the ROS burst after pathogen-associated molecular patterns (PAMPs) perception (Lee et al. 2020), through the phosphorylation and ubiquitination of RBOHD, the species of reactive oxygen species in plant immune process are regulated. Multiple evidences suggested that most ROS signaling during abiotic and biotic stress relies on the activities of two partially redundant isoforms, RBOHD and RBOHF (Miller et al. 2009; Kwak et al. 2003), increased of RBOHD mRNA levels resulted in accelerated leaf senescence. BnaC09g41020D (OPR1) could play important roles in plant defense under Al toxicity. Analogously, a number of works have shown a close relationship between OPR1 genes and some physiological processes. Previous study showed that an OPR1 isolated from Triticum aestivum confers salinity tolerance by means of scavenging oxygen species and enhancing ABA signaling (Dong and Wang 2013). A similar effect was found for OsOPR1 in response to JA, SA, and ethylene in rice (Agrawal et al. 2003). Although, proline, an oxygen-free radical scavenger, is accepted as an indicator of various stresses, but it is not involved in protecting plants against metal toxicity (Yilmaz et al. 2012). In this study it is difficult to point the exact role of proline under the Al toxicity, as in the work realized in pigeonpea (Bhamburdekar and Chavan 2011) and Lactuca sativa L. (Silva and Matos 2016).
Taken together, Aluminum (Al) is a major metal component in soils and is solubilized as phytotoxic ions (predominantly Al3+) which inhibit plant growth under low-pH conditions. Although rapeseed can produce certain defense strategies include pH regulation, metabolic adjustments, signaling cascades, and expression of different genes related to aluminum resistance, and reduce the harm of aluminum toxicity to seed germination, but the phenotypic data analysis of RRL and RDW showed that seeds germination and tender seedling growth were still inhibited. Generally, it is recognized alterations in the germination process as well as in the growth of the root, stems and leaves systems. Also, it is verified alterations in the biochemical processes, like inhibition of enzyme activity, and leading to nutrient deficiency symptoms (Kochian et al. 2005; Inostroza-Blancheteau et al. 2011). The screening of candidate genes laid a foundation for the functional identification of specific candidate genes in the genetic engineering of aluminum tolerance in rape.
Conclusion
The GWAS analysis of Al toxicity tolerance in rapeseed was conducted using RRL and RDW. The results showed that 13 SNP loci were significantly associated with these two traits. Among them, 8 SNP loci were significantly associated with RRL and located on chromosomes A03, A07, A09, A10, C05, C06, and C09, respectively. Five SNP loci were significantly associated with RDW and located on chromosomes A03, A04, A10, C05, and C07, respectively. Subsequently, fifty-nine function-known candidate genes related to Al tolerance were identified in the LD interval of these SNP loci. Four of these genes were involved in the growth regulation of organic acid, ten were involved in growth-regulating substance, eleven were related to oxidative stress, and nineteen were involved in carbon and nitrogen metabolism. All the genes could be potentially related to the Al tolerance of rapeseed. Whether they can influence Al tolerance needs to be verified by molecular assisted breeding or transgenic technology, to introduce these functional genes into rapeseed. This study laid out a foundation for further functional verification of genes and cultivation of new Al-tolerant varieties.
