Abstract
Species of new world silversides (Actinopterygii; Atherinopsidae; genus Odontesthes) possess economic relevance, biological interest and ecological importance. In the present paper we: (A) investigate the molecular diversity in marine species of Odontesthes from the South West Atlantic Ocean (SWAO), and analyse their interspecific relationships and divergence by means of DNA Barcoding, including its freshwater congeners, as well. (B) Explore the suitability of DNA Barcoding to analyse the diversity and distribution of haplotypes in Odontesthes argentinensis, the only well documented marine species from the SWAO that exhibit putative estuarine and marine populations. Molecular analysis revealed 100% of agreement between morphological identification and molecular identity. Odontesthes argentinensis, Odontesthes platensis, Odontesthes smitti, Odontesthes nigricans and Odontesthes incisa were assigned to five different barcode index numbers (BINs). Maximum-likelihood analysis showed that all marine species of Odontesthes clustered separately in a unique monophyletic phylogroup, comprising five well defined haplogroups, with genetic divergence between groups ranging from 2.75 to 7.11%. The genetic analysis including freshwater congeners showed that O. incisa clustered alone occupying a basal position. The Fst pairwise comparisons within O. argentinensis support the existence of three population groups: one conformed by Mar Chiquita Lagoon (MCh) specimens, and the others by Mar del Plata/Mar Chiquita coast and San Blas Bay coastal specimens, respectively. The AMOVA showed significant overall differentiation (Fst = 0.238; p = 0.00001) for the entire data set. The previous/present evidence is discussed, and strongly suggests that incipient speciation is occurring in O. argentinensis argentinean populations, and specimens from MCh would be considered at present as the leading candidate of a marine to freshwater incipient speciation event.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colonization of novel environments is one of the most relevant factors promoting species diversification in fishes, being the selective pressure occurring in these new habitats the one that generates divergent natural selection among populations, promotes adaptation, and finally, ecological speciation (Schluter 2000; Beheregaray and Sunnucks 2001; Betancur-R et al. 2012; Lescak et al. 2015). It is precisely in this context that the unidirectional and recurrent speciation from marine to freshwater has occurred in fishes, such as in gobies (Matthew et al. 2009; Yamasaki et al. 2015), sculpins (Yokoyama and Goto 2005), sticklebacks (Lescak et al. 2015; Takahashi et al. 2016), and salmons (Taylor et al. 1996; Moreira and Taylor 2015). In the past decade, the DNA sequencing technology introduced the possibility of using variation in short sequences of mitochondrial DNA as labels for specimens in a process known as DNA barcoding (Hebert et al. 2003). The Fish Barcode of Life Initiative (FISH-BOL) is a concerted global effort to aid in the assembly of a standardized reference sequence library for all fish species (Ward et al. 2009). The FISH-BOL employ the COI mitochondrial marker as standard for fish identification (Hebert et al. 2003). DNA barcoding results indicate that c. 93% of freshwater fish species can be discriminated by this initiative (Ward et al. 2009).
The order Atheriniformes is a monophyletic group, comprised by six families and 49 genera of generally small, silvery fishes, which belong to the Series Atherinomorpha (Dyer and Chernoff 1996). The taxonomic and systematic history of the South American silversides have been drastically changed during the last 30 years, when several subfamilies and genera were reassigned more than once in the systematic classification of Atheriniformes (Dyer 2006; Nelson et al. 2016; Helfman et al. 2009; Cousseau 2010; Betancur-R et al. 2013). The only family of marine silversides occurring in the South West Atlantic Ocean (SWAO) is Atherinopsidae, which is represented by the genus Odontesthes Evermann and Kendall, 1909 (Dyer 2006; Cousseau et al. 2004; González-Castro et al. 2016). Five species of marine Odontesthes are currently recognized from southern Brazil, Uruguay and Argentina: Odontesthes argentinensis (Valenciennes, 1835), locally called “escardón”; O. incisa (Jenyns 1841) (called “cornalito”); O. smitti (Lahille, 1929) (known as “corno”); O. platensis (Berg 1895), usually called as “panzón” and O. nigricans (Richardson 1848) (known as “pejerrey malvinense”). Most of these species have singular economic importance: they are commonly employed not only for game fishing, but also commercially exploited by the artisanal and commercial coastal fleets conducted in shallow waters of southern South America (Dyer 2006; Cousseau and Perrotta 2013; González-Castro et al. 2016).
Despite the economic relevance of the marine species of Odontesthes, few studies on the molecular taxonomy of this species group were performed (Beheregaray and Sunnucks 2001; Heras and Roldán 2011; Mabragaña et al. 2011; Bloom et al. 2013; García et al. 2014; Campanella et al. 2015; González-Castro et al. 2016; Hughes et al. 2017). The genetic relationships among the marine and freshwater species of Odontesthes employing COI are a pendent task. Previous molecular studies did not include altogether these five marine species, thus hampering the discrimination of their molecular identity and the phylogenetic relationships among them (Beheregaray and Sunnucks 2001; Heras and Roldán 2011; Mabragaña et al. 2011; García et al. 2014; González-Castro et al. 2016; Hughes et al. 2017). Aiding to this uncertainty, recent findings support (for some species of Odontesthes) that promiscuous and recent contact between incipient species blurs taxa boundaries yielding complicated taxonomy (García et al. 2014).
