Abstract
In the Neotropical region, one of the most diverse families of freshwater fishes is the monophyletic Serrasalmidae. Karyotypically, the family shows high diversity in chromosome numbers (2n = 54 to 64). However, little is discussed about whether the chromosomal changes are associated with cladogenetic events within this family. In the present study, we evaluated the role of chromosomal changes in the evolutionary diversification of Serrasalmidae. Our phylogenetic sampling included 36 species and revealed three main clades. The ancestral chromosome number reconstruction revealed the basic number 2n = 54 and a high frequency of ascending dysploid events in the most derived lineages. Our biogeographic reconstruction suggests an Amazonian origin of the family at 48–38 Mya, with independent colonization of other basins between 15 and 8 Mya. We did not find specific chromosomal changes or increased diversification rates correlated with the colonization of a new environment. On the other hand, an increase in the diversification rate was detected involving the genus Serrasalmus and Pygocentrus in the Miocene, correlated with the stasis of 2n = 60. Our data demonstrate that chromosomal rearrangements might have played an important evolutionary role in major cladogenetic events in Serrasalmidae, revealing them as a possible evolutionary driver in their diversification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The study of chromosomal changes in a macroevolutionary framework via Phylogenetic Comparative Methods is gaining substantial interest within the cytogenetics field (Martinez et al. 2015; Lume et al. 2017; Sader et al. 2019). These integrated studies discuss chromosomal polymorphisms in a temporal, spatial and phylogenetic context, clarifying the role of these karyotypic variants in cladogenetic events driving biodiversity (Jacobina et al. 2016; Cioffi et al. 2018; Costa et al. 2020). A central organizing component of genome architecture is the chromosome number (2n), and changes in this trait play a key role in evolutionary processes (Schubert 2007; Freyman and Höhna 2018). Numerical and structural chromosomal alterations may have important evolutionary consequences, affecting recombination rates, increasing reproductive isolation between lineages, and driving diversification between species (Ratomponirina et al. 1988; Yoshida and Kitano 2021). These changes are usually detected in comparative studies, in the search for karyotypic trends among phylogenetically related groups (Jacobina et al. 2016; Sader et al. 2019).
In fish, chromosomal rearrangements such as centric fusion and fission (Robertsonian translocation), and pericentric inversions have been some of the most determinant mechanisms of chromosomal alterations, differentiating both marine and freshwater lineages (Bertollo et al. 2000; Galetti et al. 2006; Jacobina et al. 2013; Sember et al. 2020). Fusions and fissions are readily identified in comparative karyotype studies, as both result in concomitant changes in chromosome morphology and chromosome number (Sember et al. 2020). On the other hand, pericentric inversions change only the chromosomal morphology, without altering chromosome number (Molina 2007; Jacobina et al. 2013). When discussing the significance of these chromosomal alterations to the diversification processes, it is common to attribute their importance to post-mating barriers, progressive isolation from populations, and incipient species (Ayala and Coluzzi 2005). Another trait that decisively impacts diversification rates is the colonization of new environments (Costa et al. 2020). This is mainly due to the morphological differentiation that often follows the arrival of a lineage in a new habitat (Friedman et al. 2020). These macroevolutionary changes are often associated with the absence of predation/competition in the new environment leading to the exploration of new niches and consequently speciation (Liem 1973; Schluter 2000). In this context, biogeographic changes may or may not be associated with chromosomal polymorphisms (Rosa et al. 2014; Costa et al. 2020; Nirchio et al. 2019).
In the Neotropical region, Characiformes fish represent one of the most diverse orders in terms of taxa, with more than 2150 species (Fricke et al. 2022). In addition, they present a high variation in chromosome number and morphology (Bertollo et al. 2000; Nakayama et al. 2012). Within this order, one of the most diverse families is the monophyletic Serrasalmidae, popularly known in Brazil as pacus, piranhas and tambaquis (Cione et al. 2009). This group, which includes about 101 valid species and 16 genera, has a high morphological diversity, with elevated bodies, laterally compressed, with abdominal spines and long dorsal fins (Kolmann, et al. 2021). Based on molecular hypotheses, the family is currently divided into two subfamilies, Colossomatinae and Serrasalminae, with Serrasalminae composed of two tribes: Myleini (comprising most of pacus species) and Serrasalmini (represented by Metynnis, Catoprion, and remaining piranha’s genera) (Mateussi et al. 2020). They inhabit almost all the continental basins of South America (Jégu 2003; Fricke et al. 2022) and a variety of lotic and lentic environments (Goulding 1980), where they perform ecological functions, and support important continental fisheries (Araujo-Lima and Goulding 1997). Ecologically, they are generally divided into two lineages, one composed of herbivores (pacus and tambaquis), and another more derived group, composed of carnivorous piranhas [Pygocentrus and Serrasalmus] (Géry 1977; Goulding 1980; Correa et al. 2007). However, in recent years, studies of diets in these species have reinforced that they are considerably more diverse than previously predicted (Kolmann et al. 2021).