References
Agrawal GK, Jwa NS, Shibato J, Han O, Iwahashi H, Rakwal R (2003) Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochem Biophys Res Commun 310(4):1073–1082. https://doi.org/10.1016/j.bbrc.2003.09.123. PMID: 14559225
Angeles-Núñez JG, Tiessen A (2010) Arabidopsis sucrose synthase 2 and 3 modulate metabolic homeostasis and direct carbon towards starch synthesis in developing seeds. Planta 232(3):701–718. https://doi.org/10.1007/s00425-010-1207-9
Anne P, Amiguet-Vercher A, Brandt B, Kalmbach L, Geldner N, Hothorn M et al (2018) CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development. https://doi.org/10.1242/dev.162354
Ascencio-Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R et al (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148(1):436–454. https://doi.org/10.1104/pp.108.121038
Atwell S, Huang YS, Vilhjálmsson Bjarni J, Willems G, Horton M, Li Y et al (2010) Genome-wide association study of 107 phenotypes in a common set of Arabidopsis thaliana inbred lines. Nature 465:627–631. https://doi.org/10.1038/nature08800. PMID: 20336072
Bai FW, Matton DP (2018) The Arabidopsis mitogen-activated protein kinase kinase kinase 20 (MKKK20) C-terminal domain interacts with MKK3 and harbors a typical DEF mammalian MAP kinase docking site. Plant Signal Behav 13(8):e1503498. https://doi.org/10.1080/15592324.2018.1503498
Bhamburdekar SB, Chavan PD (2011) Effect of some stresses on free proline content during pigeonpea (Cajanas cajan) seed germination. J Stress Physiol Biochem 7(3):235–241. https://doi.org/10.1007/s00425-011-1419-7
Blanco F, Garreton V, Frey N, Dominguez C, Perez-Acle T, Van der Straeten D et al (2005) Identification of npr1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol Biol 59(6):927–944. https://doi.org/10.1007/s11103-005-2227-x
Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD et al (2006) Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol 140:1151–1168. https://doi.org/10.1104/pp.105.074708
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Buckler ES (2007) TASSEL, software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308. PMID: 17586829
Buschmann H, Chan J, Sanchez-Pulido L, Andrade-Navarro MA, Doonan JH, Lloyd CW (2006) Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr Biol 16(19):1938–1943. https://doi.org/10.1016/j.cub.2006.08.028. PMID: 17027491
Cai SG, Wu DZ, Jabeen Z, Huang YQ, Huang YC, Zhang GP (2013) Genome-wide association analysis of aluminum tolerance in cultivated and tibetan wild barley. PLoS ONE 8:e69776. https://doi.org/10.1371/journal.pone.0069776PMCID:PMC3724880
Castilhos G, Júlia GF, Schneider ADB, Oliveira PHD, Nicoloso FT, Schetinger MRC et al (2011) Aluminum-stress response in oat genotypes with monogenic tolerance. Environ Exp Bot 74:114–121. https://doi.org/10.1016/j.envexpbot.2011.05.007
Cellier F, Conejero G, Ricaud L, Luu DT, Lepetit M, Gosti F et al (2004) Characterization of atCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J 39(6):834–846. https://doi.org/10.1111/j.1365-313X.2004.02177.x
Cha JY, Barman DN, Kim MG, Kim WY (2015) Stress defense mechanisms of NADPH-dependent thioredoxin reductases (NTRs) in plants. Plant Signal Behav 10(5):e1017698. https://doi.org/10.1080/15592324.2015.1017698
Chan A, Carianopol C, Tsai AYL, Varatharajah K, Chiu RS, Gazzarrini S (2017) Corrigendum: SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J Exp Bot 68:5981–5981. https://doi.org/10.1093/jxb/erx379
Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43(1):265–285. https://doi.org/10.1146/annurev-genet-102108-134148. PMID: 19686081
Crouzet J, Trombik T, Fraysse AS, Boutry M (2006) Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett 580(4):1123–1130. https://doi.org/10.1016/j.febslet.2005.12.043
Danquah A, de Zelicourt A, Boudsocq M, Neubauer J, Frei Dit Frey N, Leonhardt N et al (2015) Identifification and characterization of an ABA-activated map kinase cascade in Arabidopsis thaliana. Plant J Cell Mol Biol 82(2):232–244. https://doi.org/10.1111/tpj.12808. PMID: 25720833
