Abstract
Soil nutrient supply in rangelands depends on the maintenance and performance of soil microbiological communities. In this study we investigated how different rangeland management systems affects the structure and function of soil microbial communities in the clayey grassland and sandy savanna ecosystems, South Africa. These ecosystems differ in climate, soil and vegetation, with the sandy savanna ecosystem being drier, and encroached by bush. Soils were sampled under continuous and rotational grazing systems along a gradient with increasing grazing pressure. Analyses comprised of enzyme activities and phospholipid fatty acids (PLFA). The results revealed that the clayey grassland ecosystem displayed elevated enzyme activities and PLFA contents compared with the drier, sandy savanna ecosystem, irrespective of the rangeland management practices, likely because soil texture played a significant role in maintaining microbial communities. However, when microbial activity was normalized to carbon, nitrogen and microbial biomass, specific enzyme activities were significantly higher in the sandy savanna ecosystem, indicating a more efficient functioning of microbes here. Furthermore, these microbial parameters were more sensitive to grazing pressure in the clayey grassland ecosystem than other chemical or physical soil properties, whereas in the sandy savanna ecosystem this was not the case. Decreasing the grazing pressure on rangeland, as, e.g., done by commercial farmers practicing rotational grazing, appeared to stimulate microbial performance and thus microbial mediated nutrient mineralization with positive consequences on plant growth.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil microbial communities play a fundamental role in ecosystems by regulating the dynamics of organic matter decomposition and plant nutrient availability. According to Denef et al. (2009) the microbial habitat in soils can be defined as a dynamic and heterogeneous environment characterized by several abiotic and biotic processes, which can significantly transform due to changes in land-use, management or environmental conditions. The microbial communities are thus sensitive to changes in soil chemical (e.g. pH, organic carbon (C) and nitrogen (N)) and physical properties (e.g. soil texture and moisture), and can therefore be used as an important measure of sustainable land use (Bardgett et al. 1997; Patra et al. 2005; Xue et al. 2008).
Microbial biomass can be affected particularly by soil texture, directly by protecting the microbes from predation, and indirectly through the retention of soil organic matter. Generally, microbial biomass is higher in fine-textured soils due to their higher soil organic matter contents, as well as protection that soil aggregates provide from possible predators (Muller and Hoper 2004). Consequently, pore space distribution and specifically the smaller soil pores have a major impact on the abundance of bacteria and fungi and might be responsible for higher rates of carbon mineralization.
Ecologists are becoming increasingly interested in the important role that soil organisms have in regulating ecosystem processes, such as nutrient cycling and organic matter decomposition, and how decomposer organisms respond to disturbed regimes. It is clear that rangeland management affects the structure and activities of microbial communities. Microbial abundance and activity is strongly related to the quantity and quality of available plant litter, which in turn, is related to grazing intensity. Su et al. (2004) for example showed that heavy grazing pressures resulted in loss of soil organic C and N, and subsequently in a depletion of soil enzyme activities. Mofidi et al. (2012) found similar results in rangelands of Iran, where lower levels of grazing had the highest biological activity. The effect of grazing management on microbial biomass and composition (indexed by phospholipid fatty acids [PLFAs]) however, tend to be inconsistent (Oates et al. 2012). This might be due to the negative effects of heavy grazing, but positive effects of the added manure on microbial communities. Bardgett et al. (1997) found in North-Wales, for instance, that long-term removal of grazing animals from a rangeland resulted in a significant reduction in microbial biomass and activity in the surface soil layer, due to less manure in the soil. According to Degens et al. (2000), land uses that deplete soil organic C may cause declines in the catabolic diversity of soil microbial communities. Maintenance of soil organic C may thus be important for preservation of microbial diversity.
Patra et al. (2007) demonstrated that grazing affects microbial functional groups intensely by enhancing the activity of soil microbial communities and inducing changes in the size and composition of these communities. Yet, such information is limited for subtropical climates where rangeland management may be a major source of income. In particular we are not aware of information on how different grazing systems affect the size and composition of key microbial functional groups in different subtropical biomes, and this restricts our understanding of the actual effects of grazing on rangeland functioning. It also affects our ability to predict rangeland response to changes in grazing intensity or management practices, because the composition of microbial communities can determine their resistance and resilience to disturbances (Griffiths et al. 2000; Patra et al. 2005). In a study done by Griffiths et al. (2000), resilience to persistent stress was lower in soils when biodiversity was impaired.