Among marine species of Odontesthes from SWAO, O. argentinensis represents a unique and interesting study case. Several works strongly suggested that this marine fish is undergoing speciation related to colonization of estuarine habitats in South America (Beheregaray and Sunnucks 2001; Bemvenuti 2002, 2006; García et al. 2014; González-Castro et al. 2016). Beheregaray and Sunnucks (2001) suggested that ecological shifts due to colonization of estuarine habitats seem to have promoted rapid adaptive divergence and reproductive isolation in estuarine populations of O. argentinensis, which were considered as incipient ecological species. Moresco and Bemvenuti (2006) stated that, in Rio Grande do Sul (Brazil), O. argentinensis is represented by two populations: one resident population in the Patos Lagoon estuary and other in the sea. Both populations showed evidence of disjoint spawning in their respective environments. Accordingly, González-Castro et al. (2009) found ripe and spent females of O. argentinensis in the inner zone of Mar Chiquita Coastal Lagoon (Argentina) (where water is mixo-oligohaline), suggesting that spawning of O. argentinensis occurs inside the lagoon. In addition to these findings, González-Castro et al. (2016) demonstrated that O. argentinensis from Mar Chiquita Coastal Lagoon (Argentina) is meristically and morphometrically distinguishable from nearby marine populations, and appears to behave as a well-differentiated population, or even incipient ecological species.
The Neotropical fish fauna living at the southernmost extreme of South America, in Argentina, has been barcoded since 2005 (Díaz de Astarloa et al. 2008; Mabragaña et al. 2011; Rosso et al. 2012, 2017). Despite the accumulated knowledge on these species, several groups still represent a difficult task for taxonomists. In this context, the use of molecular techniques such as DNA barcoding can help providing complementary information for taxonomically conflicting species.
However, DNA Barcoding gave no full resolution to discriminate the freshwater species of Odontesthes (Rosso et al. 2012; García et al. 2014; González-Castro et al. 2016). Therefore, the effectiveness of DNA Barcoding for discriminating their marine congeners is still a pending task. In order to achieve this purpose, a formal link between a proper taxonomic discrimination with its genetic identity as well as exploring interspecific relationships among SWAO species of Odontesthes is needed. Molecular discrimination between freshwater and marine species of Odontesthes is not as straightforward as taxonomic information would anticipate (García et al. 2014; González-Castro et al. 2016; Hughes et al. 2017). Therefore, a molecular approach intended to unravel interspecific relationships among marine Odontesthes would be benefited by the inclusion of some freshwater representatives of this genus.
The aims of this work are:
(A) To investigate the molecular diversity in marine species of Odontesthes from the SWAO, and analyse their interspecific relationships and divergence by means of DNA Barcoding, including also its freshwater congeners.
(B) To explore the suitability of DNA Barcoding to analyse the diversity and distribution of haplotypes in O. argentinensis, which is the only well-documented marine species from the SWAO that exhibit estuarine and marine populations and even incipient speciation between them.
Materials and methods
Study area
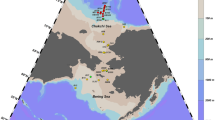
The southwestern Atlantic continental shelf is wide for almost its entire length, which extends approximately 400 miles to the Malvinas islands. Northwards, the continental shelf is narrow especially in the region of the mouth of the Rio de la Plata and along the Uruguayan coast. The shelf break occurs at about 200 m depth (Fig. 1). Throughout this region, substrates consist mainly of sand and mud, except on the southernmost shelf which has an undulated bottom topography featuring many rocky areas (Capurro 1981). Oceanic circulation in the southwestern Atlantic is characterized by the encounter between the warm, southward flowing Brazil Current and the cold, northward flowing Malvinas Current. The Atlantic coast of Buenos Aires Province (36°S to 41°S) is influenced by these two important oceanic currents. The Brazil Current flows southward along the continental margin and turns away from the coast at about 36–38°S. Interactions between the oligotrophic Brazil Current and the nutrient-rich Malvinas Current make the Atlantic coast of Buenos Aires Province an important nursery and feeding area for fishes. This is the case of Mar Chiquita Coastal Lagoon (MCh) a shallow water nursery estuary separated from the sea by a littoral line of dunes with an inlet joining it to the ocean. This lagoon is considered a World Biosphere Reserve by the Coordination Council of the Man and Biosphere Program (MaB) of UNESCO. It is approximately 60 km2, with a maximum length of 25 km parallel to the sea. Salinity fluctuates over a wide range between 0 and 36‰ (González-Castro et al. 2009), depending upon the tidal stage and the force and direction of the wind. Water temperature seasonally ranges between 3 and 25 °C.
Fish sampling
A total of 80 adult specimens belonging to the genus Odontesthes were purchased, from sport and artisanal fishermen upon landing, in five localities from Argentina (Fig. 1): (a) Mar Chiquita Coastal Lagoon (MCh), a brackish water environment, only employed for objective B); (b) Mar Chiquita coast (MCh_coast) (marine environment); (c) Mar del Plata coast (MdP) (marine environment); (d) San Blas Bay (SBB) (marine environment); (e) Comodoro Rivadavia (CR) (marine environment). Fishes were transported to the laboratory, where they were measured and taxonomically identified by means of morphological identification keys of Bemvenuti (2002), Cousseau et al. (2004) and Dyer (2006).
DNA extraction and amplification
White muscle tissue was obtained from 80 individuals of Odontesthes species (Fig. 1; Table 1), and preserved in 96% ethanol (− 20 °C) for genetic analysis. The specimens were photographed, labelled and formalin fixed (with further alcohol long-term preservation). Also, when possible, they were deposited as vouchers in the fish collection of the Instituto de Investigaciones Marinas y Costeras (IIMyC) (FCEyN-UNMDP-CONICET), Argentina.
The DNA extraction, polymerase chain reaction (PCR), and sequencing of the COI gene were performed according to standard DNA barcoding protocols (Ivanova et al. 2006), employing primer cocktails developed for fishes (Ward et al. 2005; Ivanova et al. 2007). The extraction and amplification of DNA were performed at the International Barcode of Life reference Barcode Laboratory of CONICET at IIMyC (Mar del Plata, Argentina).
Amplification of the 5′ region of COI, primers combinations, PCR reaction profile and amplicons visualization were performed according González-Castro et al. (2016). Sequencing was performed at the Canadian Centre for DNA Barcoding (CCDB) in Ontario (Canada), and in MACROGEN (Seoul, Korea). Sequencing reactions applied M13 forward and reverse primers using the Big Dye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems Inc.), and the reaction profile was comprised of an initial step of 2 min at 96 °C and 35 cycles of 30 s at 96 °C, 15 s at 55 °C, and 4 min at 60 °C. Products were directly sequenced using an ABI 3730 capillary sequencer according to manufacturer’s instructions.