De Abreu-Neto JB, Turchetto-Zolet AC, De Oliveira LFV, Bodanese Zanettini MH, Margis-Pinheiro M (2013) Heavy metal associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive of plants. FEBS J 280(7):1604–1616. https://doi.org/10.1111/febs.12159
Degenhardt J, Larsen PB, Howell SH, Kochian LV (1998) Aluminum resistance in the Arabidopsis mutantalr-104 is caused by an aluminum-induced increase in rhizosphere pH. Plant Physiol 117:19–27. https://doi.org/10.1104/pp.117.1.19. PMID: 9576770
Ditt RF, Kerr KF, Figueiredo PD, Delrow J, Comai L, Nester EW (2006) The Arabidopsis thaliana transcriptome in response to agrobacterium tumefaciens. Mol Plant Microbe Interact 19(6):665–681. https://doi.org/10.1094/MPMI-19-0665
Dixon DP, Skipsey M, Grundy NM, Edwards R (2005) Stress-induced protein s-glutathionylation in Arabidopsis. Plant Physiol 138(4):2233–2244. https://doi.org/10.1104/pp.104.058917
Doncheva S, Amenós M, Poschenrieder C, Barceló J (2005) Root cell patterning, a primary target for aluminum toxicity in maize. J Exp Bot 56:1213–1220. https://doi.org/10.1093/jxb/eri115. PMID: 15737983
Dong W, Wang M (2013) Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol 161:1217–1228
Doskocilova A, Plihal O, Volc J, Chumova J, Kourova H, Halada P et al (2011) A nodulin/glutamine synthetase-like fusion protein is implicated in the regulation of root morphogenesis and in signalling triggered by flagellin. Planta 234(3):459–476. https://doi.org/10.1007/s00425-011-1419-7. PMID: 21533644
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER, a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Enders TA, Frick EM, Strader LC (2017) An Arabidopsis kinase cascade inflfluences auxin-responsive cell expansion. Plant J 92:68–81. https://doi.org/10.1111/tpj.13635. PMID: 28710770
Estornell LH, Landberg K, Cierlik I, Sundberg E (2018) SHI/STY genes affect Pre- and Post-meiotic anther processes in auxin sensing domains in Arabidopsis. Front Plant Sci 9:150. https://doi.org/10.3389/fpls.2018.00150
Exley C (2009) Darwin, natural selection and the biological essentiality of aluminum and silicon. Trends Biochem Sci 34:589–593. https://doi.org/10.1016/j.tibs.2009.07.006. PMID: 19773172
Fourcroy P, Tissot N, Gaymard F, Briat JF, Dubos C (2016) Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe2+ transport. Mol Plant 9:485–488. https://doi.org/10.1016/j.molp.2015.09.010. PMID: 26415695
Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K et al (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48:1081–1091. https://doi.org/10.1093/pcp/pcm091
Gangl R, Tenhaken R (2016) Raffinose family oligosaccharides act as galactose storesin seeds and are required for rapid germination of Arabidopsisin the dark. Front Plant Sci 7:1115. https://doi.org/10.3389/fpls.2016.01115
Gao HH, Ye S, Wang Q, Wang LY, Wang RL, Chen LY, Tang ZL, Li JN, Zhou QY, Cui C (2019) Screening and comprehensive evaluation of aluminum-toxicity tolerance during seed germination in Brassca napus. Acta Agron Sin 45:1416–1430 (in Chinese with English abstract)
Giacomelli L, Rota-Stabelli O, Masuero D, Acheampong AK, Moretto M, Caputi L, Vrhovsek U, Moser C (2013) Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set:functional characterization and evolution of grapevine gibberellin oxidases. J Exp Bot 64(14):4403–4419. https://doi.org/10.1093/jxb/ert251
Gourley LM, Rogers SA, Ruiz-Gomez C, Clark RB (1990) Genetic aspects of aluminum tolerance in sorghum. Plant Soil 123:211–216. https://doi.org/10.1007/BF00011270
Gupta PK, Kulwal PL, Jaiswal V (2014) Association mapping in crop plants: opportunities and challenges. Adv Genet 85:109–148. https://doi.org/10.1016/B978-0-12-800271-1.00002-0. PMID: 24880734
Hamel F, Breton C, Houde M (1998) Isolation and characterization of wheat aluminum-regulated genes: possible involvement of aluminum as a pathogenesis response elicitor. Planta 205:531–538. https://doi.org/10.1007/s004250050352. PMID: 9684357
Hanada K, Sawada Y, Kuromori T, Klausnitzer R, Saito K, Toyoda T et al (2011) Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol Biol Evol 28(1):377–382. https://doi.org/10.1093/molbev/msq204
