Abstract
Soil salinization has become one of the important factors affecting the sustainable development of agriculture. Among them, the soda-alkali soil composed of sodium carbonate (Na2CO3) has a more serious impact on crops. In this study, 175 Brassica napus accessions from different sources were used as materials, combined with Brassica 60 K SNP chip data, and the root length traits of Brassica napus during germination under Na2CO3 stress (0.15%) and control conditions were analyzed for genome-wide association (GWAS). GWAS analysis detected that 5 SNPs were significantly related to root length traits under stress conditions; a single SNP could explain 10.22–12.01% of the phenotypic variation. A total of 15 candidate genes related to Na2CO3 stress resistance were identified upstream and downstream of significant SNPs, including cation exchange protein genes (CAX1), members of the zinc finger protein family (ZFHD1), peroxidase family proteins (POD), and transcription factors (MYB family and WRKY family), etc. The expression analysis of 5 candidate genes in extreme phenotypic materials showed that BnaA04g21850D (CAX1) and BnaA06g24040D (ACX5) were induced by Na2CO3 stress in both materials; BnaA06g31200D (ZFHD1) and BnaC02g37590D (MYB60) were up-regulated expression in sensitive materials; BnaA04g21990D (POD) was up-regulated expression in alkali-tolerant materials, indicating that these candidate genes may be involved in the process of rapeseed response to Na2CO3 stress. This study can provide a reference for in-depth analysis of the molecular mechanism of Na2CO3 stress resistance in rapeseed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sustainable development of world agriculture has been restricted by the growing problem of soil salinization severely. According to incomplete statistics, about 954 million hectares of soil in the world have been suffered varying degrees of salinization (Munns et al. 2008), and about 434 million hectares has composed of sodium carbonate (Na2CO3) and other alkaline salt types (Jin et al. 2008). Alkaline soil is characterized by high pH, high exchangeable sodium, dispersed physical properties and low water content (Vestin et al. 2006). Compared with salt (NaCl) stress, alkaline stress always caused much stronger inhibition of plant growth and development (He et al. 2018). Studies have shown that the degree of damage to plants from salt and alkali stress is in order of salt-alkali mixed stress > alkaline stress > salt stress (Wang 2015; Gong et al. 2016). Alkaline salts such as Na2CO3 in soda saline-alkaline soil can cause plants to suffer from ion poisoning and high pH stress, which seriously affects the normal growth of plants (Wang et al. 2015). At present, a series of advances have been made in the research on the mechanism of plant salt tolerance, and a large number of salt tolerance genetic sites and genes have been identified (Zhao et al. 2020). However, little attention has been paid on the mechanisms of plant adaptation to alkaline stress.
Rapeseed (Brassica napus L.; AACC, 2n = 38) is the third most important oil crop in the world. In recent years, with the rapid development of sequencing technology, the cost of developing single nucleotide polymorphism (SNP) markers has been greatly reduced. GWAS (Genome-wide association analysis) technology has been widely used in the genetic research of complex traits such as agronomy, quality and resistance traits of rapeseed (Li et al. 2014; Liu et al. 2016; Chen et al. 2018; Song et al. 2020; Tang et al. 2021). Among them, several genetic researches on the salt tolerance (NaCl) of rapeseed have been published. For example, 85 rapeseed inbred lines were used to investigate the salt tolerance traits of rapeseed seedlings and 62 QTLs were detected to be correlated in salt tolerance index, biological characteristics and ion balance significantly (Yong et al. 2015). In addition, according to the prediction of candidate genes and gene expression analysis, a possible salt tolerance candidate gene TSN1 was obtained (Yong et al. 2015). Zhang et al. (2017) and He et al. (2017) used GWAS analysis on salt tolerance in the germination period of rapeseed. They identified 45 and 225 SNPs that were significantly related to salt tolerance during germination in the whole genome respectively, finding out 65 salt-tolerant candidate genes near these SNPs. In the early stage of our research, GWAS analysis was carried out on the salt tolerance of Brassica napus during the germination and seedling growth stages, and many SNPs that were significantly related to the salt tolerance at the germination and seedling stages were screened, and a few of them were found to be similar to the past research, which showed that the genetic mechanism of salt tolerance in rapeseed was extremely complicated (Wan et al. 2017, 2018). However, the current theoretical research on the resistance mechanism of Brassica napus to alkaline salt stress is limiting, while the molecular mechanism involved in the resistance of rapeseed to alkaline salt stress is still unclear.
