Abstract
DNA barcoding by sequencing a standard region of cytochrome c oxidase subunit I (COI) provides an accurate, rapid method for identifying different species. In this study, we provide a molecular taxonomic assessment of demersal fishes in the Bering Sea and Chukchi Sea based on DNA barcoding, and a total of 123 mitochondrial COI partial fragments with a length of 652 bp were obtained. The consensus among all sequences was determined by alignment via a BLAST search in GenBank. Phylogenetic relationships were reconstructed based on neighbor-joining trees and barcoding gaps. The 39 species investigated in this analysis were distributed among 10 families. Five families within Scorpaeniformes including 19 species accounted for almost half of the species. The next largest group was Perciformes, with 9 species, followed by Pleuronectiformes and Gadiformes, with 5 species each, and the smallest number of species belonged to Rajiformes. At the family level, Cottidae was the largest family, followed by Zoarcidae, accounting for 8 species. The other eight families—Gadidae, Pleuronectidae, Psychrolutidae, Agonidae, Liparidae, Ammodytidae, Hexagrammidae, and Rajidae—accounted for a smaller proportion of species. In brief, our study shows that DNA barcodes are an effective tool for studying fish diversity and taxonomy in the Bering Sea and Chukchi Sea. The contribution of DNA barcoding to identifying Arctic fish species may benefit further Arctic fish studies on biodiversity, biogeography and conservation in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The correct identification of species is a prerequisite for studying fish diversity. Traditional morphology-based identification systems rely mostly on expert experience and the integrity of samples (Li et al. 2017). Furthermore, some taxa show a variety of complex characteristics, such as sexual dimorphism (Kenchington et al. 2017) or developmental variability of larvae (Batta-Lona et al. 2019; Webb et al. 2006). Therefore, identification based on morphological features is very difficult, complex and error prone. The limitations inherent in traditional identification entail the need for a new approach to species recognition. DNA barcoding is a technology for identifying species based on sequence diversity in cytochrome c oxidase subunit I (COI) (Hebert et al. 2003b). To date, there have been many studies showing the effectiveness of the COI gene for species identification in diverse animals (Hebert et al. 2003a, 2003b), including fishes (McCusker et al. 2013; Ward et al. 2005). This technology is free from excessive dependence on experience and can allow the automation and standardization of specimen identification to be realized. It provides a powerful supplement to traditional taxonomy and species identification methods. DNA barcodes can be used not only to identify whole fish but also to identify fry, roe, fish meat, fish fins, fish products or other body fragments that are difficult to identify based on morphology (Smith et al. 2008; Ward et al. 2005). Therefore, the use of DNA barcodes as an accurate and effective method of species identification is currently favored by an increasing number of researchers. Recent studies have indicated that this technology is highly reliable and efficient in many fish groups, including freshwater fishes (Keskin et al. 2013), coral reef fishes (Ward et al. 2005), ocean fishes (McCusker et al. 2013), Antarctic fishes (Li et al. 2018), and Arctic fishes (Mecklenburg et al. 2010). Moreover, it is widely used in a variety of fields, such as biodiversity assessment, fish larva identification and fishery management (Gao 2015; Panprommin et al. 2020).
The Bering Sea is located at the northernmost tip of the Pacific Ocean, while the Chukchi Sea is the marginal sea of the Arctic Ocean. The two seas are connected through the narrow Bering Strait. The seasonal ice-covered Bering and Chukchi Sea shelves are among the largest continental shelves in the world. These high-latitude shelf systems are highly productive during both the ice melt and open-water periods (Huntington et al. 2020). As seawater warms and the extent of sea ice declines, the vulnerability of the ecosystem to environmental change is thought to be high (Grebmeier et al. 2006a). As a key component of the normal operation of marine ecosystems, fish exhibit a series of basic ecological functions and play an important role in determining the carrying status of ecosystems, reflecting changes in the ecological environment (Izzo et al. 2016). There have been many reported studies of fish species and fishery resources in the Bering Sea and Chukchi Sea. For example, analyses of the composition of fish species (Meyer 1997; Norcross et al. 2010), the distribution of fish species (Busby et al. 2005; Mecklenburg et al. 2010; Rand and Logerwell 2011) and the response of fish to changes in the Arctic environment have been performed (Grebmeier et al. 2006b; Mueter et al. 2009; Robertis and Cokelet 2012). However, there have been few studies on fish taxonomy in the Bering Sea and Chukchi Sea via DNA barcoding (Mecklenburg et al. 2010). Here, we examine COI diversity among 123 fish species, with the goal of examining whether DNA barcoding can achieve clear and definite species recognition in fish. Our study provides an important scientific basis for further studies regarding biodiversity, biogeography and conservation in the Bering Sea and Chukchi Sea.