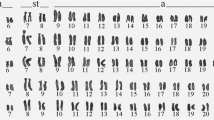
Regarding chromosomal aspects, representatives of this family have shown diversity in the chromosome (2n = 54 to 2n = 64) and fundamental (FN = 108 to FN = 122) numbers (Nirchio et al. 2003; Nakayama et al. 2001). In previous studies, many karyotypes have been described in several species of serrasalmids (Almeida-Toledo et al. 1987; Cestari and Galetti 1992; Nakayama et al. 2000, 2001, 2002, 2008, 2012; Centofante et al. 2002; Nirchio et al. 2003; Gaviria et al. 2005). A recent comparative cytogenetic analysis of Serrasalmidae based on classical and molecular cytogenetic techniques revealed the distribution of heterochromatin predominantly in pericentromeric regions in all species (Favarato et al. 2021). The cytogenetic data, when superimposed on the phylogeny of the family, revealed a tendency to increase the diploid chromosome numbers from 54 to 62 chromosomes, which occurred in a nonlinear manner and is the result of several chromosomal rearrangements (Favarato et al. 2021). However, little is discussed about whether the chromosomal changes detected are associated with the cladogenetic diversification of this family, especially considering their phylogenetic and biogeographic context. Chromosomal information from an evolutionary phylogenetic perspective can shed light on plesiomorphic and apomorphic states in different lineages (Mezzasalma et al. 2016). Thus, comparative phylogenetic methods, integrating phylogenetic and ecological traits, have sought to clarify the evolutionary systematic relationships between fish lineages (Kolman et al. 2021). In addition, they can clarify the evolutionary trends of related groups, and their role in species diversification (Aprea et al. 2013). In the present study, we evaluated the role of chromosomal changes in the evolutionary diversification of the Serrasalmidae family from a phylogenetic and biogeographic perspective. We seek to clarify and discuss the powerful evolutionary forces that boosted its diversity in the Neotropical region. We tested the hypothesis that the diversification rate of a lineage can be increased following the arrival in a new environment (e.g. Costa et al. 2020) and associated with chromosomal rearrangements (e.g. Sader et al. 2019; Costa et al. 2020; Martinez et al. 2015).
Materials and methods
Phylogenetic analyses and divergence time
A total of 36 species from the family Serrasalmidae and two outgroups (Schizodon vittatus and Steindachnerina argentea) were utilized for the phylogenetic analysis (see Supplementary Material 1). We aligned the sequences of the genes cytochrome oxidase subunit I gene COI (580pb), ribosomal DNA 16S (518pb) and 12S (318pb), and the recombination activating protein gene Rag1 (1246 pb) and Rag2 (1031 pb) using ClustalOmega as a plugin implemented in Geneious v.7.1.9 (Kearse et al. 2012; Supplementary Material 1). We then used the SeaView4 software (Gouy et al. 2010) to concatenate the different loci; the final matrix comprised 3,693 bp. A concatenated alignment containing was imported into BEAST v. 1.10.1 (Drummond and Rambaut 2007; Drummond et al. 2012). The Bayesian analysis was conducted considering all partitions simultaneously under the most general substitution model (GTR + G). Uncorrelated relaxed lognormal clock (Drummond and Rambaut 2007) and Birth–Death speciation model (Gernhard 2008) were applied. One run of 50,000,000 generations was performed, sampling every 5000 generations. In order to verify the effective sampling of all parameters and assess the convergence of independent chains, we examined their posterior distributions in TRACER v.1.6. (Rambaut et al. 2014). The MCMC sampling was considered sufficient at effective sampling sizes higher than 200. After removing 25% of samples as burn-in, the independent runs were combined and a maximum clade credibility (MCC) tree was constructed using TreeAnnotator v.1.8.2. (Drummond et al. 2012). For divergence time estimates, we used four calibration points, employing a standard deviation of 10% of the node age. Three secondary calibrations were done, one according to Betancur et al. (2015), which presented a phylogeny of 1407 ray-finned fish, dated with over 200 fossil records. For the other calibrations, we used fossils, one of pacu teeth described by DeCelles and Horton (2003) (minimum age = 38.0 Ma/offset, mean = 6.75). Another calibration points for Mylopus in the Miocene (minimum age/offset = 11.2 Ma, mean = 9.0), (Roberts 1975; Dahdul 2004). And finally, another with Megapiranha paranensis (Cione et al. 2009), to calibrate all piranha genera (minimum age = 6.8 Ma/offset, mean = 10.4).
Chromosome number reconstruction
A priori chromosomal information was obtained from the Arai checklist book (2011) and was later updated until 2022 (for more details, see supplementary material 2). The MCC tree obtained in the BEAST analysis was used for the reconstruction of the haploid chromosome number with the ChromEvol software (Glick and Mayrose 2014). This software uses a Maximum Likelihood Estimate (MLE) approach to infer ancestral chromosome numbers along a phylogeny. This is done under customizable models that use different weights for events of polyploidy (whole genome duplication), demiploidy (1.5 × genome increase) and disploidy (gain or loss of a chromosome). The analysis ran for 10,000 simulations under all eight pre-existent chromosome evolution models available on the software. The Akaike information criterion was used to assess the best-fitted model (Mayrose et al. 2010).
Ancestral area reconstruction
To investigate the historic biogeography of Serrasalmidae, we employed a model-based likelihood approach implemented in the R package BioGeoBEARS (Matzke 2013). First, the MCC tree yielded by BEAST was pruned to exclude the outgroup using the function “drop.tip” implemented in the R package phytools (Revell 2012). Our samples were drawn from the following three ecoregions: (A) Amazon and Orinoco basins, (B) São Francisco basin and Atlantic Coastal drainages and (C) Paraguay, Parana, and Uruguay basins. These areas were chosen based on the distribution of Serrasalmidae species in river basins according to Fishbase information (Froese and Pauly 2022). The distribution data was retrieved from the Global Biodiversity Facility (GBIF) database. We remove duplicates and records with obvious georeferenced errors. This procedure excluded occurrences in the ocean or outside the Neotropical region, those without country names, coordinates with zero latitude or longitude, and coordinates annotated on the coarse-scale grid without decimal precision (Supplementary Material 2).
We used the pruned maximum credibility tree for ancestral range estimation to test likelihood implementations of three different biogeographic models in BioGeoBEARS. The DEC model treats dispersal and extinction as anagenetic events (modeled as free parameters) and sympatry, subset sympatry, and vicariance as cladogenetic events (modeled as fixed parameters) (Ree and Smith 2008). The DIVA model is similar, but it allows widespread vicariance as a possible cladogenetic event (Ronquist 1997). The BAYAREA model assumes that cladogenetic events are not accompanied by changes in geographic areas (Landis et al. 2013). Each of these models was also tested with the addition of the free parameter j, which treats jump dispersal as a cladogenetic event and has been shown to improve model likelihood (Matzke 2014). We compared the results of the models with and without the parameter j using likelihood ratio tests; the model weights were calculated under the Akaike information criterion (AIC). In order to measure the numbers of dispersal, vicariance, and sympatry events, we conducted 100 stochastic mapping replicates under the best model yielded by BioGeoBEARS. Each stochastic map represents a possible biogeographic history considering the chosen model and the estimated parameters (Duplin et al. 2017).