In the present study, we performed GWAS to identify QTLs and candidate genes associated with root length under Na2CO3 stress at the germination stage with a panel of 175 rapeseed inbred lines. This research not only provides technical support for the identification of alkali-tolerant germplasm resources of Brassica napus, but also provides theoretical basis for the utilization of alkali-tolerant rapeseed resources and the cultivation of new varieties.
Materials and methods
Plant materials
A total of 175 Brassica napus accessions from different geographic origins were selected and successfully phenotyped during seed germination and early seedling growth stages under both normal and salt-stress conditions (supplementary Table S1). Five representative plants of each accession of the association panel were self-pollinated in the experimental field in Wuhan (114.35°E, 30.48°N) in 2019/2020 growing seasons.
Selection of an optimal Na2CO3 concentration
3 L of the Na2CO3 solutions (0.00%, 0.05%, 0.10%, 0.15%, 0.20% and 0.25%) were added to a box (36 × 24 × 6 cm) which was separated into 20 sections by nylon rope with two sheets of medical gauze on the top of the tray. Thirty healthy seeds from each Brassica napus accession were selected, surface-sterilized with 0.1% HgCl solution for 3 min, rinsed with distilled water for three times, and equally distributed containing two sheets of medical gauze. The germinated seeds were counted on the seventh day after sowing. The seed germination experiment was performed in a green house (20 °C).
Na2CO3 stress tolerance evaluation
3 L of the 0.15% Na2CO3 solutions (treatment) or double-distilled water (control) were added into a box (36 × 24 × 6 cm) which was separated into 60 sections by nylon rope with two sheets of medical gauze on the top of the tray. Thirty healthy seeds from each Brassica napus accession were selected, and then equally placed into the tray sections. On the eighth day, 5 seedlings from each section were selected and the root length was accurately measured with ImageJ software. The seed germination experiment was three replications in a greenhouse (20 °C).
Phenotypic data analysis
Statistical analysis of all phenotypic data was performed with the software SPSS, version 18.0 (IBM Corp, Armonk, NY, USA). Descriptive statistics was performed using the mean values of all phenotypic data from the traits of 175 rapeseed accessions.
Association mapping
The genotypes of 175 rapeseed accessions were identified using the Brassica 60 K Illumina® Infinium SNP array by Xu et al. (2016). In the early stage, the Brassica 60 K SNP chip was used to perform genotype analysis on 175 materials, and the markers with the minor allele frequency (MAF) less than 0.05 and SNP yield (call frequency) less than 80% were removed, and 19,176 high-quality SNP marker were finally obtained (Wan et al. 2017). By using General Linear Model (GLM) and Mixed Linear Model (MLM) in TASSEL5.0 software (Bradbury et al. 2007), combined with SNP chip genotype data, and Q value and kinship K value as covariates, we performed genome-wide association analysis to determine the interpretation rate (R2) of associated sites and markers for phenotypic variation. This study used the Q model in GLM and the Q + K model in MLM as the most GWAS analysis model. When P < 0.0001 or −log10P ≥ 4.0, SNP markers are considered to be significantly associated with traits (Wan et al. 2017; 2018; Zeng et al. 2021). R software was used to draw Quantile–Quantile scatter plot (QQ plot) and Manhattan plot (Manhattan plot) (Turner et al. 2014). Refering to the annotation information of the rapeseed genome (Chalhoub et al. 2014), candidate genes related to transcription factors, active oxygen scavenging and ion transporters were screened within 500 kb upstream and downstream of significant SNPs as described in previous studies (Wan et al. 2017).
Candidate genes expression analysis
According to the method above, germination stress treatment (Na2CO3 concentration of 0.10%) on the alkali-tolerant material (K) and sensitive material (G) were carried out, and the root tissues of the seedlings under normal and stress treatments were taken after 7 days. The extraction of total RNA was carried out according to the procedures of the Easystep Super Total RNA Extraction Kit. cDNA was synthesized by using RevertAid First Strand cDNA Synthesis Kit. By using SYBR Premix Ex Taq (Baosheng Bioengineering Co., Ltd., China), real-time fluorescence quantification was performed on the Bio-Rad CFX96 fluorescence quantitative PCR instrument (Bio-Rad, USA). BnActin was the internal reference gene, while the real-time fluorescence quantitative mix was Monad's MonAmp SYBR Green qPCR Mix9 (None ROX) (Monad Biotech Co., Ltd., Suzhou). Primer design was performed using Prier Premier 5 (Table 1).