Molecular analysis
Objective A
Molecular diversity: BIN analysis
The Barcode Index Number (BIN) labels operational taxonomic units (OTUs) initially generated through single linkage clustering and subsequently refined through Markov clustering (Ratnasingham and Hebert 2013). This approach creates a species-level taxonomic registry based on the analysis of patterns of nucleotide variation in the barcode region of the cytochrome c oxidase I (COI) gene. Since these OTUs show high concordance with prior morphological taxonomy, BINs can be used to verify species identifications as well as document diversity when taxonomic information is lacking. In this paper the BIN was used as a benchmark for testing the congruence between taxonomic and molecular resolution of marine species of Odontesthes in the SWAO as well as to explore the concordance of our findings with other public sequences of the same species with a known BIN. This approach, referred as the BIN discordance report within the BOLD toolbox, performs this “validation” by comparing the taxonomy of input records against all others in the same BINs. In discordant BINs, the K2P NJ trees provided by BOLD were explored in order to analyse internal relationships among sequences.
All sequence assemblies performed at CCDB, (including electropherogram (trace) files, primer sequences and specimen provenance data) were deposited in the “Odontesthes of Argentina” Project (Project-code: OdArg) on BOLD. This project also contains digital images of the morphological voucher specimens, sex and ontogenetic stage (juvenile or adult), total and standard body length as well as GPS coordinates for all collection localities.
Genetic divergence and interspecific relationships
All analyses were conducted using MEGA version 6 (Tamura et al. 2013). DNA sequences were aligned by the Muscle algorithm (Edgar 2004) and further double-checked visually. Each distinct sequence was considered a different haplotype. The number of base substitutions per site from averaging over all sequence pairs between and within species-groups was estimated. Sequences were deposited in Genbank (Table S1).
The K2P + G model was chosen for comparison purposes, as it was determined as the best-fit model under Akaike information criterion for neighbour-joining (NJ) and maximum-likelihood (ML) analyses. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.16). The analysis involved 62 nucleotide sequences. There were 652 positions in the final dataset.
A NJ analysis was performed to provide a graphic representation of divergences between species. The robustness of the obtained tree was tested using bootstrap analysis (Felsenstein 1985) with 1000 replicates.
As distance-based models erase all character-based information (DeSalle 2006), the best nucleotide substitution model was also employed to perform a maximum-likelihood (ML) analysis. Robustness of trees was tested using bootstrap analysis with 1000 replicates. To test how marine Odontesthes are genetically related to its freshwater congeners (objective a), available public sequences were downloaded from BOLD and GenBank.
Objective B
Diversity and distribution of haplotypes of putative estuarine and marine populations of O. argentinensis
For this objective, the COI sequences of 18 individuals of O. argentinensis from MCh were incorporated. Therefore, 40 individuals grouped in three putative populations were analysed: (i) MCh (N = 18); (ii) MCh_coast/MdP; grouped marine localities due to its proximity and environmental similarity) (N = 14) and (iii) SBB (N = 8).
Basic sequence properties and polymorphisms such as nucleotide (π) and haplotype (h) diversities were examined with DNASP 5.10 (Librado and Rozas 2009). Genetic variation was partitioned into three components: among groups (Fct), among populations within groups (Fsc), and among individuals within populations (Fst) The genetic population structure was assessed by analysis of molecular variance (AMOVA) based on traditional F-statistics as implemented in Arlequin 3.5 (Excoffier and Lischer 2010). A phylogenetic network based on differences in nucleotide sequences was constructed according to the median-joining method using the software Network (Bandelt et al. 1999).
Past demographic histories of populations were tested for population size changes with Tajima’s D and Fu’s F neutrality tests as implemented in DNASP (Tajima 1989; Fu 1997). They represent the deviation from neutrality, which is based on the expectation of a constant population size at mutation-drift equilibrium. A negative Tajima’s D denotes an excess of low frequency polymorphisms relative to expectation, indicating population size expansion and/or positive selection (Tajima 1989). A positive Tajima’s D implies low levels of both, low and high frequency polymorphisms, indicating a decrease in population size and/or balancing selection (Fu 1997).
Results
Taxonomic and molecular diversity
The silversides analysed were morphologically identified unambiguously as belonging to five different marine species of the genus Odontesthes (Table 1). The molecular analysis revealed a 100% of agreement between each morphological identification and its respective molecular identity (COI sequence) (Table 1).
Odontesthes argentinensis, O. platensis, O. smitti, O. nigricans and O. incisa were assigned to five different BINs (Table 2). Nevertheless, when compared with public sequences available in BOLD, the scenario become less discrete. The BIN discordance report showed a lack of concordance between taxonomic identification of some specimens from the SWAO and species of Odontesthes contained in two BINs. A thorough examination of each BIN showed that only three species of Odontesthes (O. platensis, O incisa, and O. nigricans) possess private BINs (i.e. no other species are included in these BINs). Conversely, the BINs corresponding to O. argentinensis and O. smitti from the SWAO included more than one species. Particularly, the BIN AAB5756 mostly included O. smitti from Argentina, but also their marine relatives from Chile O. regia and O. gracilis, and a freshwater species, O. hatcheri. On the other hand, the BIN AAB5755 included the marine O. argentinensis and five freshwater species, O. bonariensis, O. perugiae, O. humensis, O. mauleanum and O. hatcheri. Even when both discordant BINs amalgamate several species, their K2P NJ tree topologies showed different pictures of clustering within the BINs. The BIN AAB5756 displayed four evident clusters (Fig. S2). Two distant clusters included specimens of O. regia and O. gracilis from Perú and Chile. A cluster mostly containing O. smitti from Argentina and other one conformed by O. hatcheri from Argentina and Chile constituted the two remaining clusters of BIN AAB5756. Conversely, the BIN AAB5755 did not show any evident pattern in sequence assemblage (S3 Figure).