Diversification rate analysis
Shifts in diversification rates were calculated using speciation/extinction model type analysis in BAMM (Rabosky et al. 2014). For this, we used the same pruned phylogeny of the previous analysis. The missing taxa per tip (subgenus) in the phylogenetic tree was estimated according to the total number of species reported for each genus on the FishBase database (Froese and Pauly 2022). We divided the genera in three clades based on the MCC tree to state the percentage of species informed by clade: Clade I, subfamily Colossomatinae (Colossoma + Mylossoma + Piaractus), Clade II, tribe Myleini (Myleus + Myloplus + Mylesinus) and Clade III (subclade a Metynnis) and (subclade b Catoprion + Pristobrycon + Pygopristis + Pygocentrus + Serrasalmus), tribe Serrasalmini.
Priors for the BAMM control file were generated using the dated phylogenetic tree input into the function set BAMM priors in the package BAMM tools v. 2.5.0 implemented in R. The control file was set for 1,000,000 generations and the analysis was run twice as recommended, returning similar results. Resulting MCMC Log likelihoods were tested against generation number using the CODA package (Plummer et al. 2006) implemented in R. All remaining outputs contained in the event data file were analysed using BAMMtools in R.
Results
Phylogenetic analyses and divergence time
The MCC tree yielded by BEAST (Fig. 1) showed the family Serrasalmidae as monophyletic with a high support (pp = 0.97) with estimated origin around 48 Mya. Within the family, the genera Serrasalmus and Pygocentrus were paraphyletic, forming a well-supported clade (pp = 1) that originated approx. 16.9 Mya. The individuals of genera Catoprion, Pygopristis and Prystobrycon formed a monophyletic group with approx. 21.5 My (pp = 0.99). The monophyletic genus Metynnis (pp = 0.99) was found to be the sister of these subclades, with an estimated origin of approx. 25.9 Mya a. These three subclades together were named here as Clade III. The Clade II (pp = 0.99) was formed by genera Myleus and Mylesinus, diverging around 28.9 Mya. Lastly, Clade I was composed of the monophyletic genus Piaractus (pp = 0.99), the paraphyletic genus Mylossoma and the representant from genus Colossoma (pp = 0.98) and sister to the remaining Serrasalmidae clades (Fig. 1).
Phylogenetic comparative methods
We reconstructed the haploid chromosome number across the phylogeny of subfamily Serrasalminae based on Maximum Likelihood using ChromEvol. After all runs were completed, the AIC scores were assessed for choosing the best-fitted model. The models M1 (CONST_RATE), M2 (CONST_RATE_DEMI), and M4 (CONST_RATE_NO_DUPL) presented lower AIC scores, all very similar (91.32, 91.32, and 89.32 respectively). These three models presented almost identical results when plotted on the phylogeny, therefore the simplest model (M1) was chosen for the discussion of the results (Fig. 2). The ancestral haploid chromosome number of the subfamily was inferred to be n = 28 (pp = 0.5), with a very close probability of n = 29 (pp = 0.43). All the main clades presented different ancestral chromosome numbers. Clade III presented n = 30 (pp = 0.87), with independent disploidy events responsible for n = 31 in Pristobrycon and Pygopristis, and n = 32 in a few Serrasalmus species. Clade II presented n = 29 (pp = 0.95), without further number changes. Clade I presented n = 27 (pp = 0.51) with a close possibility of n = 28 (pp = 0.43). According to the results, chromosome evolution on this group was mainly directed by ascending and descending disploidy (exp. of 7 and 5.6 respectively), with no incidences of polyploidy or demipolyploidy.
Ancestral chromosome number reconstruction on Serrasalminae using the model M1 of ChromEvol. Pies at the nodes represent the posterior probability of inferred chromosome number, with the most probable number written at the center of the pie. Numbers above the branches represent the probability of one of the four chromosome number change events (gains, losses, duplication or demiduplication) given by maximum likelihood estimation
We used BAMM to detect heterogeneity in evolutionary rates across Serrasalmidae phylogeny. The 95% credible set of rate shift configurations sampled with BAMM presented three distinct shift configurations. The set with the highest probability (f = 0.4, Fig. 3) presented one shift of diversification on the ancestral node of the clade formed by Serrasalmus and Pygocentrus at around 11 Mya.
We tested whether this increase in the rate of diversification in clade III was associated with biogeographic colonization of new environments. To this, we used BioGeoBEARS for ancestral area reconstruction based on the Bayesian topology. The DIVALIKE model presented the lowest AIC score (90.87), being considered the best-fitted model (Fig. 1). The cladogenetic events were majorly sympatry events (83.9%) with only a few vicariance events (16.1%) mainly at the more recent nodes of the phylogeny. The whole subfamily and each of the main clades was confirmed to have originated in the Amazon region of Brazil, with the first incursions to other regions occurring ~20 Mya on Clade III.
Discussion
Our phylogenetic analyses provided molecular support for the recognition of three main clades in Serrasalmidae, congruent with previous studies (Ortí et al. 2008; Cione et al. 2009; Thompson et al. 2014; Mateussi et al. 2020; Favarato et al. 2021). The previous phylogenies currently subdivide Serrasalmidae into two subfamilies: Colossomatinae [Clade I] and Serrasalminae (split into the tribes Myleini [CladeII] and Serrasalmini [Clade III]) (Mateussi et al. 2020). When evaluating the ancestral karyotype in our topology, it was revealed that during the cladogenesis of this family, there were two distinct events of chromosomal rearrangements. The first one, leading to a descending dysploidy to n = 27 (2n = 54) in the subfamily Colossomatinae (Clade I), and the other, an ascending dysploidy in the subfamily Serrasalminae with Myleini and Serrasalmini tribes (Clade II and III), showing n = 29 (2n = 58), n = 30 (2n = 60), 31 (2n = 62) and n 32 (2n = 64) respectively. Regarding the first diverging lineage of the family, some studies have suggested n = 27 as the most plesiomorphic karyotype, due to its presence in older groups, such as the genera Mylossoma, Brachypomus, Colossoma and Piaractus (Nakayma et al. 2012). The chromosome number 2n = 54 was also detected in other representatives of the order Characiformes, such as the families Anostomidae and Prochilodontidae, which have a high degree of chromosomal conservation (Vicari et al. 2006; Aguilar and Galleti 2008). However, upon conducting the ancestral chromosomal reconstruction, we did not obtain evidence that would support this hypothesis. During the early divergence of the Serrasalmidae family, we observed two distinct trends: one characterized by chromosomal conservation and the other by variation, resulting in an increase in the chromosomal number.