Results and analysis
Different Na2CO3 concentrations of stress on the germination of rapeseed seeds
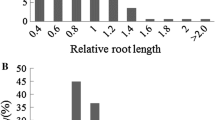
The results showed that when the concentration was lower (0.05%), there was no significant difference in root length under 0.05% Na2CO3 concentration; when the concentration was 0.10%, the root length growth and development were significantly different, while the difference between the materials was not significant; when the concentration reached 0.15%, the root length of rapeseed seedlings was severely inhibited, and the variation between materials was large; when the concentrations were 0.20% and 0.25%, the rapeseed seeds could not germinate (Figs. 1 and 2). Therefore, the Na2CO3 concentration of 0.15% could be used as the appropriate concentration to evaluate the alkali resistance of rapeseed during the germination period.
Phenotypic variation analysis of root length characters in Brassica napus under Na2CO3 stress
The extensive phenotype variation was observed for the root length under normal and Na2CO3 conditions (Tale 2). Comparing the coefficient of variation of root length traits of 175 rapeseed varieties under the control and treatment conditions, Na2CO3 stress had a wide range of phenotypic variation on the degree of inhibition of root growth of rapeseed germplasm with different genotype backgrounds (Table 2). The root length under the control condition showed a continuous normal distribution, and the root length under treatment conditions showed a partial normal distribution, indicating that the root length traits under the control and treatment conditions were controlled by polygenes, which conformed to the genetic characteristics of quantitative traits (Fig. 3).
GWAS results of root length traits
The QQ chart analysis results showed (Fig. 4a) that the root length (CRL) under the control had a larger false positive under the Q model, and the P value of the makers detected under the Q + K model was closer to the expected value, which could reduce false positives and make correlation analysis results more reliable. Root length under Na2CO3 stress (TRL) had the P values of the markers that detected to be close to the expected value under the Q model and the Q + K model. In summary, the Q + K model was selected under CRL, while the Q model and Q + K model were selected under TRL for correlation analysis.
Association mapping of three salt tolerance indices in the panel of 214 rapeseed inbred lines. a Quantile–Quantile plot of root length traits of rapeseed under control and treatment conditions; b GLM-Q model for root length traits of rapeseed under treatment conditions; c MLM-Q + K model for root length traits of rapeseed under treatment conditions; d MLM-Q + K model for root length traits of rapeseed under control conditions. The horizontal line depicts the significance threshold [–log10 P = 4.0]
The correlation analysis between CRL and TRL was performed using the above model (Fig. 4b–d). As a result, 13 SNPs were identified, located on chromosomes A4, A6, C1, C2, C4, C6, C7 and C9, which can explain 10.22–12.01% genetic variation (Table 3). Among them, 8 SNPs were significantly associated with CRL, located on chromosomes A4, C1, C2, C4, C6, C7 and C9 (Table 3). Five SNPs were significantly associated with TRL, located on chromosomes A4, A6 and C2 respectively. Three SNPs were detected by Q model and Q + K model at the same time (Table 3). The same or similar genetic SNPs were not identified under the control and stress conditions, indicating that there are large genetic differences in root length growth under different environments.
Candidate gene screening and expression verification
Brassica napus genes, which were the orthologous to A. thaliana stress tolerance-related genes located within 500 kb of significant SNPs, were regarded as possible candidate genes for Na2CO3 tolerance based on annotation information available in the B. napus reference genome (Chalhoub et al. 2014). Fifteen candidate genes were found within 500 k of the upstream and downstream SNPs significantly associated with TRL (Table 4), such as cation exchange protein gene BnaA04g21850D (CAX1), peroxidase superfamily proteins BnaA04g21990, BnaA04g22000, etc. The expression levels of some candidate genes in alkali-tolerant materials (K) and sensitive materials (G) were verified (Fig. 5). The results showed that BnaA04g21850D (CAX1) and BnaA06g24040D (ACX5) were induced by Na2CO3 stress in both materials; BnaA06g31200D (ZFHD1) and BnaC02g37590D (MYB 60) only up-regulated expression in sensitive materials; BnaA04g21990D (POD) only up-regulated expression in alkali-tolerant materials, indicating that these candidate genes may be involved in the process of rapeseed response to Na2CO3 stress.