Furthermore, understanding the microbial mechanisms regulating C and N cycling is important to fully appreciate how grazing impacts rangeland functioning. To gain access to C, soil microbes synthesize extracellular enzymes that decompose organic matter. These enzymes are seen as the proximal drivers of decomposition and are essential for the stabilization of soil organic matter (Six et al. 2006). Since enzyme production requires energy, shifts in enzyme activity reflect not only the quantity of C in a system, but the availability of relatively labile C. According to a study done by Hargreaves and Hofmockel (2014), when enzyme activities were expressed per unit microbial biomass, shifts in these activities indicated that microbial communities distributed more energy to enzyme production than to growth and this is indicative of lower C use efficiency. Managing rangeland systems can therefore promote plant–microbial linkages by changing the physiological capacity of the microbial community.
In summary, there is still limited knowledge on the effects of grazing and rangeland management on microbial community composition of soil in semi-arid ecosystems. The objective in this study was therefore to assess soil microbial community structure and function along grazing gradients within a clayey grassland and a sandy savanna ecosystem. This gradient approach allowed us to test how spatial disturbance due to high grazing pressure, alters soil microbial properties in relation to undisturbed grassland. The study was performed in rangeland systems with either rotational or continuous grazing. As rotational grazing provides resting camps, soil microbial properties may be restored prior to the next grazing cycle, while this is not promoted in continuous grazing systems. In this regard we may obtain insights into how specific soil microbial properties respond to grazing pressure. Soil microbial parameters selected comprised specific soil enzymes that are considered to be representative of the functional component of soil microorganisms, as well as PLFAs as a measure of structural microbial diversity. The enzyme β-glucosidase, for instance, is an enzyme involved in the C-cycle, whereas urease is an important enzyme in the N-cycle. Both acid- and alkaline-phosphatase are good indicators of soil fertility, and the dehydrogenase enzyme is a very good proxy for total microbiological activity in soil (Kandeler 2007; Nannipieri et al. 2011). This study is supplementary to two other studies done by Kotzé et al. (2013) on the impacts of rangeland management on the properties of clayey soils along grazing gradients in the semi-arid grassland biome of South Africa, and by Sandhage-Hofmann et al. (2015) on rangeland management effects on soil properties in the savanna biome, South Africa.
Materials and methods
Research areas
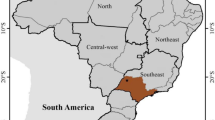
Two research areas had been selected, one in a clayey grassland ecosystem and one in a sandy savanna ecosystem of South Africa (Fig. 1).
The research area for the clayey grassland ecosystem is located near Thaba Nchu, in the Free State Province, South Africa (latitude 28°–29°S, longitude 26°–27°E; altitude 1400–1600 m above sea level). The semi-arid climate is relatively dry, has a low, variable and unpredictable rainfall (mean annual precipitation of 553 mm, with about 70% of the rain occurring in summer between January and March), and a high evaporation of 1832 mm a−1 (Basson 1997). Non-perennial streams exist in the area. The soils are all classified as Lixisols (WRB 2007), and they have more than 15% clay in the A-horizons and more than 25% clay in the B-horizons, with no obvious signs of wind or water erosion. This research area belongs to the “Moist Cool Highveld Grassland Type”, which is part of the Grassland Biome (Bredenkamp et al. 1996). This grassland type is widespread and covers the central eastern part of the Highveld in the Free State Province. A single layer of perennial C4 bunchgrasses such as Themeda triandra, Eragrostis curvula and Cynodon dactylon dominate this particular grassland type, and trees are sparse. The amount of grass cover depends beside the low rainfall and evapotranspiration on the degree of grazing. Frost, fire and grazing support the grass dominance and prevent establishment of trees. The land is characterised by maize and wheat production in the northwest and stock farming and subsistence farming in the east and south. The recommended animal stocking rate for the area of Thaba Nchu is 6 ha LSU−1 (Department of Agriculture and Rural Development 2003).