Throughout our chromosome number reconstruction, it was possible to identify ascending dysploid events (increasing the chromosome numbers) in the most derived genera, such as Mylesinus, Myleus and Myloplus, which present 2n = 58. This increase becomes accentuated in representatives of the most diverse genera, Pygopristis, Pygocentrus and Serrasalmus with 2n = 60, 62 and 64. The differential morphology of karyotypes with more derived chromosome numbers, such as an increase in acrocentric chromosomes, indicates that chromosomal fission rearrangements drive the karyoevolution of the Serrasalmidae family (Nakayama et al. 2012). Centric fissions lead to karyotype diversity within a population, consequently increasing the probability of genetic isolation and speciation (Perry et al. 2004). Indeed, these processes appear to have been one of the main mechanisms in cladogenetic events in this family.
Our biogeographic reconstruction and molecular dating also allowed us to understand the phylogenetic relationships in a temporal context and discuss geographic distribution across space and time. Regarding the geographic distribution, most of the representatives of Serrasalmidae occur in the Amazon basins, which suggests that this region is the center of origin of the group. On the other hand, much is discussed about the evolutionary origin of the family, which has often been controversial. Some authors suggest an older origin, around 66–56 Ma, during the Middle Paleocene, but with the beginning of its diversification around 45 Ma, in the late Eocene (Thompson et al. 2014; Burns and Sidlauskas 2019). Another study pointed to a younger age, between 42 and 38 Ma (Kolmann et al. 2021). Our analyses point to a scenario between 48 and 38 Ma, within the threshold of previous studies, associated with the uplift of the Andes (Armijo et al. 2015). In this context, the diversification scenario has been congruent with these studies (Burns and Sidlauskas 2019; Kolmann et al. 2021).
The cladogenetic events that separate the three major clades are related to the late Eocene and early Oligocene (38 to 30 Mya) in our study, which coincides with the great division of the West–East Amazon drainage, with the origin of the Purus Arc and a period of mega-wetland formation in the proto-Orinoco-Amazonas (Lundberg et al. 1998; Albert and Reis 2011). The uplift of this arc is due to an orogenetic response to the initial elevation of the Andes Mountain range, which consequently may have caused allopatric speciation in some fish species (Armijo et al. 2015). This context of allopatry is reinforced by the presence of fossils of C. macropomum, known as pacus, in the Magdalena River basin in Colombia, which today is an inhabitant of the Amazon and Orinoco rivers. This has suggested that this taxon inhabited ancient systems that connected the Amazon and Magdalena River basins, today separated by the Andes Mountain range (Lundberg et al. 2009). Ecological factors have also been associated with the diversification of frugivorous pacus, which coincides with the diversification of fruit plants during the Eocene (Correa et al. 2015). However, with regard to chromosomal aspects, the subfamily Cossolomatinae was the group that presented descending dysploidy (2n = 54), which has not presented an increase in chromosomal diversification, contrary to the subfamily Serrasalminae, constituted by an ascending to dysploidy with 2n = 58 to 64.
During the Lower Eocene and Upper Miocene (30–20 Mya), the uplift of the Andes in the North and Central region culminated in a change in the course of several rivers (Lundberg et al. 1998). These orogenetic processes significantly altered the hydrography in South America, leading to fracturing processes in several basins, with redirection of river courses and headwater capture events (Hoorn et al. 2010; Evenstar et al. 2015). These processes may have provided ecological opportunities and colonization of Serrasalmideos for new habitats, providing speciation events (Melo et al. 2018, 2022; Roxo et al. 2019; Ochoa et al. 2020). Together with these cladogenetic mechanisms, they may have triggered chromosomal rearrangements with a tendency towards ascending dysploidy. However, our biogeographic reconstruction demonstrated that dispersal/ vicariance events apparently did not accompany karyotypic changes.
As the ancestral chromosome reconstruction shows, ascending dysploidy lead to higher chromosome numbers at the most derived lineages of Serrasalminae, culminating in the 2n = 60 karyotype being predominant in the Serrasalmus and Pygocentrus genera. In addition to this, our data revealed an increase in the diversification rate during the Miocene (11–8 Mya), involving these genera. These findings are in agreement with other studies, which also pointed to rapid and recent radiation involving this group of piranhas, which have diversified greatly in the plains of South America (Hubert and Renno 2006; Hubert et al. 2007). The apparent correlation between the 2n = 60 karyotypes and the increase in diversification rate, coupled with the tendency for ascending dysploidy in the family, may point to a scenario in which high chromosome numbers are associated with species diversification and evolutionary success. However, causation would be hard to infer until further and more detailed genomic studies for these groups are provided.
During the late Miocene and Pliocene, hydrological and paleogeographic events may have driven changes in the diversification rates of this group. In relation to other river basins, the Amazon separated from the Paraná-Paraguay system by 10 Ma, leading to the separation of the ichthyofauna in these systems (Hubert and Renno 2006). Headwater capture events have also been identified between the Upper Paraná and São Francisco basins around 10 Mya (Hubert and Renno 2006). Dispersions such as separations between watersheds, provided by tectonic activations and changes in the course of rivers (geodispersion), may have provided a certain advantage for carnivorous piranhas, in dispersing species and conquering new habitats, reducing predation and competition. These processes can lead to increased rates of speciation and diversification, as happened in the subfamily Hipostominae (Cardoso et al. 2012). In addition to orogenetic movements, sea level fluctuations may also have further contributed to promoting this diversification (Hubert and Renno 2006). In the last 10 Mya, the sea level has varied from 35 m above to 122 m below the current level and may also have contributed to accelerate this process (Hubert et al. 2007). In this context, in addition to allopatric speciation, processes of sympatric speciation have also been detected in some sister lineages of piranhas, associated with habitat heterogeneity. As is the case of S. compressus n = 30 and S. hollandi n = 32, which live in the Madeira River and diverged in the last 2 Ma (Hubert et al. 2007).