Discussion
The root system is the direct organ of the plant to feel the soil environment, and the salt, alkali, acid, heavy metal and other unhealthy environments in the soil can have an adverse effect on the growth (Crawford 1990). Therefore, studying the response of roots to stress and analyzing the genetic rules of root traits are of great significance for plants to adapt to adverse environments. Previous studies have shown that rapeseed can tolerate higher concentrations of NaCl stress in both the germination and seedling stages. For example, the salt tolerance (NaCl) concentration of rapeseed germination was about 1.0%, and the half-lethal concentration was about 1.3% (Wan et al. 2018). The results of this study showed that the alkali salt (Na2CO3) stress had a more obvious inhibitory effect on rapeseed seed germination. The 0.10% Na2CO3 concentration could severely inhibit the germination of rapeseed seedlings while rapeseed cannot germinate under 0.20% Na2CO3 concentration. Comparing the stress of neutral salt (NaCl) and alkaline salt (Na2CO3), it was found that neutral salt had a stronger inhibitory effect on the growth of shoot parts, while alkaline salt had a stronger inhibitory effect on root growth. Compared with neutral salt, the excessive pH caused by alkaline salt would seriously inhibit the normal metabolic activities of the root system, and then affected the growth and development of the rapeseed root system. Therefore, enhancing the tolerance of rapeseed to alkaline environment was the key to improving the overall salt-alkali tolerance of rapeseed.
In recent years, the GWAS were applied to identified abiotic stress-related genetic loci and candidate genes in rapeseed (He et al. 2017; Zhang et al. 2017; Wan et al. 2017, 2018), but the genetic mechanism of rapeseed to adapt to alkaline stress is still unclear. In this study, GWAS was used to successfully identify 5 SNPs that are significantly related to rapeseed root length under Na2CO3 stress. Currently, no previous genetic locus related to Na2CO3 stress resistance has been reported, thus, the genetic locus identified in this study can be considered as a new genetic locus. Interestingly, the SNP Bn-A04-p16303678 (located on A4 chromosome 16,484,408) identified in our study was 134 kb away from the SNP (located on A4 chromosome 16,618,792) related to root length under NaCl stress identified by He et al. (2017), suggesting that they are likely to be the same locus, indicating that rapeseed may have a certain commonality in responding to Na2CO3 stress and NaCl stress. Except for this site, no other locus was overlapped, revealing the difference in salt tolerance and alkali tolerance of rapeseed in the genetic level.
High salt ion concentration and pH are some of the main characteristics of saline-alkali soil (Vestin et al. 2006). Plants growing in saline-alkali soil were subjected to osmotic stress, that seriously affected plant growth and development, and even lead to its death (He et al. 2018; Ma et al. 2020). In order to cope with salt-alkali stress, plants have evolved a variety of mitigation strategies, such as synthesizing osmotic regulators, improving the antioxidant capacity of enzymes and selective absorption of ions (Zhu et al. 1998; Ashraf and Foolad 2005; Zhang et al. 2016). Some resistance genes have also been identified one after another, such as the key gene of proline synthesis P5CS (Funck et al. 2020), the ion transporter family gene OsHKT (Chuamnakthong et al. 2019), transcription factors such as MYBs (He et al. 2018) and so on. In this study, 15 candidate genes were screened near significant SNPs. Among these candidate genes, several transcription factors were screened, including WRKY family (WRKY20 and WRKY33), MYB family (MYB60 and MYB96) and ZFHD1. Studies have shown that these transcription factors are all induced and expressed by adversity stress and participate in the process