The research area of the sandy savanna ecosystem is located near Kuruman at the border of the Northern Cape and North-West Province of South Africa, and is situated on the fringe of the Kalahari (latitude 27°–28°S, longitude 22°–24°E; altitude 1050–1200 m above sea level). The arid climate in Kuruman receives rainfall mostly in the summer months of October to March with a mean annual rainfall of 355 mm and temperature of 17.5 °C, and a very high evaporation level of 2050 mm a−1. The soils are deep Arenosols with aeolian origin, underlain by calcrete (WRB 2007), typically containing less than 10% clay. The vegetation in the area is dominated by the Kalahari thornveld and shrub bushveld (Tainton 1999) and has been more specifically described as the Kalahari Mixed Thornveld A16 (Mucina and Rutherford 2006), characterized by a fairly well developed tree stratum with Acacia erioloba, Acacia mellifera, Acacia haematoxylon (≤2 m height) and some Boscia albitrunca as the dominant trees. The shrub layer is dominated by individuals of Acacia mellifera, Acacia hebeclada, Lycium hirsutum, Grewia flava and Acacia haematoxylon. The grass cover contains species such as Stipagrostis uniplumis, Eragrostis lehmanniana, and Schmidtia kalahariensis. The majority of the Northern Cape Province is used for stock farming including cattle, sheep or goat farming as well as mining whilst only about 4% is reserved for conservation (Hoffman and Cowling 1990). In this research area, the estimated grazing capacity of the rangelands is 13 ha LSU−1 (Department of Agriculture and Rural Development 2003). Overgrazing is seen as one of the main causes of land degradation in the Northern Cape, with alien plant invasions posing a threat to the rich flora of the area. This is also one of the worst affected areas in terms of bush encroachment, which implies that large areas of grazing land have been lost, species diversity have been reduced and habitats have been transformed (DEAT 2002). Various land-use activities all contribute to a loss of vegetation cover, soil erosion and ultimately land degradation.
Rangeland management
Communal and commercial livestock ranching are the most common rangeland management systems in both the clayey grassland and sandy savanna ecosystems (Smet and Ward 2006; Tefera et al. 2010). These management systems differ mainly in ownership, as well as in managing of the grazing resources. The commercial farms selected in the research areas are all well developed and mainly market-orientated, while managed typically using a rotational grazing system at moderate stocking densities. Size and animal type (sheep, cattle) vary with time, but management structure remains more or less constant. The farms are divided into camps, where soils are allowed to restore their vegetation after intense grazing.
The communal farms in both ecosystems belonged to the former homeland Bophuthatswana, which is developed as an integral part of the Betterment Villages on land previously owned by commercial farmers (Jacobs 2003). The communal production systems selected in the research areas are based on pastoralism and members of a community share grazing areas. The rangeland is a common pool resource with no restrictions in stocking rates and a continuous grazing system. There are often unclear boundaries, with open access rights to grazing areas. Livestock seem to be of little economic importance, either as income source for people (most income is derived from social grants) or as contributing significantly towards nutrition.
Vegetation and soil sampling
Two rangeland management systems as described above, were selected for this study to compare rotational and continuous grazing systems. In the clayey grassland ecosystem four independent farms were selected as replicates for every management system, whereas in the sandy savanna ecosystem three farms were selected as replicates.
For each farm a representative grazing gradient was selected, along which the rangeland quality was increasingly degraded with increasing grazing pressure. In the clayey grassland ecosystem the gradients started nearby an artificial water point, regardless whether vegetation was present or not. In the sandy savanna ecosystem the gradients also started nearby an artificial water point, but only where vegetation first appeared. Additionally, composite samples were taken in direct proximity to the water points, where no vegetation grew. In both ecosystems these plots were defined exclusively through grass quality conditions, using a similar technique as Van der Westhuizen et al. (2005) to identify indicator grass species for the purpose of rangeland condition assessment, independent of bare patches or bush encroachment. Indicator grass species defined on-site by plant experts for the purpose of rangeland condition assessment, were identified. Rangeland condition ranged from poor, moderate to good conditions. For more details on inorganic site properties and plant functional types, see also Kotzé et al. (2013), Linstädter et al. (2014), Sandhage-Hofmann et al. (2015), and Table 1.
In the clayey grassland ecosystem, the dominant grass species that was used as indicator species was Themeda triandra for good grazing condition, Eragrostis spp. for moderate grazing condition, and Aristida spp. and Cynodon dactylon for poor grazing condition. The dominant grass indicator species in the sandy savanna ecosystem were Stipagrostis spp. for good grazing condition, Eragrostis spp. for moderate grazing condition and Aristida spp. and Schmidtia kalahariensis for poor grazing condition. Acacia species were also dominant in the sandy savanna ecosystem, partly due to bush encroachment. Bare patches were common in both ecosystems in poor grazing conditions, although they also occurred under moderate and good grazing conditions.