Hatzig SV, Frisch M, Breuer F, Nesi N, Ducournau S, Wagner MH et al (2015) Genome-wide association mapping unravels the genetic control of seed germination and vigor in Brassica napus. Front Plant Sci 6:221. https://doi.org/10.3389/fpls.2015.00221. PMID: 25914704
He YJ, Wu DM, Wei DY, Fu Y, Cui YX, Dong HL et al (2017) Gwas, qtl mapping, and gene expression analyses in Brassica napus reveal genetic control of branching morphogenesis. Sci Rep UK 7:15971. https://doi.org/10.1038/s41598-017-15976-4. PMID: 29162897
Huang J, Zhang JH, Li WZ, Hu W, Duan LC, Feng Y et al (2013a) Genome-wide association analysis of ten chilling tolerance indices at the germination and seedling stages in maize. J Integr Plant Biol 55:735–744. https://doi.org/10.1111/jipb.12051. PMID: 23551400
Huang SM, Deng LB, Guan M, Li JN, Lu K, Wang HZ et al (2013b) Identification of genome-wide single nucleotide polymorphisms in allopolyploid crop Brassica napus. BMC Genom 14:717. https://doi.org/10.1186/1471-2164-14-717. PMID: 24138473
Inostroza-Blancheteau C, Aquea F, Reyes-Díaz M, Alberdi M, Arce-Johnson P (2011) Identification of aluminum-regulated genes by cDNA-AFLP analysis of roots in two contrasting genotypes of highbush blueberry (Vaccinium corymbosum L.). Mol Biotechnol 49(1):32–41. https://doi.org/10.1007/s12033-010-9373-3. PMID: 21225377
Jain M, Nagar P, Sharma A, Batth R, Aggarwal S, Kumari S et al (2018) GLYI and D-LDH play key role in methylglyoxal detoxification and abiotic stress tolerance. Sci Rep 8:5451. https://doi.org/10.1038/s41598-018-23806-4
Jiang L, Chen ZP, Gao QC, Ci LK, Cao SQ, Han Y et al (2016) Loss-of-function mutations in the APX1 gene result in enhanced selenium tolerance in Arabidopsis thaliana. Plant Cell Environ 39(10):2133–2144. https://doi.org/10.1111/pce.12762
Jost R, Altschmied L, Bloem E, Bogs J, Gershenzon J, Hahnel U et al (2005) Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth Res 86(3):491–508. https://doi.org/10.1007/s11120-005-7386-8
Kaya H, Takeda S, Kobayashi MJ, Kimura S, Iizuka A, Imai A et al (2019) Comparative analyses of the reactive oxygen species-producing enzymatic activity of Arabidopsis NADPH oxidases. Plant J 98(2):291–300. https://doi.org/10.1111/tpj.14212
Khare D, Mitsuda N, Lee S, Song WY, Hwang D, Ohme-Takagi M et al (2017) Root avoidance of toxic metals requires the GeBP-LIKE 4 transcription factor in Arabidopsis thaliana. New Phytol 213(3):1257–1273. https://doi.org/10.1111/nph.14242
Kihara T, Ohno T, Koyama H, Sawafuji T, Hara T (2003) Characterization of NADP-isocitrate dehydrogenase expression in a carrot mutant cell line with enhanced citrate excretion. Plant Soil 248:145–153. https://doi.org/10.1023/a:1022383426356
Kim J, Shiu SH, Thoma S, Li WH, Patterson SE (2006) Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol 7(9):87. https://doi.org/10.1186/gb-2006-7-9r87
Kim J, Sharkhuu A, Jin JB, Li PH, Jeong JC, Baek D et al (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145(3):722–735. https://doi.org/10.1104/pp.107.104935
Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260. https://doi.org/10.1146/annurev.pp.46.060195.001321
Kochian LV, Hoekenga OA, Piñeros MA (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Physiol Plant Mol Biol 55:459–493. https://doi.org/10.1146/annurev.arplant.55.031903.141655
Kochian LV, Piñeros MA, Hoekenga OA (2005) The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274:175–195. https://doi.org/10.1007/1-4020-4099-7_9
Kochian LV, Piñeros MA, Liu J, Magalhaes JV (2015) Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol 66:57–598. https://doi.org/10.1146/annurev-arplant-043014-114822. PMID: 25621514
Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL et al (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22(2623–2633):701. PMID: 12773379
Lee DH, Lal NK, Lin ZJD, Ma SS, Liu J, Castro B et al (2020) Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat Commun 11(1):1838. https://doi.org/10.1038/s41467-020-15601-5. PMID: 32296066