of regulating the response of plants to environmental stress (Tran et al. 2005). The results of this study showed that both MYB60 and ZFHD1 were only up-regulated in sensitive material, but there was no significant difference in their expression levels in resistant materials. This difference in expression levels may be one of the reasons for the difference in alkali tolerance in rapeseed. In addition, candidate genes also include CAX1, LTP, ACX5, SAD1 and POD. CAX1 can encode a bubbly Ca2+ transporter, which plays an important role in the process of plant roots dealing with salt damage (Navarro-León et al. 2020). The results of qRT-PCR showed that CAX1 was induced by Na2CO3 stress in both resistant and sensitive rapeseeds, indicating that this gene had a certain function in regulating the resistance of rapeseed to Na2CO3 stress. Non-specific lipid transporter (nsLTP) is an important type of small alkaline secreted protein in plants, which plays an important role in plant resistance to adversity and stress (Lindorff-Larsen and Winther 2001). Overexpression of a pepper lipid transfer protein gene CaLTP1 in Arabidopsis can significantly enhance the salt tolerance of the plant (Jung et al. 2005). Under salt stress conditions, the root length of the over-expression line is significantly longer than that of the wild type (Jülke and Ludwig-Müller 2016). It is likely to also participate in the process of regulating the resistance of rapeseed roots to Na2CO3 stress. Jasmonic acids (JAs) are a new type of plant growth regulators that are widely present in higher plants. They play the important role in regulating plant growth and development, photosynthetic characteristics, and stress resistance (Susanne 2002). ACX5 encodes acyl-CoA oxidase, which is involved in the biosynthesis of jasmonate (Schilmiller and Howe 2007). The results of this study showed that ACX5 was up-regulated when resistant rapeseed and sensitive rapeseed were subjected to Na2CO3 stress, indicating that the JA metabolic pathway may play a certain role in the Na2CO3 rapeseed response to stress. SAD1 can encode a polypeptide of Sm-like snRNP protein. Previous studies have shown that compared with wild-type Arabidopsis, SAD1 mutants are more sensitive to ABA and salt stress, and are importantly reflected in the growth and development of roots. This study was based on rapeseed root traits (Xiong et al. 2001; Seifert et al. 2014). Besides, among the candidate genes, there were several peroxidase superfamily protein genes, which were distributed near significant SNPs on A4, A6 and C2 chromosomes. qRT-PCR showed that under Na2CO3 stress, the expression of this gene was upregulated in alkali tolerant rapeseed, but not in sensitive rapeseed. The difference in expression of this gene might be one of the reasons for the difference in alkali tolerance.
Brassica napus has become an ideal material for the development and utilization of saline-alkali land due to its high biological yield, strong salt-tolerant ability, and wide range of suitable planting areas. With the further in-depth study of salt and alkali tolerance in Brassica napus, using conventional hybridization, molecular marker-assisted selection, gene editing and other methods, excellent genes can be transferred into existing high-quality rapeseed varieties or genetic editing technology. The key genes for salt tolerance were improved directionally to create rapeseed materials with stronger salt and alkali tolerance, and new varieties with high yield, high quality and high resistance were cultivated.
Conclusion
GWAS identified a total of 5 genetic locus that are significantly related to root length traits under Na2CO3 stress conditions, and 15 candidate genes were screened near SNP sites, and expression levels of some candidate genes were verified. These candidate genes may be regulating rapeseed. Na2CO3 plays an important role in resistance to stress.