Materials and methods
Specimen collection and morphological identification
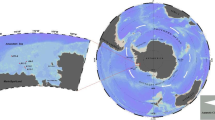
The samples were collected during the 6th (2014), 8th (2017) and 9th (2018) Chinese National Arctic Research Expeditions (CHINAREs). The surveyed sea areas were the Bering Sea continental slope, the Bering Sea continental shelf, the Chukchi Sea continental shelf and the Chukchi Sea continental slope area (the boundary of the Bering Strait is located at 65°05'N) (Fig. 1). All specimens were collected with a triangular bottom trawl net (20 mm mesh size; 6.5 m long, 2.2 m wide and 0.65 m high) in surveys conducted on the R/V Xuelong icebreaker. The time of each network operation was 10–60 min, with a speed of 3–4 kn. Specimens were fished from each station, and specimens from the same station were packaged together. Following morphological identification by visual inspection, all fish were classified by taxonomic specialists. The systemic classification and naming procedures were based primarily on "Fishes of the World (4th edition)" and the "Latin-Chinese Dictionary of Fish Names by Classification System". Muscle samples of fish were obtained and preserved in 95% ethanol for DNA extraction after morphological characterization and specimen identification. Then, the whole fish were preserved in a 95% ethanol solution and stored as voucher specimens at the Third Institute of Oceanography, Ministry of Natural Resources.
DNA extraction, amplification and sequencing
A total of 123 genomic DNA samples were extracted, including 16 from the 6th Arctic expedition, 24 from the 8th Arctic expedition, and 83 from the 9th Arctic expedition. Back muscle tissue of each fish was obtained and preserved in a centrifuge tube with 95% ethanol. A TransGen kit (Easy Pure Marine Animal Genomic DNA Kit) was used to extract the genomic DNA of the Arctic fish, which was then stored at 4 °C for later use. The primers used to amplify the COI gene fragment were F1:5′-TCAACCAACCACAAAGACATTGGCAC-3′ and R1:5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′ according to Ward et al. (2005). The PCR system had a volume of 25 μL, containing 2.5 μL of 10 × PCR buffer (including Mg2+), 2 μL of dNTPs (2 mmol•L–1), 1 μL of each primer, 0.25 μL of Taq DNA polymerase, 1 μL of the extracted DNA, and deionized water to the final of 25 μL. The thermal cycling program included an initial denaturation step of 4 min at 95 °C followed by 30 cycles of 0.5 min at 94 °C, annealing for 0.5 min at 52 °C, and extension for 0.5 min at 72 °C, with a final step of 10 min at 72 °C. Negative controls were included in all amplification reactions to confirm the absence of contaminants. The PCR products were visualized on 1.0% agarose stained with gel green (Biotium, Hayward, CA, USA), and successful amplification products were sent to Personalbio for purification and sequencing.
Data analysis
The original data obtained by sequencing were manually compared with the corresponding sequencing peak map to check for errors to ensure the accuracy of the data. The DNASTAR Lasergene software package was used to edit and align the sequences. All high-quality sequences were compared with the NCBI BLAST program to determine the species identity of the samples. Sequence similarity greater than 98% was the criterion for identification at the species level, and a similarity lower than 98% was used for identification at the genus level (Wong and Hanner 2008). Neighbor-joining (NJ) analysis implemented in MEGA 7.0 based on the K2P model with 1000 bootstrap replicates was employed to both calculate the genetic distances and examine the relationships among taxa.