All the discussed scenarios highlight that historical and ecological processes seem to have shaped this family's genetic and phylogenetic diversity, involving several types of chromosomal rearrangements. In this context, ascending dysploidy seems to have driven the karyotypic evolution of some lineages towards higher chromosome numbers,resultin in highly diversified persistent lineages across different hydrographic basins of South America. These data, combined with previous studies (Correa et al. 2015), demonstrate that the rate of diversification in Serrasalmidae is not correlated with biogeographic changes, but that it could possibly be linked to ecological processes that lead to morphological changes (Kolmann et al. 2021), as well as karyoevolutionary differentiation by dysploidy. Generally, dysploidy does not necessarily imply changes in DNA content, only in the structure of chromosomal rearrangements that can occur in the genome. These processes, so far, have been considered to have a neutral effect in relation to the diversification of evolutionary processes over the long term (Escudero et al. 2014).
Final considerations
Our data demonstrate that chromosomal rearrangements played an important evolutionary role in major cladogenetic events in Serrasalmidae, revealing them as a powerful evolutionary driver in family diversification. Our results support the hypothesis that ascending dysploidy acted as one of the main drivers in the chromosomal evolution of the Serrasalminae family and seems to be more correlated with diversification patterns than biogeographic history. In this context, we suggest the importance of integrating cytogenetic studies to evaluate the systematic aspects of the Serrasalmidae family. In addition, we highlight the importance of correctly interpreting the karyotype in a phylogenetic and biogeographic context.
Change history
12 September 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10709-023-00195-9
References
Aguilar CTD, Galetti PM Jr (2008) Chromosome mapping of 5S rRNA genes differentiates Brazilian populations of Leporellus vittatus (Anostomidae, Characiformes). Genet Mol Biol 31:188–194. https://doi.org/10.1590/S1415-47572008000200004
Albert JS, Reis R (2011) Historical biogeography of Neotropical freshwater fishes. Univ of California Press, Berkeley
Almeida-Toledo LF, Foresti F, Toledo-Filho SA, Bernardino G, Ferrari W, Alcantara RCG (1987) Cytogenetic studies in colossoma macropomum, colossoma mitrei and their interespecific hybrid. Selection, hybridization and genetic engineering in aquaculture. Springer-Verlag, Berlin
Aprea G, Andreone F, Fulgione D, Petraccioli A, Odierna G (2013) Chromosomal rearrangements occurred repeatedly and independently during species diversification in Malagasy geckos, genus Paroedura. African Zoology 48(1):96–108. https://doi.org/10.1080/15627020.2013.11407572
Arai R (2011) Fish karyotypes: a check list. Springer Science & Business Media, Berlin
Araújo-Lima CARM, Goulding M (1997) So fruitful fish: ecology, conservation and aquaculture of the Amazon’s Tambaqui. Columbia University, New York, p 157
Armijo R, Lacassin R, Coudurier-Curveur A, Carrizo D (2015) Coupled tectonic evolution of Andean orogeny and global climate. Earth Sci Rev 143:1–35. https://doi.org/10.1016/j.earscirev.2015.01.005
Ayala FJ, Coluzzi M (2005) Chromosome speciation: humans, drosophila, and mosquitoes. Proc National Acad Sci 102(suppl1):6535–6542. https://doi.org/10.1073/pnas.0501847102
Bertollo LA, Born GG, Dergam JA, Fenocchio AS, Moreira-Filho O (2000) A biodiversity approach in the neotropical Erythrinidae fish, Hoplias malabaricus. Karyotypic survey, geographic distribution of cytotypes and cytotaxonomic considerations. Chromosome Res 8:603–613. https://doi.org/10.1023/A:1009233907558
Betancur RR, Orti G, Pyron RA (2015) Fossil-based comparative analyses reveal ancient marine ancestry erased by extinction in ray-finned fishes. Ecol Lett 18(5):441–450. https://doi.org/10.1111/ele.12423
Burns MD, Sidlauskas BL (2019) Ancient and contingent body shape diversification in a hyperdiverse continental fish radiation. Evolution 73(3):569–587. https://doi.org/10.1111/evo.13658
Cardoso YP, Almiron A, Casciotta J, Aichino D, Lizarralde MS, Montoya-Burgos JI (2012) Origin of species diversity in the catfish genus Hypostomus (Siluriformes: Loricariidae) inhabiting the Paraná river basin, with the description of a new species. Zootaxa 3453(1):69–83. https://doi.org/10.11646/zootaxa.3453.1.5
Centofante L, Porto JIR, Feldberg E (2002) Chromosomal polymorphism in Serrasalmus spilopleura Kner, 1858 (Characidae, Serrasalminae) from central Amazon basin. Caryologia 55(1):37–45. https://doi.org/10.1080/00087114.2002.10589256
Cestari MM (1992) Chromosome evolution in the genus Serrasalmus and cytotaxonomic considerations. Brazil J Genet 15(3):555–567
Cestari MM, Galetti PM Jr (1992) Chromosome studies of Serrasalmus spilopleura (Characidae, Serrasalminae) from the Parana-Paraguay rivers: evolutionary and cytotaxonomic considerations. Copeia. https://doi.org/10.2307/1446541
Cioffi MDB, Moreira-Filho O, Ráb P, Sember A, Molina WF, Bertollo LAC (2018) Conventional cytogenetic approaches-useful and indispensable tools in discovering fish biodiversity. Cur Genetic Med Rep 6(4):176–186. https://doi.org/10.1007/s40142-018-0148-7
Cione AL, Dahdul WM, Lundberg JG, Machado-Allison A (2009) Megapiranha paranensis, a new genus and species of Serrasalmidae (Characiformes, Teleostei) from the upper Miocene of Argentina. J Vertebr Paleontol 29(2):350–358. https://doi.org/10.1671/039.029.0221
Correa SB, Winemiller KO, Lopez-Fernandez H, Galetti M (2007) Evolutionary perspectives on seed consumption and dispersal by fishes. Bioscience 57(9):748–756. https://doi.org/10.1641/B570907