The length of the grazing gradients differed between and within the different rangeland management systems. Therefore, in the sandy savanna ecosystem the gradients included 6 single plots, each 10 × 10 m in size, with distances between these plots being kept constant within each sampling site. Not all three grazing conditions were necessarily present in the grazing gradient, therefore a variation in replicates exist.
For vegetation sampling, two to three experienced observers independently assessed total plant cover, hereafter called grass cover, within each plot. The grass cover included all species of the grass layer, e.g. grasses and herbs, as well as dwarf shrubs in the clayey grassland ecosystem. In the sandy savanna ecosystem the area covered by large shrubs and trees (e.g. plants belonging to the tree layer) was not included. Biomass was estimated by taking 10 randomly distributed sword height measurements (0.5 m2) in two strips (0.5 m wide × 10 m long) per plot with a rising plate meter. To calibrate these measurements, the two strips were mowed and the biomass was weighed after drying at 68 °C for 48 h.
For soil sampling, composite samples were collected along the grazing gradient using a 50 mm diameter hand auger in the clayey grassland ecosystem. Each composite sample comprised of 10 randomly taken subsamples thoroughly mixed, in the poor, moderate and good grazing conditions, respectively. In the sandy savanna ecosystem, composite soil samples were taken along a centreline in each plot, every 2 m using a 50 mm diameter hand auger, and combined into a composite sample. Soil samples for selected microbiological properties were taken only in the 0–5 cm soil layer. Due to the sensitivity of soil samples for microbial analyses, samples were sieved (<2 mm) in the field, and stored in a portable freezer (approximately −30 °C), before being transported to the laboratory and prepared for the various analyses.
Results for soil physical and chemical properties measured in the clayey grassland and sandy savanna ecosystems are summarized in Table 1, since they were already reported in detail by Kotzé et al. (2013), Sandhage-Hofmann et al. (2015), respectively. These soil properties differed significantly (p < 0.001) between the two ecosystems, except for pH (p = 0.477).
Soil microbiological analyses
Enzyme activities
Specific enzymes (β-glucosidase, urease, acid- and alkaline-phosphatase, dehydrogenase) were chosen because of their importance to nutrient cycling in ecosystems; they were determined colorimetrically using enzyme-specific procedures (described below). Once the colour was developed, a spectrophotometer or microplate reader, ELx800 (BioTek Instruments Inc., USA) was used in determining the absorbance at the specific wavelength. Enzyme assays were done in duplicate, with a control for each sample. Dehydrogenase is the only method that required keeping the soil moist for the assay; however, this was done for all the assays for standardization of results (Tabatabai 1994). Due to this, all data was corrected for water content during the data analysis.
The enzyme β-glucosidase was determined through a methodological adaption of Dick et al. (1996). Briefly, 1 g of field-moist soil was incubated at 37 °C for 1 h with toluene, modified universal buffer pH 6.0, and ρ-nitrophenyl-β-d-glucopyranoside (ρNG) solution), shaken with calcium chloride and tris(hydroxymethyl) aminomethane before filtering through a Whatman no. 2v filter paper. Absorbance of released ρ-nitrophenol (ρNP) was tested with a microplate reader at 405 nm.
Urease activity was determined by the non-buffered method described by Kandeler and Gerber (1988) for urease enzyme determination. Firstly, 5 g of field-moist soil was incubated at 37 °C for 2 h with an urea solution and then shaken with potassium chloride. The resulting suspension was filtered through Whatman no. 2v filter paper. Before the absorbance was measured with a microplate reader at 690 nm, the filtrate was prepared with sodium salicylate/sodium hydroxide solution and sodium dichloroisocyanurate solution for the colour development.
Both acid- and alkaline-phosphatase enzymes were determined by an assay that was adapted from Tabatabai (1994). Briefly, 1 g of field-moist soil was incubated at 37 °C for 1 h with toluene, modified universal buffer (pH 6.5 for acid phosphatase and 11 for alkaline phosphatase), and ρNP. Then, after adding calcium chloride and sodium hydroxide, it was filtered immediately through Whatman no. 2v filter paper. Absorbance, also of ρNP, was measured with a microplate reader at 405 nm.
The assay from Von Mersi and Schinner (1991) was used to determine dehydrogenase, where 1 g of field-moist soil was mixed with tris-hydroxymethyl aminomethane (THAM) and iodonitrotetrazolium violet-formazan (INT) solutions. This enzyme assay required incubation at 40 °C in the dark for 2 h. The samples were mixed with an extraction solution and kept in the dark for another 30 min. Absorbance of the reaction product INT was tested with a glass cuvette on the spectrophotometer at 464 nm.