Results
Morphological analysis
A total of 123 specimens were collected during three CHINAREs. Most of them were adults and could be directly distinguished. However, there were also some juvenile and incomplete specimens, which were difficult to identify based on morphological characteristics. These specimens were identified as Limanda sp., Hippoglossoides sp., Lycodes sp., Ammodytes sp., Hemilepidotus sp. and Liparis sp., etc. (Table 1).
Amplification and sequencing
A total of 123 mitochondrial COI gene DNA fragments were successfully amplified using primers. No stop codons, deletions or insertions were observed in any of the sequences after alignment. The length of the amplified COI gene was 652 bp. The number of haplotypes identified in each species ranged from 1 to 6. Overall nucleotide frequencies were C (27.7%), T (30.8%), A (23.4%), and G (18.2%). The 123 COI sequences were deposited in the GenBank database under the accession number MW435025—MW435147.
Species identification by phylogenetic analysis of COI sequences
The phylogenetic tree constructed by the NJ method is shown in Fig. 2. The same morphological species of fish formed cohesive units. All high-quality sequences were identified by BLAST searches and comparisons in GenBank, and the similarity was higher than 98% (Table 1). A total of 39 fish species belonging to 5 orders, 10 families and 23 genera were identified through DNA barcoding analysis in this survey. Among these species, 19 species of Scorpaeniformes accounted for 48.72% of the total species. Additionally, 9 species of Perciformes accounted for 23.08% of the total number of species, and 5 species from each of Pleuronectiformes and Gadiformes accounted for 12.82% of the total number of species. The smallest number of species was found in Rajiformes, which included only one species. At the family level, the number of Cottidae species was largest, at 9, accounting for 23.08% of the total number of species, followed by Zoarcidae, with 8 species, accounting for 20.51%. The other eight families—Gadidae, Pleuronectidae, Psychrolutidae, Agonidae, Liparidae, Ammodytidae, Hexagrammidae, and Rajidae—accounted for smaller proportions. At the genus level, the number of species from the genus Lycodes was greatest, at 8. Based on the NJ tree, all species from the same family were clustered together, indicating that the families were all monophyletic except for Cottidae, in which Hemilepidotus papilio was sister to other genera, thus, the NJ analysis recovered the family Cottidae as polyphyletic. Besides, Psychrolutidae was nested within Cottidae, indicating the complicated relationship between the two families.
The NJ tree showed that the barcoding results were not all consistent with the conventional taxonomy. However, 61.54% of species were identified successfully, showing at least 98% similarity. Icelus spiniger was distinctly different from I. spatula, with specimens identified as I. spatula morphologically sharing 100% nucleotide sequence similarity with the I. spiniger reference. Hippoglossoides elassodon was identified as H. robustus morphologically but presented 100% similarity to the H. elassodon reference. In similar cases, Lycodes palearis was identified as L. adolfi, Anisarchus medius was identified as A. macrops, Myoxocephalus Scorpius was identified as Artediellus atlanticus, and Gymnocanthus tricuspis was identified as G. detrisus. The sequences of Liparis bathyarcticus and L. tunicatus were distinctly different from the L. tanakae references. Some juvenile fish and incomplete individuals that were initially recorded as unidentified “sp.” were identified effectively based on barcoding. Limanda sp. shared 99% nucleotide sequence similarity with the reference L. aspera, indicating that it was L. aspera. Hippoglossoides sp. was identified as H. dubius with 99% nucleotide sequence similarity. Zoarcidae sp. was identified as L. polaris with 99% nucleotide sequence similarity. Ammodytes sp. was identified as A. hexapterus with 100% nucleotide sequence similarity. Hemilepidotus sp. was identified as H. papilio with 99% nucleotide sequence similarity. Liparis sp. was identified as L. gibbus with 99% nucleotide sequence similarity.
Genetic distance and barcoding gaps
The intraspecific distances ranged from 0 to 0.35%, and the minimum interspecific distances of the species were greater than 2% except for Liparis tunicatus vs L. fabricii (1.43%), Hippoglossoides elassodon vs H. robustus (0.62%), H. elassodon vs H. dubius (0.54%), H. robustus vs H. dubius (0.54%), and Icelus spatula vs I. spiniger (0.69%). Nevertheless, the minimum interspecific distance of all species was still greater than the maximum intraspecific distances. Thus, it was obvious that there were barcode gaps in the genetic distance between intraspecific distances and interspecific distance (Fig. 3).