Correa SB, Araujo JK, Penha JM, Cunha CN, Stevenson PR, Anderson JT (2015) Overfishing disrupts an ancient mutualism between frugivorous fishes and plants in Neotropical wetlands. Biol Cons 191:159–167. https://doi.org/10.1016/j.biocon.2015.06.019
Costa L, Jimenez H, Carvalho R, Carvalho-Sobrinho J, Escobar I, Souza G (2020) Divide to conquer: evolutionary history of Allioideae Tribes (Amaryllidaceae) is linked to distinct trends of karyotype evolution. Front Plant Sci 11:320. https://doi.org/10.3389/fpls.2020.00320
Dahdul WM (2004) Fossil serrasalmine fishes (Teleostei: Characiformes) from the Lower Miocene of north-western Venezuela. Spec Pap Palaeontol 71:23–28
DeCelles PG, Horton BK (2003) Early to middle Tertiary foreland basin development and the history of Andean crustal shortening in Bolivia. Geol Soc Am Bull 115(1):58–77. https://doi.org/10.1130/0016-7606(2003)115%3C0058:ETMTFB%3E2.0.CO;2
Drummond AJ, Rambaut A (2007) BEAST: bayesian evolutionary analysis by sampling trees. BioMedCentral Evol Biol 7:214. https://doi.org/10.1186/1471-2148-7-214
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29(8):1969–1973. https://doi.org/10.1093/molbev/mss075
Dupin J, Matzke NJ, Särkinen T, Knapp S, Olmstead RG, Bohs L, Smith SD (2017) Bayesian estimation of the global biogeographical history of the Solanaceae. J Biogeogr 44(4):887–899. https://doi.org/10.1111/jbi.12898
Escudero M, Martín-Bravo S, Mayrose I, Fernández-Mazuecos M, Fiz-Palacios O, Hipp AL, Luceño M (2014) Karyotypic changes through dysploidy persist longer over evolutionary time than polyploid changes. PloS One 9(1):e85266. https://doi.org/10.1371/journal.pone.0085266
Evenstar LA, Stuart FM, Hartley AJ, Tattitch B (2015) Slow Cenozoic uplift of the western Andean Cordillera indicated by cosmogenic 3He in alluvial boulders from the pacific planation surface. Geophys Res Lett 42(20):8448–8455. https://doi.org/10.1002/2015GL065959
Favarato RM, Ribeiro LB, Campos A, Porto JIR, Nakayama CM, Ota RP, Feldberg E (2021) Comparative cytogenetics of Serrasalmidae (Teleostei: Characiformes): the relationship between chromosomal evolution and molecular phylogenies. Plos one 16(10):e0258003. https://doi.org/10.1371/journal.pone.0258003
Freyman WA, Höhna S (2018) Cladogenetic and anagenetic models of chromosome number evolution: a Bayesian model averaging approach. Syst Biol 67(2):195–215. https://doi.org/10.1093/sysbio/syx065
Fricke R., Eschmeyer WN, Van der Laan R (2022) Eschmeyer’s catalog of fishes: Genera, species, references. Available at: https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
Friedman NR, Bennet BL, Fischer G et al (2020) Macroevolutionary integration of phenotypes within and across ant worker castes. Ecol Evol 10:9371–9383. https://doi.org/10.1002/ece3.6623
Froese R, Pauly D (2022) FishBase. World Wide Web electronic publication. Available at: www.fishbase.org. Acecessed 02 Feb 2022
Galetti PM Jr, Molina WF, Affonso PRA, Aguilar CT (2006) Assessing genetic diversity of Brazilian reef fishes by chromosomal and DNA markers. Genetica 126(1):161–177. https://doi.org/10.1007/s10709-005-1446-z
Gaviria JI, Nirchio M, Granado Á, Estrada A (2005) Karyotype and nucleolar organizer regions of Pygocentrus cariba (Serrasalminae) from Caicara Del Orinoco. Venezuela Interciencia 30(1):44–47
Gernhard T (2008) The conditioned reconstructed process. J Theor Biol 253(4):769–778. https://doi.org/10.1016/j.jtbi.2008.04.005
Géry J (1977) Characoids of the world. Tropical Fish Hobbyist Publications, Neptune City, p 672
Glick L, Mayrose I (2014) ChromEvol: assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Mol Biol Evol 31(7):1914–1922. https://doi.org/10.1093/molbev/msu122
Goulding M (1980) The fishes and the forest: explorations in Amazonian natural history. University of California Press, Berkeley, p 280
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27(2):221–224. https://doi.org/10.1093/molbev/msp259
Hoorn C, Wesselingh FP, Steege HT, Bermudez MA, Mora A, Sevink J et al (2010) Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330(6006):927–931. https://doi.org/10.1126/science.1194585
Hubert N, Renno JF (2006) Historical biogeography of South American freshwater fishes. J Biogeogr 33(8):1414–1436. https://doi.org/10.1111/j.1365-2699.2006.01518.x
Hubert N, Duponchelle F, Nunez J, Garcia-Davila CARMEN, Paugy D, Renno JF (2007) Phylogeography of the piranha genera Serrasalmus and Pygocentrus: implications for the diversification of the Neotropical ichthyofauna. Mol Ecol 16(10):2115–2136. https://doi.org/10.1111/j.1365-294X.2007.03267.x
Jacobina UP, Vicari MR, Martinez PA, de Belo CM, Bertollo LAC, Molina WF (2013) Atlantic moonfishes: independent pathways of karyotypic and morphological differentiation. Helgol Mar Res 67(3):499–506. https://doi.org/10.1007/s10152-012-0338-8
Jacobina UP, Martinez PA, Torres RA, Souza G (2016) Trends on the karyotype acrocentrization within Carangidae (Perciformes): a new phylogenetic evidence about a traditional marine paradigm. Zebrafish 13(1):45–53. https://doi.org/10.1089/zeb.2015.1143
Jégu M (2003) Subfamily Serrasalminae (Pacus and piranhas). 182–196. CLOFFSCA-Check list of the freshwater fishes of South and Central America. Edipucrs, Porto Alegre.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kolmann MA, Hughes LC, Hernandez LP, Arcila D, Betancur RR, Sabaj MH, Ortí G (2021) Phylogenomics of piranhas and pacus (Serrasalmidae) uncovers how dietary convergence and parallelism obfuscate traditional morphological taxonomy. Syst Biol 70(3):576–592. https://doi.org/10.1093/sysbio/syaa065