Haplotype diversity in marine species of SWAO
Partial COI sequences of 652 bp were successfully amplified for 80 specimens of Odontesthes (Table 1), including all the marine specimens (N = 62) and those estuarine (N = 18) specimens of O. argentinensis, employed only for the objective B. Among the 62 COI samples of marine silversides, 29 haplotypes were recovered (Table 3), with 68 variable (polymorphic) sites of which 54 were parsimony informative characters. The Haplotype diversity (h) estimated was 0.938 (s.d = 0.015).
No shared haplotypes were found between any of the species analysed. Although the sample size of the studied species was nearly identical (N = 9–11 specimens per species, with the exception of O. argentinensis, with N = 22) it is noticeable the different number of haplotypes/specimens obtained. At this respect, O. incisa with just nine specimens sampled showed eight haplotypes, with the consequent highest haplotypes/specimens rate (0.89). Conversely, O. platensis presented only two haplotypes for ten specimens (Table 3).
Genetic divergence
An overall mean distance of 0.0388 (0.0076 s.e.) was obtained. The number of base substitutions per site from averaging over all sequence pairs between and within species-groups is shown (Table 4). The highest divergence values for marine species were 0.0711 (O. argentinensis−O. incisa) and 0.065 (O. argentinensis−O. nigricans), whereas the lowest value was recorded for O. smitti−O. platensis (0.0275).
Interspecific relationships
The NJ and ML analyses (based on K2P + G model) generated trees with nearly identical topologies, where all marine species of Odontesthes clustered separately in a unique monophyletic phylogroup (100% of robustness), comprising five well defined haplogroups (bootstrap values ranged between 99 and 100%). There was no evidence for cryptic diversity (Fig. 2). Tree topology showed two major branches that constitute the Odontesthes phylogroups. One of them was conformed only by O. incisa (the basal taxon), while the other branch was further subdivided in two: one composed by O. nigricans and the other by O. smitti, O. platensis and O. argentinensis (Fig. 2). Odontesthes argentinensis and O. platensis were the most derivative haplogroups (71% bootstrap support) (Fig. 2).
ML tree (using the K2P + G model of molecular evolution) of the COI sequences for the marine species of Odontesthes obtained by MEGA. Bootstraps values are indicated on nodes. Outgroups: A. breviceps (Atherina breviceps); A. boyeri (Atherina boyeri); M. curema (Mugil curema); M. liza (Mugil liza). Specimen code of marine Odontesthes as in Table S1 (Supporting Information)
The more comprehensive phylogenetic analysis, which included freshwater congeneric species, showed that Odontesthes clustered in two branches, where O. incisa remains in a basal position (100% robustness) constituting alone one of those branches (Fig. 3). The freshwater species of Odontesthes (O. bonariensis, O. perugiae, and one O. hatcheri specimen) clustered within the marine species clade of O. argentinensis. Odontesthes smitti samples constituted a cluster with O. regia, O. gracilis and the remain O. hatcheri specimens included in the analysis (83% robustness), where the highest divergence observed is between O. smitti and O. hatcheri samples (0.016). Unexpectedly, O. platensis was more related to the Chilean freshwater/estuarine species O. mauleanum and O. brevianalis although with a genetic divergence of 0.033. No genetic divergence was observed between both Chilean Odontesthes species.
ML tree (using the K2P + G model of molecular evolution) of the COI sequences for the species of Odontesthes (marine and freshwater) obtained by MEGA. Bootstraps values are indicated on nodes. Size of triangles is proportional to number of haplotypes. Mugil curema and Mugil liza were used as outgroups
Diversity and distribution of haplotypes in putative estuarine and marine populations of O. argentinensis
The 40 COI sequences analysed, corresponding to O. argentinensis from estuarine (MCh, N = 18) and marine specimens (MdP/MCh_coast, N = 14; SBB, N = 8) showed 21 variable sites, equivalent to 15 haplotypes.
The haplotype network showed a star-shaped topology (Fig. 4). The most frequent haplotypes were Hp10 (represented by 13 individuals) and Hp6 (represented by 11 individuals). Haplotype 10 differs from Hp6 in a single step mutation, and was almost exclusive from Mch (12 individuals), including only one specimen from MdP/MCh_coast. Instead, Hp6 was the central haplotype and included individuals from the three putative populations, being mostly constituted by MdP/MCh_coast specimens (MdP/MCh_coast, N = 8; SBB, N = 2; MCh, N = 1). Moreover, Hp6 was shortly interconnected by only one step mutation to most (nine) haplotypes. Haplotype Hp11 also included more than one individual, shared by two individuals of MCh and one from MdP/MCh_coast. Haplotypes Hp1, Hp2, Hp3, Hp4 and Hp5 were private haplotypes for SBB. Haplotypes Hp14 and Hp15 were private for MCh and Hp7, Hp8, Hp9, Hp12 and Hp13 for MdP/MCh_coast.
Haplotype network (constructed with NETWORK software) based on COI sequences for the putative populations of Odontesthes argentinensis. Black dots correspond to missing haplotypes and circle size is proportional to haplotype frequency. Numbers correspond to haplotype. Different colours indicate the collection sites: green, Mar Chiquita coastal lagoon; blue, Mar del Plata/Mar Chiquita coasts; yellow, San Blas Bay
Genetic variability was high for the pooled samples, displaying a value of 0.824 for haplotype diversity (h) and 0.00302 for nucleotide diversity (π). Haplotype diversity (h) were 0.5556 in MCh, 0.73333 in MdP/MCh_coast and 0.9286 in SBB, while π vary from 0.0483 (MCh), 0.0698 MdP/MCh_coast and 0.1666 (SBB). AMOVA showed significant overall differentiation (Fst = 0.238; p = 0.00001) for the entire data set. Pairwise Fst showed significant values in all cases, and varied from 0.11 to 0.31. The highest value was found for the pairwise comparison between MCh and SBB populations, while the lowest value was detected between SBB and MdP/MCh_coast (Table 5). The AMOVA revealed that 9% of the genetic variance was found among groups (FCT = 0.089; p = 0.652). The differences among populations within groups accounted for 15% (FSC = 0.164; p = 0.036) of the total variation, while 76% of the variance could be attributed to among individuals within-population variability (Fst = 0.238; p = 0.000). The results of Tajima’s D and Fu’s Fs tests showed that only the population of MdP/MCh_coast presented significant negative values [(− 2.18 (p = 0.0.004); − 4.30 (p = 0.001), respectively)].