Discussion
Correct species identification is the foundation for revealing fish diversity. Traditional morphological identification methods require the experience of high-level classification experts and sample preservation integrity. Among the samples utilized in this study, there were some juveniles and damaged individuals, and it was difficult to carry out morphological identification. Thus, many samples could only be identified to the family or genus level, such as Limanda sp., Hippoglossoides sp., Lycodes sp., Ammodytes sp., Hemilepidotus sp. and Liparis sp. At the same time, only a few species with similar morphological features could not be clearly distinguished. However, after we conducted a molecular evaluation of the fish in the Bering Sea and the Chukchi Sea, COI-based DNA barcoding was proven to be effective for identifying Arctic fish species, with 100% of species exhibiting monophyletic DNA clusters. Among the 39 species investigated in this study, only 24 species (61.54%) were identified correctly by morphological examination. However, all species were accurately identified with COI sequences. The effectiveness (number of species exhibiting monophyletic clusters) of the DNA barcoding analysis in our study was demonstrated to be higher than those found in other barcoding studies of fishes (Hubert et al. 2008; Steinke et al. 2009; Ward et al. 2005).
The existence of barcode gaps increases the effectiveness of DNA barcodes for identifying species. In this study, all the obtained minimum interspecific distances were greater than the maximum intraspecific genetic distances. A value of 2% has been suggested as a threshold value between species and genus divergence (Ward 2009). However, the genetic distances between species of Hippoglossoides, Icelus and Liparis were lower than 2% (0.54% to 1.43%), which was probably associated with recent speciation (McCusker et al. 2013). The NJ tree illustrated short genetic distances between congeneric species of the genera Hippoglossoides, Icelus and Liparis. The members of these three genera exhibited the shortest interspecific distances, which was consistent with recent molecular phylogenies (McCusker et al. 2013). Despite the low interspecific distance between these congeneric species, based on the phylogenetic tree, the species within each genus clustered closely together, formed their own branches and showed a high support rate (97–100%). McCusker et al. (2013) reported that barcoding was still effective (species were monophyletic) under conditions of low genetic divergence. Nevertheless, it is recommended that other molecular markers be combined in barcodes in future efforts aimed at the molecular identification of these species (Qu et al. 2018). Overall, 123 fish collected from the Bering Sea and the Chukchi Sea could be identified to the species level using COI sequences.
The phylogenetic tree constructed based on the obtained sequences showed cluster formation; clustering in the phylogenetic tree can help detect problems and is a valuable tool, especially for closely related species without obvious morphological differences (Dettai et al. 2011). Although barcode analysis is mostly used to delimit species boundaries, there are obvious phylogenetic signals within COI sequence information (Hebert et al. 2003a; Ward et al. 2005). In the phylogenetic tree based on the NJ method obtained in this study, different individuals of each species were clustered together. However, it should be noted that at the family level, H. papilio of Cottidae and Agonidae formed a separate branch while Psychrolutidae was nested within Cottidae. This may be the reason why Mecklenburg et al. (Mecklenburg et al. 2010) indicated that the internal relationships of the Cottidae are obscure and not well defined. However, H. papilio was represented in the specimens by only one specimen, and additional specimens will certainly be necessary to further clarify the relationship between H. papilio and Cottidae.
Conclusions
This study shows that DNA barcoding is an accurate and efficient method of species identification. A total of 123 fish collected from the Bering Sea and the Chukchi Sea were identified by DNA barcoding. Thirty-nine species from ten families were characterized; all species were identified correctly. We also observed low interspecific divergence (< 2%), probably associated with recent speciation. It is recommended that other molecular markers be included to develop unique DNA barcodes that are suitable for Arctic fish. In follow-up studies, it is necessary to combine morphology-based identification systems with DNA barcoding to identify species because morphological identification alone may not be sufficiently robust. In addition, our work provides important information for further studies regarding the biodiversity, biogeography and conservation of Arctic fishes.