Li M, Qin R, Jiang W, Liu D (2015) Cytogenetical effects of aluminum on root meristem cells of Helianthus annuus L. Bot Sci 93:1–8. https://doi.org/10.17129/botsci.230
Li F, Chen BY, Xu K, Gao GZ, Yan GX, Qiao JW et al (2016) A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci 242:169–177. https://doi.org/10.1016/j.plantsci.2015.05.012. PMID: 26566834
Li Y, Cai H, Liu P, Wang C, Gao H, Wu C et al (2017) Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem Biophys Res Commun 484:292–297. https://doi.org/10.1016/j.bbrc.2017.01.104. PMID: 28131829
Li DH, Dossa K, Zhang YX, Wei X, Wang LH, Zhang YJ et al (2018) GWAS uncovers differential genetic bases for drought and salt tolerances in sesame at the germination stage. Genes 9:87. https://doi.org/10.3390/genes9020087. PMID: 29443881
Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD et al (2010) Acyl-lipid metabolism. Arabidopsis Book 8:e0133. https://doi.org/10.1199/tab.0133
Liu S, Wang J, Wang L, Wang X, Xue Y, Wu P et al (2009) Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res 19:1110–1119. https://doi.org/10.1038/cr.2009.70. PMID: 19546891
Liu S, Fan CC, Li JN, Cai GQ, Yang QY, Wu J et al (2016) A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor Appl Genet 129:1203–1215. https://doi.org/10.1007/s00122-016-2697-z. PMID: 26912143
Lou HQ, Fan W, Jin JF, Xu JM, Chen WW, Yang JL et al (2020) A NAC-type transcription factor confers aluminum resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ 43:463–478. https://doi.org/10.1111/pce.13676. PMID: 31713247
Lu K, Wang TY, Xu XF, Tang ZL, Qu CM, He B et al (2016) Genome-wide association analysis of the height of podding and thickness of pod canopy in Brassica napus. Acta Agron Sin 42:344–352 (in Chinese with English abstract)
Luo Q, Zhao Z, Li DK, Zhang Y, Yang Y (2016) Overexpression of NaKR3 enhances salt tolerance in Arabidopsis. Genet Mol Res 15(1):15016378. https://doi.org/10.4238/gmr.15016378
Magadlela A, Morcillo RJL, Kleinert A, Venter M, Steenkamp E, Valentine A (2019) Glutamate dehydrogenase is essential in the acclimation of Virgilia divaricata, a legume indigenous to the nutrient-poor Mediterranean-type ecosystems of the Cape Fynbos. J Plant Physiol 43:153053. https://doi.org/10.1016/j.jplph.2019.153053
Magalhaes JV, Liu J, Guimarães CT, Lana UGP, Alves VMC, Wang YH et al (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in Sorghum. Nat Genet 39:1156–1161. https://doi.org/10.1038/ng2074. PMID: 17721535
Malinova I, Mahlow S, Alseekh S, Orawetz T, Fernie AR, Baumann O et al (2014) Double knockout mutants of Arabidopsis grown under normal conditions reveal that the plastidial phosphorylase isozyme participates in transitory starch metabolism. Plant Physiol 164(2):907–921. https://doi.org/10.1104/pp.113.227843
Marchi L, Polverini E, Degola F, Baruffini E, Restivo FM (2014) Glutamate dehydrogenase isoenzyme 3 (GDH3) of Arabidopsis thaliana is less thermostable than GDH1 and GDH2 isoenzymes. Plant Physiol Biochem 83:225–231. https://doi.org/10.1016/j.plaphy.2014.08.003. PMID: 25180813
Marciano DPRO, Ramos FT, Alvim MN, Magalhaes JR, Franca MGC (2010) Nitric oxide reduces the stress effects of aluminum on the process of germination and early root growth of rice. J Plant Nutr Soil Sci 173:885–891. https://doi.org/10.1002/jpln.200900312
Maser P, Thomine S, Schroeder J, Ward JM, Hirschi K, Sze H et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126(4):1646–1667. https://doi.org/10.1104/pp.126.4.1646
Mattiello L, Da SF, Menossi M (2012) Linking microarray data to QTLs highlights new genes related to al tolerance in maize. Plant Sci 191–192:8–15. https://doi.org/10.1016/j.plantsci.2012.04.009. PMID: 22682560
McCoy JG, Arabshahi A, Bitto E, Bingman CA, Ruzicka FJ, Frey PA et al (2006) Structure and mechanism of an ADP-glucose phosphorylase from Arabidopsis thaliana. Biochemistry 45(10):3154–3162. https://doi.org/10.1021/bi052232m. PMID: 16519510
Melotto M (2008) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 55:979–988. https://doi.org/10.1111/j.1365-313X.2008.03566.x. PMID: 18547396