Discussion
Taxonomic and molecular diversity
The use of DNA Barcoding allowed the discrimination of all marine species of the genus Odontesthes from the SWAO with a 100% of concordance with the prior taxonomic identification. However, when this data was contrasted with the large amount of sequences stored in BOLD, these findings were only partially supported. Particularly, some freshwater species of Odontesthes, such as O. bonariensis, O. perugiae, O. humensis, O. mauleanum and O. hatcheri, received the same BIN (AAB5755) as its marine counterpart O. argentinensis. Rueda et al. (2017) assessed the impact of the introduction of the nonnative O. bonariensis in Patagonia (where the patagonic silverside, O. hatcheri inhabits). These authors analysed microsatellite markers and mitochondrial DNA and quantified the incidence of hybridization between these two species. They concluded that in several areas, introductions resulted in extensive hybridization, with high frequencies of F2 and backcrossed hybrids in natural populations. This could be the explanation of our results (O. hatcheri sharing BIN with O. bonariensis). Accordingly and as expected, in our phylogenetic analyses some of these freshwater species clustered tightly within the clade of O. argentinensis. The lack of resolution of the COI gene to discriminate between these species of Odontesthes was already reported (García et al. 2014). Particularly, it was already noted that additional molecular (Hughes et al. 2017) or morphometric (González-Castro et al. 2016) approaches are needed in order to unambiguously discriminate between the marine O. argentinensis and the freshwater O. bonariensis. Conversely, the lack of resolution between marine species as O. smitti, O. regia and O. gracilis using the COI gene was not reported until this study. The samples of O. smitti from the SWAO received the BIN AAB5756 which also hosts sequences labeled as O. regia, O. gracilis, O. hatcheri, O. argentinensis and O. nigricans. A private BIN for O. argentinensis is not available, but disparate different taxonomic features between O. argentinensis and O. smitti (Cousseau et al. 2004; Dyer 2006; Cousseau and Perrotta 2013) strongly suggested that sequences of O. argentinensis receiving the BIN AAB5756 are misidentifications. Our results also confirm that specimens of O. nigricans assigned to the BIN AAB5756 represents a misidentification, since a private BIN (AAF4482) was assigned by BOLD for the SWAO specimens of this species identified in this study. The lack of resolution of the COI gene at the species level for the genus Odontesthes is not universal. Our survey revealed that at least three species of this genus (O. incisa, O. platensis and O. nigricans) can be unambiguously identified by means of this molecular marker.
Irrespective of the concordance between taxonomic identification and genetic identity using the COI gene as a molecular marker, our results showed a strong structure in the COI sequence composition for species of Odontesthes from the SWAO as revealed by the large genetic divergence between-BINs. The observed genetic divergences among COI spanned the expected range for congeneric species of Atheriniform fishes (Heras and Roldán 2011).
Haplotype diversity in marine species of SWAO
The high haplotype diversity (0.824) observed in the present work may partially obey to the high haplotype/specimen ratio observed in some species. This ratio was particularly high in the small O. incisa with almost one haplotype per specimen analysed. These results suggest that a population molecular approach for this species should be conducted in a near future, in order to go deep in its genetic-biogeographic structure.
Interspecific relationships
Among the five well-defined haplogroups constituted in the Maximum Likelihood analysis, O. incisa was the basal taxon, suggesting that this small silverside would be the most ancient-marine Odontesthes species from the SWAO. Campanella et al. (2015) obtained similar results based on a time-calibrated phylogeny, suggesting that this small marine silverside would be the ancient species of the genus.
Two opposite hypotheses are currently on debate about the continental or marine origin of new world silversides (Odontesthes): Dyer postulated a continental origin of the genus, while Lahille and White proposed a marine origin (Dyer 1998; Lahille 1929; White 1986). So far, the fossils records are scarce: Bogan et al. (2009) found fossil remains of Odontesthes (Middle Pleistocene) corresponding to freshwater deposits. The ancient records are referred to Miocene age, which would correspond to freshwater deposits (Dyer 1998; Bogan et al. 2009). The reconstruction of habitat occupancy for Atheriniformes performed by Campanella et al. (2015) showed that, within Atherinopsidae (new world silversides), the ancestors of subfamilies Atherinosinae (from which Odontesthes belong) and Menidiinae were reconstructed as marine, supported by a high probability (76%).
Diversity and distribution of haplotypes in putative estuarine and marine populations of O. argentinensis, in the context of recurrent marine to freshwater speciation
The diversity and distribution of haplotypes strongly suggests that the three localities compared correspond to different populations of O. argentinensis, which could be under incipient speciation events. Each locality presented several private and distinctive haplotypes. Moreover, MCh exhibited a, highly abundant, almost-unique haplotype (Hp10; 12/13 individuals) that remark its differences from the other populations analyzed. The Hp6, predominantly marine, would be the origin of freshwater MCh Hp10.