References
Ashraf M, Foolad MR (2005) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59:206–216. https://doi.org/10.1016/j.envexpbot.2005.12.006
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Chalhoub B, Denoeud F, Liu S et al (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953. https://doi.org/10.1126/science.1253435
Chen L, Wan H, Qian J, Guo J, Sun C, Jing W, Yi B, Ma C, Tu J, Song L, Fu T, Shen J (2018) Genome-wide association study of cadmium accumulation at the seedling stage in rapeseed (Brassica napus L.). Front Plant Sci 9:375. https://doi.org/10.3389/fpls.2018.00375
Chuamnakthong S, Mampei M, Ueda A (2019) Characterization of Na+ exclusion mechanism in rice under saline-alkaline stress conditions. Plant Sci 287:110171. https://doi.org/10.1016/j.plantsci.2019.110171
Crawford R (1990) Importance of root to shoot communication in the responses to environmental stress. Phytochemistry 30:3499
Funck D, Baumgarten L, Stift M, Wirén NV, Schnemann L (2020) Differential contribution of P5CS isoforms to stress tolerance in Arabidopsis. Front Plant Sci 11:565134. https://doi.org/10.3389/fpls.2020.565134
Gong B, Wang X, Wei M, Yang F, Li Y, Shi Q (2016) Over-expression of S-adenosylmethionine synthetase 1 enhances tomato callus tolerance to alkali stress through polyamine and hydrogen peroxide cross-linked networks. Plant Cell Tiss Org Cult 124:377–391. https://doi.org/10.1007/s11240-015-0901-5
He Y, Wu D, You J, Qian W (2017) Genome-wide association analysis of salt tolerance related traits in Brassica napus and candidate gene prediction. Sci Agric Sin 50:1189–1201. https://doi.org/10.3864/j.issn.0578-1752.2017.07.002
He Y, Yang X, Xu C, Guo D, Niu L, Wang Y, Li J, Yan F, Wang Q (2018) Overexpression of a novel transcriptional repressor GmMYB3a negatively regulates salt-alkali tolerance and stress-related genes in soybean. Biochem BiophRes Co. https://doi.org/10.1016/j.bbrc.2018.03.026
Jin H, Kim HR, Plaha P, Liu SK, Park JY, Piao YZ, Yang ZJ, Jiang GB, Kwak SS, An G, Son M, Jin YH, Sohn JH, Lim YP (2008) Expression profiling of the genes induced by Na2CO3 and NaCl stresses in leaves and roots of Leymus chinensis. Plant Sci 175:784–792. https://doi.org/10.1016/j.plantsci.2008.07.016
Jülke S, Ludwig-Müller J (2016) Response of Arabidopsis thaliana roots with altered lipid transfer protein (LTP) gene expression to the clubroot disease and salt stress. Plants 5:2. https://doi.org/10.3390/plants5010002
Jung HW, Kim KD, Hwang BK (2005) Identification of pathogen-responsive regions in the promoter of a pepper lipid transfer protein gene (caltpi) and the enhanced resistance of the caltpi transgenic arabidopsis against pathogen and environmental stresses. Planta 221:361–373
Li F, Chen B, Xu K, Wu J, Song W, Ian B, Harper A, Martin T, Liu S, Gao G, Wang N, Yan G, Qiao J, Li J, Li H, Xiao X, Zhang T, Wu X (2014) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res 2014:355–367. https://doi.org/10.1093/dnares/dsu002
Lindorff-Larsen K, Winther JR (2001) Surprisingly high stability of barley lipid transfer protein, LTP1, towards denaturant, heat and proteases. Febs Lett. https://doi.org/10.1016/S0014-5793(00)02424-8
Liu S, Fan C, Li J, Cai G, Yang Q, Wu J, Yi X, Zhang C, Zhou Y (2016) A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor Appl Genet 129:1203–1215. https://doi.org/10.1007/s00122-016-2697-z
Ma H, Meng C, Zhang K, Wang K, Fan H, Li Y (2020) Study on physiological mechanism of using cottonseed meal to improve salt-alkali tolerance of cotton. J Plant Growth Regul 488:145–148. https://doi.org/10.1007/s00344-020-10083-7
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Navarro-León E, López-Moreno FJ, Torre-González ADL, Ruiz JM, Blasco B (2020) Study of salt-stress tolerance and defensive mechanisms in Brassica rapa CAX1a TILLING mutants. Environ Exp Bot 175:104061. https://doi.org/10.1016/j.envexpbot.2020.104061
Schilmiller AL, Howe K (2007) Functional diversification of Acyl-Coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol 143(2):812–824
Seifert GJ, Xue H, Tuba A (2014) The Arabidopsis thaliana FASCICLIN LIKE ARABINOGALACTAN PROTEIN 4 gene acts synergistically with abscisic acid signalling to control root growth. Ann Bot 6:1125–1133. https://doi.org/10.1093/aob/mcu010