Landis MJ, Matzke NJ, Moore BR, Huelsenbeck JP (2013) Bayesian analysis of biogeography when the number of areas is large. Syst Biol 62(6):789–804. https://doi.org/10.1093/sysbio/syt040
Liem KF (1973) Evolutionary strategies and morphological innovations: cichlid pharyngeal jaws. Syst Zool 22(4):425–441. https://doi.org/10.2307/2412950
Lundberg JG, Marshall LG, Guerrero J, Horton B, Malabarba MCSL, Wesselingh F (1998) The stage for Neotropical fish diversification: a history of tropical South American rivers. Phylogeny Classif Neotropical Fishes 27:13–48
Lundberg JG, Sabaj Pérez MH, Dahdul WM, Aguilera OA (2009) The Amazonian neogene fish fauna. Amazonia. https://doi.org/10.1002/9781444306408.ch17
Martinez PA, Zurano JP, Amado TF, Penone C, Betancur-RR BCJ, Jacobina UP (2015) Chromosomal diversity in tropical reef fishes is related to body size and depth range. Mol Phylogenet Evol 93:1–4. https://doi.org/10.1016/j.ympev.2015.07.002
Mateussi NT, Melo BF, Ota RP, Roxo FF, Ochoa LE, Foresti F, Oliveira C (2020) Phylogenomics of the Neotropical fish family Serrasalmidae with a novel intrafamilial classification (Teleostei: Characiformes). Molecular phylogenetics and evolution 153:106945. https://doi.org/10.1016/j.ympev.2020.106945
Matzke NJ (2014) Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst Biol 63(6):951–970. https://doi.org/10.1093/sysbio/syu056
Matzke NJ (2013) BioGeoBEARS: Biogeography with Bayesian (and likelihood) evolutionary analysis in R Scripts. University of California, Berkeley. Available at: http://CRAN.R-project.org/package=BioGeoBEARS
Mayrose I, Barker MS, Otto SP (2010) Probabilistic models of chromosome number evolution and the inference of polyploidy. Syst Biol 59(2):132–144. https://doi.org/10.1093/sysbio/syp083
Melo BF, Sidlauskas BL, Hoekzema K, Vari RP, Dillman CB, Oliveira C (2018) Molecular phylogenetics of Neotropical detritivorous fishes of the family Curimatidae (Teleostei: Characiformes). Mol Phylogenet Evol 127:800–812. https://doi.org/10.1016/j.ympev.2018.06.027
Melo BF, Sidlauskas BL, Near TJ, Roxo FF, Ghezelayagh A, Ochoa LE et al (2022) Accelerated diversification explains the exceptional species richness of tropical characoid fishes. Syst Biol 71(1):78–92. https://doi.org/10.1093/sysbio/syab040
Mezzasalma M, Andreone F, Glaw F, Petraccioli A, Odierna G, Guarino FM (2016) A karyological study of three typhlopid species with some inferences on chromosome evolution in blindsnakes (Scolecophidia). Zoologischer Anzeiger-A J Comparative Zool 264:34–40. https://doi.org/10.1016/j.jcz.2016.07.001
Molina WF (2007) Chromosomal changes and stasis in marine fish groups. Fish cytogenetics. CRC Press, Boca Raton, pp 69–110
Nakayama CM, Porto JIR, Feldberg E (2000) Ocorrência de dois citótipos em Serrasalmus spilopleura Kner, 1858 (Characiformes, Serrasalmidae) da região de confluência dos rios Negro e Solimões, Amazonas, Brasil. Acta Amazon 30:149–149. https://doi.org/10.1590/1809-43922000301154
Nakayama C, Jegú M, Porto JIR (2001) Feldberg E (2001) Karyological evidence for a cryptic species of piranha within Serrasalmus rhombeus (Characidae, Serrasalminae) in the Amazon. Copeia 3:866–869. https://doi.org/10.1643/0045-8511(2001)001[0866:KEFACS]2.0.CO;2
Nakayama CM, Rebelo Porto JI, Feldberg E (2002) A comparative cytogenetic study of five piranha species (Serrasalmus, Serrasalminae) from the Amazon basin. Genetica 114(3):231–236. https://doi.org/10.1023/A:1016275505655
Nakayama CM, Feldberg E, Bertollo LAC (2008) Mapping of ribosomal genes and chromosomal markers in three species of the genus Serrasalmus (Characidae, Serrasalminae) from the Amazon basin. Genet Mol Biol 31:868–873. https://doi.org/10.1590/S1415-47572008005000018
Nakayama CM, Feldberg E, Bertollo LAC (2012) Karyotype differentiation and cytotaxonomic considerations in species of Serrasalmidae (Characiformes) from the Amazon basin. Neotropical Ichthyol 10:53–58. https://doi.org/10.1590/S1679-62252012000100005
Nirchio M, Fenocchio AS, Swarça AC, Pérez JE, Granado A, Estrada A, Ron E (2003) Cytogenetic characterization of hybrids offspring between Colossoma macropomum (Cuvier, 1818) and Piaractus brachypomus (Cuvier, 1817) from Caicara del Orinoco. Venezuela Caryologia 56(4):405–411. https://doi.org/10.1080/00087114.2003.10589351
Nirchio M, Gaviria JI, Siccha-Ramirez ZR, Oliveira C, Foresti F, Milana V, Rossi AR (2019) Chromosomal polymorphism and molecular variability in the pearly razorfish Xyrichtys novacula (Labriformes, Labridae): taxonomic and biogeographic implications. Genetica 147:47–56. https://doi.org/10.1007/s10709-019-00051-9
Ochoa LE, Datovo A, DoNascimiento C, Roxo FF, Sabaj MH, Chang J et al (2020) Phylogenomic analysis of trichomycterid catfishes (Teleostei: Siluriformes) inferred from ultraconserved elements. Sci Rep 10(1):1–15. https://doi.org/10.1038/s41598-020-59519-w
Ortí G, Sivasundar A, Dietz K, Jégu M (2008) Phylogeny of the Serrasalmidae (Characiformes) based on mitochondrial DNA sequences. Genet Mol Biol 31:343–351. https://doi.org/10.1590/S1415-47572008000200030
Perry JO, Slater HR, Choo KH (2004) Centric fission—simple and complex mechanisms. Chromosome Res 12(6):627–640. https://doi.org/10.1023/B:CHRO.0000036594.38997.59
Plummer M, Best N, Cowles K, Vines K (2006) CODA: Convergence diagnosis and output analysis for MCMC. R Package Version, 6, 7–11. Available at: https://www.r-project.org/doc/Rnews/
Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW et al (2014) BAMM tools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol Evol 5(7):701–707. https://doi.org/10.1111/2041-210X.12199
Rambaut A (2014) FigTree v1. 4.2, a graphical viewer of phylogenetic trees. Available at: http://tree.bio.ed.ac.uk/software/figtree/right.angle.bracket.