Odontesthes argentinensis is a widely distributed western Atlantic coastal species, occurring in marine and estuarine environments from the Sao Paulo State, in Brazil, southwards Chubut province, Argentina (Dyer 2006). There is at present increasing published evidence, which strongly suggest that its estuarine-populations are under incipient-speciation events, associated to significant behavioural and ecological divergence, relative to its incursion/adaptation to these brackish environments (Beheregaray and Sunnucks 2001; Moresco and Bemvenuti 2006; Heras and Roldán 2011; Llompart et al. 2013; Díaz et al. 2016). In this regard, specimens of O. argentinensis inhabiting MCh would be an example of this process: González-Castro et al. (2009) suggested that there is a reproductively isolated population of O. argentinensis in MCh. Moreover, González-Castro et al. (2016) showed that estuarine (MCh) and marine populations of O. argentinensis studied are meristically and morphometrically distinguishable and appear to behave as well differentiated populations, or even incipient ecological species. In agreement of these facts, the present results (Fst pairwise comparisons, Table 5) support the existence of three populations-groups: one comprised by specimens of O. argentinensis from MCh Lagoon, and the others by specimens from MdP/MCh coast and SBB, respectively.
A significant excess of low-frequency haplotypes and thereby negative and significant values of both Tajima’s and Fu’s neutrality tests were observed in MdP/MCh coast population indicating a departure from neutrality. Neutrality tests yielded to different historical demographic scenarios, in which population-expansion could be proposed for O. argentinensis populations. This substantial genetic divergence occurs on a very small geographic scale as already found in other studies (Bemvenuti 2000; García et al. 2014).
Recurrent marine to freshwater speciation in fish has been widely documented. It was predicted that the new selective pressure occurring in novel habitats is responsible of the subsequent divergent natural selection. This, in turn, would promote adaptation to the new habitat and at last, the ecological incipient speciation (Schluter 2000; Beheregaray and Sunnucks 2001; Betancur-R et al. 2012; Lescak et al. 2015). Yamasaki et al. (2015) demonstrated considerable species diversity of Rhinogobius gobies by parallel life history divergence through colonization of and adaptation to various freshwater habitats. Takahashi et al. (2016) suggested that a diversification of cold-water adapted euryhaline fish, such as Pungitius and Cottus, was promoted by the isolation of lineages in discontinuous freshwater. Genomic analyses of the threespine stickleback fish, originated after the 1964 Great Alaska Earthquake, support the recent and repeated independent colonization of freshwater habitats by oceanic ancestors in these 50 years (Lescak et al. 2015). These authors find evidence of recurrent gene flow between oceanic and freshwater ecotypes where they co-occur.
All these previous/present evidence strongly suggest that incipient speciation is occurring in O. argentinensis populations of SWAO, and specimens from MCh would be considered at present as the leading candidate of a marine to freshwater incipient speciation event. In southern South America, highstands of the sea level during the Holocene may have had striking influence in the diversification of regional fauna. Particularly, it was suggested that the sea-level fluctuation during the Holocene has induced the emergence of new geographical extensions constrained by some conditioning factors as salinity regimes, habitats and substrates (Isla 2012). In this scenario, changes in salinity dynamics, landscape and habitats can induce changes in the distribution, isolation and therefore the speciation of some taxa (Isla 2012). Also, to recognize the natural trend in the inlet evolution of MCh, Isla (1997) analyzed historical maps, nautical charts and ancient aerial photographs. This survey revealed a complete isolation of MCh from the Ocean by the year 1748, as stated by Cardiel (1748) who mentioned 300 steps between the coastal lagoon and the ocean beach. An English chart drawn by Kitchin in 1772 (Isla 1997) described the inlet as “misfit for boats”, denoting the first record of an opening (small, but adequate for fish entry) of the Lagoon to the sea. In the following years, the inlet migrated to the north until it became blocked again (Storni 1915). Many times, man had to open the inlet: the farmers used to wait for strong winds from the north that piled up water to the southern shore of the lagoon and practiced a channel that rapidly became broader and deeper (Isla 1997).
These evidences suggests that the first entry in MCh of O. argentinensis could have happened either during the sea-level fluctuation of the Holocene (prox. 7000 years ago) or in more recent times (last three hundred years), when the first opening of the inlet was documented. Irrespective of that, the current evidence about marine to freshwater speciation in fishes suggests that both scenarios may have promoted in MCh the incipient speciation of O. argentinensis from marine origin we record in the present paper. Upcoming research employing higher-mutation rate nuclear and mitochondrial markers (RAD seq and d-Loop), as well as additional sampling locations, will expand the knowledge on this issue.