Data availability
All data generated in this study are included in this published article [and its supplementary information] and in GenBank (NCBI) [https://www.ncbi.nlm.nih.gov/genbank] where sequences with accession numbers MW435025—MW435147 were deposited.
References
Batta-Lona PG, Galindo-Sanchez CE, Arteaga MC, Robles-Flores J, Jimenez-Rosenberg SPA (2019) DNA barcoding and morphological taxonomy: identification of lanternfish (Myctophidae) larvae in the Gulf of Mexico. Mitochondrial DNA A DNA Mapp Seq Anal 30(2):375–383. https://doi.org/10.1080/24701394.2018.1538364
Busby MS, Mier KL, Brodeur RD (2005) Habitat associations of demersal fishes and crabs in the Pribilof Islands region of the Bering Sea. Fish Res 75(1):15–28. https://doi.org/10.1016/j.fishres.2005.05.012
Dettai A et al (2011) The actinopterygian diversity of the CEAMARC cruises: barcoding and molecular taxonomy as a multi-level tool for new findings. Deep Sea Res Part II 58(1–2):250–263. https://doi.org/10.1016/j.dsr2.2010.05.021
Gao L (2015) Applications of DNA barcoding in biodiversity inventory and assessment. Biodivers Sci 23(3):286–287. https://doi.org/10.17520/biods.2015070
Grebmeier JM, Cooper LW, Feder HM, Sirenko BI (2006a) Ecosystem dynamics of the Pacific-influenced Northern Bering and Chukchi Seas in the Amerasian Arctic. Prog Oceanogr 71(2–4):331–361. https://doi.org/10.1016/j.pocean.2006.10.001
Grebmeier JM, Cooper LW, Feder HM, Sirenko BI (2006b) Ecosystem dynamics of the Pacific-influenced Northern Bering and Chukchi Seas in the Amerasian Arctic. Prog Oceanogr 71(2/4):331–361. https://doi.org/10.1016/j.pocean.2006.10.001
Hebert PD, Cywinska A, Ball SL, deWaard JR (2003a) Biological identifications through DNA barcodes. Proc Biol Sci 270(1512):313–321. https://doi.org/10.1098/rspb.2002.2218
Hebert PD, Ratnasingham S, deWaard JR (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci 270(Suppl 1):S96–S99. https://doi.org/10.1098/rsbl.2003.0025
Hubert N et al (2008) Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE 3(6):e2490. https://doi.org/10.1371/journal.pone.0002490
Huntington HP et al (2020) Evidence suggests potential transformation of the Pacific Arctic ecosystem is underway. Nat Clim Chang 10(4):342–348. https://doi.org/10.1038/s41558-020-0695-2
Izzo C et al (2016) Fish as proxies of ecological and environmental change. Rev Fish Biol Fish 26(3):265–286. https://doi.org/10.1007/s11160-016-9424-3
Kenchington EL, Baillie SM, Kenchington TJ, Bentzen P (2017) Barcoding Atlantic Canada’s mesopelagic and upper bathypelagic marine fishes. PLoS ONE 12(9):e0185173. https://doi.org/10.1371/journal.pone.0185173
Keskin E, Agdamar S, Tarkan AS (2013) DNA barcoding common non-native freshwater fish species in Turkey: low genetic diversity but high population structuring. Mitochondrial DNA 24(3):276–287. https://doi.org/10.3109/19401736.2012.748041
Li Y, Zhang L, Zhang R, Song P, Wang L, Zhang L, Lin L (2017) Identification of several fish larvae based on DNA barcoding in the investigated waters of Cangnan. J Ocean Univ China (nat Sci) 47(12):72–79. https://doi.org/10.16441/j.cnki.hdxb.20160343
Li Y, Zhang L, Song P, Zhang R, Wang L, Lin L (2018) Fish diversity and molecular taxonomy in the Prydz Bay during the 29th CHINARE. Acta Oceanol Sin 37(8):15–20. https://doi.org/10.1007/s13131-018-1228-y
McCusker MR, Denti D, Van Guelpen L, Kenchington E, Bentzen P (2013) Barcoding Atlantic Canada’s commonly encountered marine fishes. Mol Ecol Resour 13(2):177–188. https://doi.org/10.1111/1755-0998.12043