Meng LJ, Wang BX, Zhao XQ, Ponce K, Qian Q, Ye GY (2017) Association mapping of ferrous, zinc and aluminum tolerance at the seedling stage in Indica rice using MAGIC populations. Front Plant Sci 8:1822. https://doi.org/10.3389/fpls.2017.01822PMID:29123537. PMID: 29123537
Meng L, Zhang T, Geng S, Scott PB, Li H, Chen S (2019) Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. J Proteom 196:81–91. https://doi.org/10.1016/j.jprot.2019.02.001
Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V et al (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal. https://doi.org/10.1126/scisignal.2000448. PMID: 19690331
Min X, Jin X, Liu W, Wei X, Zhang Z, Ndayambaza B, Wang Y (2019) Transcriptome-wide characterization and functional analysis of MATE transporters in response to aluminum toxicity in Medicago sativa L. Peer J 7:e6302. https://doi.org/10.7717/peerj.6302. PMID: 30723620
Murgia I, Tarantino D, Soave C, Morandini P (2011) Arabidopsis CYP82C4 expression is dependent on Fe availability and circadian rhythm and correlates with genes involved in the early Fe deficiency response. J Plant Physiol 168:894–902. https://doi.org/10.1016/j.jplph.2010.11.020. PMID: 21315474
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126:467–475. https://doi.org/10.1016/j.cell.2006.05.050
Nguyen VT, Burow MD, Nguyen HT, Le BT, Le TD, Paterson AH (2001) Molecular mapping of genes conferring aluminum tolerance in rice (oryza sativa L.). Theor Appl Genet 102:1002–1010. https://doi.org/10.1007/s001220000472
Ohyama K, Ogawa M, Matsubayashi Y (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J 55(1):152–160. https://doi.org/10.1111/j.1365-313X.2008.03464.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. PMID:10835412
Qian P, Sun R, Basharat B, Bullet A, Zhou W (2014) Effects of hydrogen sulfide on growth, antioxidative capacity, and ultrastructural changes in oilseed rape seedlings under aluminum toxicity. J Plant Growth Regul 33:526–538. https://doi.org/10.1007/s00344-013-9402-0
Ren YB, Cao JS, Miao M, Meng Y, Fan TT, Cao SQ et al (2018) DFR1-mediated inhibition of proline degradation pathway regulates drought and freezing tolerance in Arabidopsis. Cell Rep 23(13):3960–3974. https://doi.org/10.1016/j.celrep.2018.04.011
Rieu I, Eriksson S, Powers SJ, Gong F, Griffiths J, Woolley L et al (2008a) Genetic analysis reveals that C-19-GA 2-Oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20:2420–2436. https://doi.org/10.1105/tpc.108.058818
Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Powers J, Gong SJ, Linhartova F et al (2008b) The gibberellin biosynthetic genes ATGA20ox1 and ATGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53(3):488–504. https://doi.org/10.1111/j.1365-313X.2007.03356.x
Rockwell NC, Wolfger H, Kuchler K, Thorner J (2009) ABC transporter Pdr10 regulates the membrane microenvironment of Pdr12 in saccharomyces cerevisiae. J Membr Biol 229(1):27–52. https://doi.org/10.1007/s00232-009-9173-5. PMID: 19452121
Ryan PR (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Biol 52:527–560. https://doi.org/10.1146/annurev.arplant.52.1.527. PMID: 11337408
Ryan PR, Delhaize E (2010) The convergent evolution of aluminum resistance in plants exploits a convenient currency. Funct Plant Biol 37:275–284. https://doi.org/10.1071/FP09261
Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E (2009) A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol 149:340–351. https://doi.org/10.1104/pp.108.129155. PMID: 19005085
Sade H, Meriga B, Surapu V, Gadi J, Sunita MS, Suravajhala P, Kavi Kishor PB (2016) Toxicity and tolerance of aluminum in plants: tailoring plants to suit to acid soils. Biol Met 29(2):187–210. https://doi.org/10.1007/s10534-016-9910-z. PMID: 26796895
Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Millar AH et al (2009) The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes result in altered metabolic sensitivity to oxidative stress. Plant J 58:53–68. https://doi.org/10.1111/j.1365-313x.2008.03761.x