References
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Beheregaray LB, Sunnucks P (2001) Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Mol Ecol 10:2849–2866
Bemvenuti MA (2000) Diferenciação geográfica do peixe-rei Odontesthes argentinensis (Atherinopsidae), no extremo sul do Brasil, através da morfometria multivariada. Atlántica 22:71–79
Bemvenuti MA (2002) Diferenciação morfológica das espécies de peixes-rei, Odontesthes Evermann & Kendall (Osteichthyes, Atherinopsidae) no extremo sul do Brasil: morfometria multivariada. Rev Bras Zool 19:251–287
Bemvenuti MA (2006) Silversides in South Brazil: morphological and ecological aspects. Biocell 30:111–118
Betancur-R R, Orti G, Stein AM, Marceniuk AP, Pyron A (2012) Apparent signal of competition limiting diversification after ecological transitions from marine to freshwater habitats. Ecol Lett 15:822–830
Betancur-R R, Broughton RE, Wiley EO et al (2013) The tree of life and a new classification of bony fishes. PLoS Curr. https://doi.org/10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288
Bloom DD, Weir JT, Piller KR, Lovejoy NR (2013) Do freshwater fishes diversify faster than marine fishes? A test using state-dependent diversification analyses and molecular phylogenetics of New World silversides (Atherinopsidae). Evolution 67:2040–2057
Bogan S, de los Reyes ML, Cenizo MM (2009) Primeros registros fósiles de pejerreyes (Telostei: Atheriniformes) en el Pleistoceno Medio de la provincia de Buenos Aires, Argentina. Rev Mus Argentino Cienc Nat 11(2):185–192
Campanella D, Hughes LC, Unmack PJ, Bloom DD, Piller KR, Ortí G (2015) Multi-locus fossil-calibrated phylogeny of Atheriniformes (Teleostei, Ovalentaria). Mol Phylogenet Evol 86:8–23
Capurro LRA (1981) Características físicas del Atlántico Sudoccidental. In: Boltovskoy D (ed) Atlas de1 Zooplancton del Atlántico Sudoccidental y métodos de trabajo con el zooplancton marino. Publicaciones Especiales INIDEP, Mar de1 Plata, pp 219–225
Cardiel J (1748) Diario del Viaje y Misión a1 Río del Sauce (Río Negro) por Fines de Marzo de 1748. Inst Invest Geogr Buenos Aires, 1930, p 278
Cousseau MB (2010) Ictiología. Aspectos Fundamentales. La vida de los peces sudamericanos. EUDEM, Mar del Plata
Cousseau MB, Perrotta RG (2013) Peces marinos de Argentina: biología, distribución, pesca. INIDEP, Mar del Plata
Cousseau MB, Gosztonyi AE, Elías I, Re ME (2004) Estado actual del conocimiento de los peces de la plataforma continental argentina y adyacencias. In: Sánchez RP, Bezzi SI (eds) El mar argentino y sus recursos pesqueros Tomo 4. Los peces marinos de interés pesquero. Caracterización biológica y evaluación del estado de explotación. INIDEP, Mar del Plata, pp 17–38
DeSalle R (2006) Species discovery versus species identification in DNA barcoding efforts: response to Rubinoff. Conserv Biol 20:1545–1547
Díaz J, Villanova GV, Brancolini F et al (2016) First DNA Barcode reference library for the identification of South American freshwater fish from the Lower Paraná River. PLoS ONE 11(7):e0157419. https://doi.org/10.1371/journal.pone.0157419
Díaz de Astarloa JM, Mabragaña E, Hanner R, Figueroa DE (2008) Morphological and molecular evidence for a new species of longnose skate (Rajiformes: Rajidae: Dipturus) from Argentinean waters based on DNA barcoding. Zootaxa 1921:35–46
Dyer BS (1998) Phylogenetic systematics and historical biogeography of the Neotropical silverside family Atherinopsidae (Teleostei, Atheriniformes). In: Malabarba LR, Reis RE, Vari RP, Lucena ZM, Lucena CAS (eds) Phylogeny and classification of neotropical fishes. Edipucrs, Porto Alegre, pp 519–536
Dyer BS (2006) Systematic revision of the South American silversides (Teleostei, Atheriniformes). Biocell 30:69–88
Dyer BS, Chernoff B (1996) Phylogenetic relationships among atheriniform fishes (Teleostei: Atherinomorpha). Zool J Linn Soc 117:1–69
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol Ecol Resour 10:564–567
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
García G, Ríos N, Gutiérrez V, Guerra-Varela J, Bouza-Fernández C, Gómez-Pardo B, Martínez-Portela P (2014) Promiscuous speciation with gene flow in silverside fish genus Odontesthes (Atheriniformes, Atherinopsidae) from South Western atlantic ocean basins. PLoS ONE 8:e104659
González-Castro M, Díaz de Astarloa JM, Cousseau MB et al (2009) Fish composition in a south-western Atlantic temperate coastal lagoon: spatial-temporal variation and relationships with environmental variables. J Mar Biol Assoc UK 89:593–604
González-Castro M, Rosso JJ, Mabragaña E, Díaz de Astarloa JM (2016) Surfing among species, populations and morphotypes: inferring boundaries between two species of new world silversides (Atherinopsidae). CR Biol 399(1):10–29
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identification through DNA barcodes. Proc R Soc Lond B Biol 270:313–321
Helfman S, Collette BB, Facey DE, Bowen BW (2009) The diversity of fishes, biology, evolution and ecology, 2nd edn. Wiley-Blackwell, Oxford
Heras S, Roldán MI (2011) Phylogenetic inference in Odontesthes and Atherina (Teleostei: Atheriniformes) with insights into ecological adaptation. CR Biol 334:273–281
Hughes LC, Somoza GM, Nguyen BN, Bernot JP, González-Castro M, de Astarloa JMD, Ortí G (2017) Transcriptomic differentiation underlying marine-to-freshwater transitions in the South American silversides Odontesthes argentinensis and O. bonariensis (Atheriniformes). Ecol Evol 7:5258–5268. https://doi.org/10.1002/ece3.3133
Isla F (1997) Seasonal behaviour of Mar Chiquita tidal inlet in relation to adjacent beaches, Argentina. J Coastal Res 13(4):1221–1232
Isla F (2012) Highstands of the sea level and the speciation of coastal communities: opportunities for the new territories in Southern South America. Biodivers Chile 7:45–59
Ivanova NV, deWaard JR, Hebert PDN (2006) An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes 6:998–1002
Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes 7:544–548
Lahille F (1929) El pejerrey. Boletín del Ministerio de Agricultura de la Nación 28(3):260–395
Lescak EA, Bassham SL, Catchen J, Gelmond O, Sherbick ML, von Hippel FA (2015) Evolution of stickleback in 50 years on earthquake-uplifted islands. PNAS 112(52):E7204–E7212. https://doi.org/10.1073/pnas.1512020112