Mecklenburg CW, Møller PR, Steinke D (2010) Biodiversity of arctic marine fishes: taxonomy and zoogeography. Mar Biodivers 41(1):109–140. https://doi.org/10.1007/s12526-010-0070-z
Meyer RM (1997) Demersal fish assemblages of the northeastern Chukchi Sea. Alaska Fish Bull 95(2):195–208
Mueter FJ et al (2009) Ecosystem responses to recent oceanographic variability in high-latitude Northern Hemisphere ecosystems. Prog Oceanogr 81(1–4):93–110. https://doi.org/10.1016/j.pocean.2009.04.018
Norcross BL, Holladay BA, Busby MS, Mier KL (2010) Demersal and larval fish assemblages in the Chukchi Sea. Deep Sea Res Part II 57(1–2):57–70. https://doi.org/10.1016/j.dsr2.2009.08.006
Panprommin D, Soontornprasit K, Tuncharoen S, Iamchuen N (2020) The utility of DNA barcoding for the species identification of Larval Fish in the Lower Ing River, Thailand. Turk J Fish Aquat Sci 20(9):671–679. https://doi.org/10.4194/1303-2712-v20_9_02
Qu M, Tang W, Liu Q, Wang D, Ding S (2018) Genetic diversity within grouper species and a method for interspecific hybrid identification using DNA barcoding and RYR3 marker. Mol Phylogenet Evol 121:46–51. https://doi.org/10.1016/j.ympev.2017.12.031
Rand KM, Logerwell EA (2011) The first demersal trawl survey of benthic fish and invertebrates in the Beaufort Sea since the late 1970s. Polar Biol 34(4):475–488. https://doi.org/10.1007/s00300-010-0900-2
Robertis AD, Cokelet ED (2012) Distribution of fish and macrozooplankton in ice-covered and open-water areas of the eastern Bering Sea. Deep-Sea Research Part II 65–70:217–229 doi: https://doi.org/10.1016/j.dsr2.2012.02.005
Smith PJ, McVeagh SM, Steinke D (2008) DNA barcoding for the identification of smoked fish products. J Fish Biol 72(2):464–471. https://doi.org/10.1111/j.1095-8649.2007.01745.x
Steinke D, Zemlak TS, Boutillier JA, Hebert PDN (2009) DNA barcoding of Pacific Canada’s fishes. Mar Biol 156(12):2641–2647. https://doi.org/10.1007/s00227-009-1284-0
Ward RD (2009) DNA barcode divergence among species and genera of birds and fishes. Mol Ecol Resour 9(4):1077–1085. https://doi.org/10.1111/j.1755-0998.2009.02541.x
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci 360(1462):1847–1857. https://doi.org/10.1098/rstb.2005.1716
Webb KE, Barnes DKA, Clark MS, Bowden DA (2006) DNA barcoding: a molecular tool to identify Antarctic marine larvae. Deep Sea Res Part II 53(8–10):1053–1060. https://doi.org/10.1016/j.dsr2.2006.02.013
Wong HK, Hanner RH (2008) DNA barcoding detects market substitution in North American seafood. Food Res Int 41(8):828–837. https://doi.org/10.1016/j.foodres.2008.07.005
Acknowledgements
This work was funded by the National Natural Science Foundation of China [Grant No. 41806217 and 41876176], the Scientific Research Foundation of Third Institute of Oceanography, Ministry of Natural Resources [Grant No. 2018010], and Chinese Arctic and Antarctic Administration, State Oceanic Administration.
Author information
Authors and Affiliations
Contributions
HL designed experiments and wrote the manuscript, FY performed data analyses, XW wrote the manuscript, YL, NZ, RZ, CL and HZ performed experiments, and, LL and PS conceived experiments. All the authors discussed the results and gave their opinions on the manuscript.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, H., Yang, F., Wang, X. et al. Demersal fish diversity and molecular taxonomy in the Bering Sea and Chukchi Sea. Conservation Genet Resour 14, 85–97 (2022). https://doi.org/10.1007/s12686-021-01241-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-021-01241-4