Sardar A, Nandi AK, Chattopadhyay D (2017) CBL-interacting protein kinase 6 negatively regulates immune response to pseudomonas syringae in Arabidopsis. J Exp Bot 68(13):3573–3584. https://doi.org/10.1093/jxb/erx170. PMID: 28541442
Sarry JE, Kuhn L, Ducruix C, Lafaye A, Junot C, Hugouvieux V et al (2006) The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 6(7):2180–2198. https://doi.org/10.1002/pmic.200500543
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR et al (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37:645–653. https://doi.org/10.1111/j.1365-313x.2003.01991.x. PMID: 14871306
Shi YY, Gao LL, Wu ZC, Zhang XJ, Wang MM, Zhang CS et al (2017) Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol 17:92. https://doi.org/10.1186/s12870-017-1044-0. PMID: 28558653
Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9:34. https://doi.org/10.1016/j.molp.2015.08.010. PMID: 26343970
Silva P, Matos M (2016) Assessment of the impact of Aluminum on germination, early growth and free proline content in Lactuca sativa L. Ecotoxicol Environ Saf 131:151–156. https://doi.org/10.1016/j.ecoenv.2016.05.014. PMID: 27229755
Smart SVDBC (2002) The plant PDR family of ABC transporters. Planta 216:95–106. https://doi.org/10.1007/s00425-002-0889-z. PMID: 12430018
Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D et al (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70(6):978–990. https://doi.org/10.1111/j.1365-313X.2012.04946.x. PMID: 22348445
Strader LC, Bartel B (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21:1992–2007. https://doi.org/10.1105/tpc.109.065821. PMID: 19648296
Suzuki M, Sato Y, Wu S, Kang BH, Mccarty DR (2015) Conserved functions of the mate transporter big embryo1 in regulation of lateral organ size and initiation rate. Plant Cell 27(8):2288–2300. https://doi.org/10.1105/tpc.15.00290
Taguchi G, Ubukata T, Nozue H, Kobayashi Y, Takahi M, Yamamoto H et al (2010) Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J 63(6):1031–1041. https://doi.org/10.1111/j.1365-313X.2010.04298.x
Tan M, Liao F, Hou L, Wang J, Wei L, Jian H et al (2017) Genome-wide association analysis of seed germination percentage and germination index in Brassica napus L. under salt and drought stresses. Euphytica 213:40. https://doi.org/10.1007/s10681-016-1832-x
Tang QY, Zhang CX (2013) Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci 20:254–260. https://doi.org/10.1111/j.1744-7917.2012.01519.x. PMID: 23955865
Tao YH, Niu YN, Wang Y, Chen TX, Amir NS, Zhang J et al (2018) Genome-wide association mapping of aluminum toxicity tolerance and fine mapping of a candidate gene for nrat1 in rice. PLoS ONE 13:e0198589. https://doi.org/10.1371/journal.pone.0198589. PMID: 29894520
Tehseen M, Cairns N, Sherson S, Cobbett CS (2010) Metallochaperone-like genes in Arabidopsis thaliana. Metallomics Integr Biomet Sci 2(8):556–564. https://doi.org/10.1039/c003484c
Unterseer S, Bauer E, Haberer G, Seidel M, Knaak C, Ouzunova M et al (2014) A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k snp genotyping array. BMC Genom 15:823. https://doi.org/10.1186/1471-2164-15-823. PMID: 25266061
Velasquez SM, Ricardi MM, Poulsen CP, Oikawa A, Dilokpimol A, Halim A (2015) Complex regulation of prolyl-4-hydroxylases impacts root hair expansion. Mol Plant 8(5):734–746. https://doi.org/10.1016/j.molp.2014.11.017. PMID: 25655826
Wang X, Zhang Y, Ma QB, Zhang ZL, Xue Y, Bao SL et al (2007) SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J 26(7):1934–1941. https://doi.org/10.1038/sj.emboj.7601647. PMID: 17363895
Wang R, Liu X, Liang S, Ge Q, Li Y, Shao J et al (2015) A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis. J Exp Bot 66:6327. https://doi.org/10.1093/jxb/erv344. PMID: 26160579
Wang K, Guo Q, Froehlich JE, Hersh HL, Zienkiewicz A, Howe GA et al (2018) Two abscisic acid responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell 30(5):1006–1022. https://doi.org/10.1105/tpc.18.00250
Wei LJ, Jian HJ, Lu K, Filardo F, Yin NW, Liu LZ et al (2016) Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 14:1368–1380. https://doi.org/10.1111/pbi.12501. PMID: 26563848