Song J, Guan Z, Hu J, Guo C, Yang Z, Wang S, Liu D, Wang B, Lu S, Zhou R, Xie W, Cheng Y, Zhang Y, Liu K, Yang Q, Chen L, Guo L (2020) Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants 6:1–12. https://doi.org/10.1038/s41477-019-0577-7
Susanne B (2002) Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214:497–504. https://doi.org/10.1007/s00425-001-0688-y
Tang S, Zhao H, Lu S, Yu L, Zhang G, Zhang Y, Yang Q, Zhou Y, Wang X, Ma W, Xie W, Guo L (2021) Genome-and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol Plant 14:470–487. https://doi.org/10.1016/j.molp.2020.12.003
Tran LSP, Nakashima K, Sakuma Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K (2005) Isolation and functional analysis of Arabidopsis stress-inducible zinc finger homeodomain transcription factor ZFHD1: role of the ZFHD1 and NAC transcription factors in drought-inducible expression of the early responsive to dehydration stress 1 gene. Plant Cell Physiol. https://doi.org/10.14841/jspp.2005.0.599.0
Turner SD (2014) qqman: an R package for visualizing GWAS results using QQ and manhattan plots. BioRxiv 1:005165. https://doi.org/10.1101/005165
Vestin J, Nambu K, Hees P, Bylund D (2006) The influence of alkaline and non-alkaline parent material on soil chemistry. Geoderma 135:97–106
Wan H, Chen L, Guo J, Li Q, Wen J, Yi B, Ma C, Tu J, Fu T, Shen J (2017) Genome-wide association study reveals the genetic architecture underlying salt tolerance-related traits in rapeseed (Brassica napus L.). Front Plant Sci 8:593. https://doi.org/10.1101/005165
Wan H, Wei Y, Qian J, Gao Y, Guo J, Li Q, Wen J, Yi B, Ma C, Tu J, Fu T, Shen J (2018) Association mapping of salt tolerance traits at germination stage of rapeseed (Brassica napus L.). Euphytica 214:190. https://doi.org/10.1007/s10681-018-2272-6
Wang L, Fang C, Wang K (2015) Physiological responses of to long-term salt, alkali and mixed salt-alkali stresses. J Plant Nutr 38:526–540. https://doi.org/10.1080/01904167.2014.937874
Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1:771–781. https://doi.org/10.1016/S1534-5807(01)00087-9
Xu L, Hu K, Zhang Z, Guan C, Chen S, Wei H, Li J, Wen J, Yi B, Ma C, Tu J, Fu T (2016) Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res 23:43–52. https://doi.org/10.1093/dnares/dsv035
Yong HY, Wang C, Bancroft I, Li F, Wu X, Kitashiba H, Nishio T (2015) Identification of a gene controlling variation in the salt tolerance of rapeseed (Brassica napus L.). Planta 242:313–326. https://doi.org/10.1007/s00425-015-2310-8
Zeng C, Wan H, Wu X, Dai X, Chen J, Ji Q, Qian F (2021) Genome-wide association study of low nitrogen tolerance traits at the seedling stage of rapeseed. Bio Plant 65:10
Zhang M, Smith JAC, Harberd NP, Jiang C (2016) The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol Biol 9:651–659. https://doi.org/10.1007/s11103-016-0488-1
Zhang R et al (2017) Genome-wide association study of root length and hypocotyl length at germination stage under saline conditions in Brassica napus. Sci Agric Sin 50(1):15–27. https://doi.org/10.3864/j.issn.0578-1752.2017.01.002
Zhao C, Zhang H, Song C, Zhu JK, Shabala S (2020) Mechanisms of plant responses and adaptation to soil salinity. Innovation 1:100017. https://doi.org/10.1016/j.xinn.2020.100017
Zhu J, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell 10:1181–1192. https://doi.org/10.2307/3870720
Funding
The study was supported by the China Agriculture Research System (CARS-12) and the Open Fund Project of Engineering Technology Research Center for Protection, Development and Utilization of Characteristic Biological Resources in the Hanjiang River Basin of Hubei Province (06450003).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: HW, CZ, JS and JW. Performed the experiments and analyzed the data: JC, HZ and JT. Data acquisition:JC, HZ, CL, JR, JT, and JL. Wrote the paper: JC. Contributed to writing the manuscript: HZ, CL, JT and HW. Revised the manuscript: HW, CZ and JS.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The experiments shown in the manuscripts submitted for publication comply with the current laws of the country in which they were performed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, J., Zhang, H., Tong, J. et al. Genome-wide association analysis of root length traits in Brassica napus at germination stage under sodium carbonate stress. Euphytica 217, 197 (2021). https://doi.org/10.1007/s10681-021-02928-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-021-02928-3