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Llompart FM, Colautti DC, Maiztegui T, Cruz-Jimenez AM, Baigún CRM (2013) Biological traits and growth patterns of pejerrey Odontesthes argentinensis. J Fish Biol 82:458–474
Mabragaña E, Díaz de Astarloa JM, Hanner R, Zhang J, González-Castro M (2011) DNA Barcoding identifies Argentine fishes from marine and brackish waters. PLoS ONE 6:e28655
Matthew E, Neilson ME, Stepien CA (2009) Evolution and phylogeography of the tubenose goby genus Proterorhinus (Gobiidae: Teleostei): evidence for new cryptic species. Biol J Linn Soc 96:664–684
Moreira AL, Taylor EB (2015) The origin and genetic divergence of black” kokanee, a novel reproductive ecotype of Oncorhynchus nerka. Can J Fish Aquat Sci 72:1584–1595
Moresco A, Bemvenuti MA (2006) Biologia reprodutiva do peixe-rei Odontesthes argentinensis (Valenciennes) (Atherinopsidae) da região marinha costeira do sul do Brasil. Rev Bras Zool 23:1168–1174
Nelson JS, Grande TC, Wilson MVH (2016) Fishes of the world, 5th edn. Wiley, New Jersey
Ratnasingham PDN (2013) Hebert P (2013) A DNA-based registry for all animal species: the barcode index number (BIN) system. PLoS ONE 8:e66213
Rosso JJ, Mabragaña E, González-Castro M, de Astarloa JMD (2012) DNA barcoding Neotropical fishes: news from the Pampa Plain, Argentina. Mol Ecol Res 12:999–1011
Rosso JJ, Rueda EC, Sanchez S et al (2017) Basin-scale distribution and haplotype partitioning in different genetic lineages of the Neotropical migratory fish Salminus brasiliensis. Aquat Conserv. https://doi.org/10.1002/aqc.2830
Rueda EC, Mullaney KA, Conte-Grand C, Evelyn MH, Cussac V, Ortí G (2017) Displacement of native Patagonian freshwater silverside populations (Odontesthes hatcheri, Atherinopsidae) by introgressive hybridization with introduced O. bonariensis. Biol Invasions 19:971–988. https://doi.org/10.1007/s10530-016-1295-y
Schluter D (2000) The ecology of adaptive radiation. Oxford University Press, Oxford
Storni S (1915) Informe sobre el levantamiento hidrográfico de la Laguna Mar Chiquita y alrededores. Anuario Hidrográfico 1915:294–299
Tajima F (1989) Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Takahashi H, Moller PR, Shedko SV, Ramatulla T, Joen SR, Zhang CG, Sideleva VG, Takata K, Sakai H, Goto A, Nishida M (2016) Species phylogeny and diversification process of Northeast Asian Pungitius revealed by AFLP and mtDNA markers. Mol Phylogenet Evol 99:44–52
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Taylor EB, Foote CJ, Wood CC (1996) Molecular genetic evidence for parallel life-history evolution within a Pacific salmon (sockeye salmon and kokanee, Oncorhynchusnerka). Evolution 50:401–416
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc B 360:1847–1857
Ward RD, Hanner R, Hebert PDN (2009) The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol 74:329–356
White BN (1986) The Isthmian link, antitropicality and American biogeography: distributional history of the Atherinopsinae (Pisces: Atherinidae). Syst Zool 35(2):176–194
Yamasaki YY, Nishida M, Suzuki T, Mukai T, Watanabe K (2015) Phylogeny, hybridization, and life history evolution of Rhinogobius gobies in Japan, inferred from multiple nuclear gene sequences. Mol Phylogenet Evol 90:20–33
Yokoyama R, Goto A (2005) Evolutionary history of freshwater sculpins, genusCottus (Teleostei; Cottidae) and related taxa, as inferred from mitochondrial DNA phylogeny. Mol Phylogenet Evol 36:654–668
Acknowledgements
This work was supported by CONICET (PIP No. 11220130100339), MINCYT (PICT-2014-0665) and also personal funds of MGC. The authors would like to thank: Julio Mangiarotti (forest guard of Mar Chiquita Biosphere Reserve), Daniel Giménez (silversides sport-game fishing expert); Pablo Rizzo, Cristian Di Paolo and Marcelo Pons (sport game fishing guides of Mar Chiquita); Daniel Blanco, Santiago Gaudioso and Juan Pablo Gaudioso (San Gabriel and Juan y Juan fishing-points of Mar Chiquita); Carlos Martin (fisherman of Mar del Plata); Mónica Iza and Florencia Celesia (Mar Chiquita Town Hall) and Mar Chiquita Town Hall authorities (Flavia Laguné and Carlos Alberto Ronda).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interests.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed consent
Not aplicable. No studies with human participants were performed by any of the authors.
Research involving human participants and/or animals
This article does not contain any studies with human participants performed by any of the authors.
Statement on the welfare of animals
Fish under study are not protected (local restrictions, IUCN or CITES listed species) under wildlife conservation. No experimentation was conducted on live specimens in this study, as in fact they were no longer alive when were obtained from sport (Mar Chiquita Coastal lagoon, Mar Chiquita coast and San Blas Bay) and artisanal fishermen (Mar del Plata coast, Comodoro Rivadavia coast) upon landing. The locations involved in the study were not part of any protected area, except for Mar Chiquita Coastal lagoon; however, as stated above, fishes obtained in this lagoon came from sport game fishermen.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2019_66_MOESM1_ESM.doc
Supplementary material 1 Table S1. Specimen code of individuals employed for Phylogenetic analysis (ML, Fig. 2) of marine Odontesthes from Argentina (DOC 94 kb)
10709_2019_66_MOESM2_ESM.pdf
Supplementary material 2 Figure S2. K2P Neighbour-Joining Tree of COI sequences available in BOLD included in the BIN AAB5756 (PDF 8 kb)
10709_2019_66_MOESM3_ESM.pdf
Supplementary material 3 Figure S3. K2P Neighbour-Joining Tree of COI sequences available in BOLD included in the BIN AAB5755 (PDF 13 kb)
Rights and permissions
About this article
Cite this article
González-Castro, M., Rosso, J.J., Delpiani, S.M. et al. Inferring boundaries among fish species of the new world silversides (Atherinopsidae; genus Odontesthes): new evidences of incipient speciation between marine and brackish populations of Odontesthes argentinensis. Genetica 147, 217–229 (2019). https://doi.org/10.1007/s10709-019-00066-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-019-00066-2