Ratomponirina C, Brun B, Rumpler Y (1988) Synaptonemal complexes in Robertsonian translocation heterozygous in lemurs. In: Brandham PE (ed) Kew Chromosome Conference III. HMSO Publications Center, London, pp 65–73
Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol 57(1):4–14. https://doi.org/10.1080/10635150701883881
Revell LJ (2012) phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x
Roberts TR (1975) Characoid fish teeth from Miocene deposits in the Cuenca Basin, Ecuador. J Zool 175(2):259–271. https://doi.org/10.1111/j.1469-7998.1975.tb01400.x
Ronquist F (1997) Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst Biol 46(1):195–203. https://doi.org/10.1093/sysbio/46.1.195
Rosa R, Vicari MR, Dias AL, Giuliano-Caetano L (2014) New insights into the biogeographic and karyotypic evolution of Hoplias malabaricus. Zebrafish 11(3):198–206. https://doi.org/10.1089/zeb.2013.0953
Roxo FF, Ochoa LE, Sabaj MH, Lujan NK, Covain R, Silva GS et al (2019) Phylogenomic reappraisal of the Neotropical catfish family Loricariidae (Teleostei: Siluriformes) using ultraconserved elements. Mol Phylogenet Evol 135:148–165. https://doi.org/10.1016/j.ympev.2019.02.017
Sader MA, Amorim BS, Costa L, Souza G, Pedrosa-Harand A (2019) The role of chromosome changes in the diversification of Passiflora L. (Passifloraceae). Syst Biodiversity 17(1):7–21. https://doi.org/10.1080/14772000.2018.1546777
Schluter D (2000) Ecological character displacement in adaptive radiation. Am Naturalist 156(S4):S4–S16. https://doi.org/10.1086/303412
Schubert I (2007) Chromosome evolution. Curr Opin Plant Biol 10(2):109–115. https://doi.org/10.1016/j.pbi.2007.01.001
Sember A, Oliveira EA, Ráb P, Bertollo LAC, Freitas NL, Viana PF et al (2020) Centric fusions behind the karyotype evolution of neotropical nannostomus pencilfishes (characiforme, lebiasinidae): first insights from a molecular cytogenetic perspective. Genes 11(1):91. https://doi.org/10.3390/genes11010091
Thompson AW, Betancur-RR L-F, Ortí G (2014) A time-calibrated, multi-locus phylogeny of piranhas and pacus (Characiformes: Serrasalmidae) and a comparison of species tree methods. Mol Phylogenet Evol 81:242–257. https://doi.org/10.1016/j.ympev.2014.06.018
Van-Lume B, Esposito T, Diniz-Filho JAF, Gagnon E, Lewis GP, Souza G (2017) Heterochromatic and cytomolecular diversification in the Caesalpinia group (Leguminosae): relationships between phylogenetic and cytogeographical data Perspectives in Plant Ecology. Evol Syst 29:51–63. https://doi.org/10.1016/j.ppees.2017.11.004
Vicari MR, Moreira-Filho O, Artoni RF, Bertollo LAC (2006) ZZ/ZW sex chromosome system in an undescribed species of the genus Apareiodon (Characiformes, Parodontidae). Cytogenet Genome Res 114(2):163–168. https://doi.org/10.1159/000093333
Yoshida K, Kitano J (2021) Tempo and mode in karyotype evolution revealed by a probabilistic model incorporating both chromosome number and morphology. PLoS Genetics 17(4):e1009502. https://doi.org/10.1371/journal.pgen.1009502
Acknowledgements
We wish to thank Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE) for the scholarship granted to LC (BFP-0040-2.03/22). The authors wish to thank the Brazilian agencies Conselho Nacional de Desenvolvimento Cientiífico e Tecnológico (CNPq) for a productivity fellowship for GS (process number PQ - 312852/2021-5).
Author information
Authors and Affiliations
Contributions
All authors wrote the main and reviewed text the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The co-author name “Gustavo Souza” is corrected.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jacobina, U.P., Pontes, A.I., Costa, L. et al. Macroevolutionary consequences of karyotypic changes in the neotropical Serrasalmidae fishes (Ostariophysi, Characiformes) diversification. Genetica 151, 311–321 (2023). https://doi.org/10.1007/s10709-023-00191-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-023-00191-z