Wild M, Daviere JM, Cheminant S, Regnault T, Baumberger N, Heintz D et al (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24(8):3307–3319. https://doi.org/10.1105/tpc.112.101428
Wuddineh WA, Mazarei M, Zhang JY, Poovaiah CR, Mann DGJ, Ziebell A et al (2015) Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol J 13(5):636–647. https://doi.org/10.1111/pbi.12287. PMID: 25400275
Xu LP, Hu KN, Zhang ZQ, Guan CY, Chen S, Hua W et al (2016) Genome-wide association study reveal the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res 23:43. https://doi.org/10.1093/dnares/dsv035. PMID: 26659471
Xu JM, Wang ZQ, Jin JF, Chen WW, Fan W, Zheng SJ, Yang JL (2019) FeSTAR2 interacted by FeSTAR1 alters its subcellular location and regulates Al tolerance in buckwheat. Plant Soil 436:489–501
Xue YF, Chen BJ, Wang R, Win AN, Li JN, Chai YR (2018) Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and its parental species. Appl Biochem Biotechnol 184:582. https://doi.org/10.1007/s12010-017-2563-8. PMID: 28799009
Yan CX, Yan ZY, Wang YZ, Yan XY, Han YZ (2014) Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J Exp Bot 65(20):5933–5944. https://doi.org/10.1093/jxb/eru334
Yang SH, Yang HJ, Grisafi P, Sanchatjate S, Fink GR, Sun Q et al (2010) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45(2):166–179. https://doi.org/10.1111/j.1365-313X.2005.02585.x. PMID: 16367962
Yang JL, Fan W, Zheng SJ (2019) Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J Zhejiang Univ ENCE B 20(6):513–527. https://doi.org/10.1631/jzus.B1900188. PMID: 31090277
Yilmaz HM, Yakar M, Mutluoglu O, Kavurmaci MM, Yurt K (2012) Monitoring of soil erosion in cappadocia region(selime-aksaray-turkey). Environ Earth Sci 66(1):75–81. https://doi.org/10.1007/s12665-011-1208-4
Yoo CM, Quan L, Blancaflor EB (2012) Divergence and redundancy in CSLD2 and CSLD3 function during Arabidopsis thaliana root hair and female gametophyte development. Front Plant Sci 3:111. https://doi.org/10.3389/fpls.2012.00111
Yuriko K, Takashi I, Kazuhiko K, Orito Y, Hiroyuki K (2007) Characterisation of lanthanum toxicity for root growth of, Arabidopsis thaliana, from the aspect of natural genetic variation. Funct Plant Biol 34:984–994. https://doi.org/10.1071/FP07133
Zhai LM, Sun CH, Feng Y, Li DY, Chai XF, Wang L et al (2018) AtROP6 is involved in reactive oxygen species signaling in response to iron-deficiency stress in Arabidopsis thaliana. FEBS Lett 592(20):3446–3459. https://doi.org/10.1002/1873-3468.13257. PMID: 30238451
Zheng H, Zhang F, Wang S, Su Y, Ji X, Jiang P et al (2018a) MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsisthaliana. Plant Cell 30:67–82. https://doi.org/10.1105/tpc.17.00830. PMID: 29255112
Zheng X, Chen L, Li X (2018b) Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress. Bot Stud. https://doi.org/10.1186/s40529-018-0238-6
Zhou SX, Chen MT, Zhang YB, Gao Q, Noman A, Wang Q et al (2019) OsMKK3, a stress-responsive protein kinase, positively regulates rice resistance to nilaparvata lugens via phytohormone dynamics. Int J Mol Sci 20(12):3023. https://doi.org/10.3390/ijms20123023 . PMID: 3
Acknowledgements
This work was supported by the National Key Research and Development Program (2018YFD0100500), and the Chongqing Municipal Science and Technology Innovation Project (cstc2019jscx-msxm1538).
Author information
Authors and Affiliations
Contributions
Data curation: HG, SY. Formal analysis: HG. Funding acquisition: CC, QZ. Investigation: HG, SY, LW, RW, WL, JW, LM, FY. Project administration: CC, QZ. Supervision: CC, QZ. Writing—original draft: HG. Writing—review and editing: CC, QZ, JW.
Corresponding authors
Ethics declarations
Conflict of interest
There were no conflict of interest in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gao, H., Ye, S., Wu, J. et al. Genome-wide association analysis of aluminum tolerance related traits in rapeseed (Brassica napus L.) during germination. Genet Resour Crop Evol 68, 335–357 (2021). https://doi.org/10.1007/s10722-020-00989-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-00989-2