Phospholipid fatty acids
A modified version of the method described by Marschner (2007) was used to determine PLFA. Briefly, 2 g of frozen field-moist soil was mixed with a citrate buffer, chloroform, methanol, and Bligh and Dyer reagent. The samples were shaken for 2 h, then vortexed and centrifuged at 2500 rpm. The soil-free supernatant was moved to a new tube and the soil was washed with more Bligh and Dyer reagent, and once it was vortexed and centrifuged at 2500 rpm again, more supernatant was transferred to the new tube. Chloroform and citrate buffer were added and then the organic phase was removed. The samples were dried under a N2 stream, conditioned with chloroform, and run through an elution chamber. The phospholipids were collected and dried again under a N2 stream. An internal standard of methyl nonadecanoate (Sigma N5377-1G) was added along with methanol:toluol, hexane:chloroform, acetic acid, and deionized water. The organic phase was collected and dried under a N2 stream. Then the purified PLFAs, which had been methanolyzed into fatty acid methyl esters, were read on a Varian 430-GC gas chromatograph (Agilent Technologies, USA). The identification of individual PLFA markers was performed as described by Reichel et al. (2013). A total of thirty-two different PLFAs including saturated, monounsaturated, polyunsaturated, cyclopropyl, and methyl fatty acids were identified. Twelve PLFAs (14:0, i15:0, a15:0, 15:0, 2OH14:0, i16:0, 16:0, i17:0, 17:0, 18:2ω9,12, 18:1ω9c, 18:0) consistently present in the samples were used for data analysis. The fatty acid signatures 14:0, i15:0, a15:0, 15:0, 2OH14:0, i16:0, i17:0, 17:0 and 18:0, which are considered to be of bacterial origin, were used as biomarkers for bacterial biomass. The fatty acids 18:1ω9c and 18:2ω9,12 were used as an indicator for fungal biomass (Frostegard and Baath 1996). We used fatty acids i15:0, a15:0, i16:0, and i17:0 to represent Gram-positive bacteria (Gram+), whereas 14:0, 15:0, 2OH14:0 and 17:0 were used to represent Gram-negative bacteria (Gram−) (Djukic et al. 2010). The microbial biomass was indicated by total concentration of all PLFA markers (Total PLFA). PLFA-derived ratios of Gram+/Gram− and fungi/bacteria were calculated using summed marker concentrations for each microbial group.
Statistical analyses
Statistical analyses were performed on all measured soil properties using two-way ANOVA in order to determine statistically significant differences between, as well as within the farms of the clayey grassland and sandy savanna ecosystems. Due to strong differences found between the two ecosystems, statistical analyses were subsequently completed for each separate ecosystem, testing for differences between the main effects: rangeland management systems and grazing conditions; as well as interactions among them. For comparison of means we used a post hoc Tukey-HSD test with a p < 0.05 level of significance. To explore variation in soil microbial community composition among study sites, the mole percentages (nmol %) of twelve individual PLFAs were subjected to principal component analysis (PCA) after standardizing to unit variance. All data were tested for normal distribution as well as homogeneity before statistical analyses were performed using SPSS version 12.0 (SPSS Inc. Chicago, USA).
Results
As was indicated in Table 1, the clayey grassland ecosystem had significant higher average organic matter and nutrient contents than the sandy savanna ecosystem. As a result, there were better conditions for grass and associated microbial growth in the sampled clayey grassland ecosystem than in the sandy savanna ecosystem. All measured biological parameters were also significantly higher in the clayey grassland compared to the sandy savanna ecosystem (Table 2). As could be expected, both ecosystems had relatively high coefficient of variance (CV) values of between 38 and 92%, with the sandy savanna ecosystem having the highest CV values for most biological parameters, compared to the clayey grassland ecosystem.
Grass cover and biomass
In the clayey grassland ecosystem, rotational grazing had significantly higher grass cover and biomass than continuous grazing (grass cover: F(2,12) = 10.521, p = 0.007, biomass: F(2,12) = 15.082, p = 0.002) (Table 3). Also in the sandy savanna ecosystem, the amount of biomass in rotational grazing was significantly higher than in continuous grazing: F(2,9) = 33.660, p = 0.000. Total biomass also significantly increased from poor, to moderate and good grazing conditions in this ecosystem: F(2,9) = 25.546, p = 0.001 (Table 3). Interaction between biomass and grass cover was not significant.
Enzyme activities
All soil microbial enzyme activities measured were significantly higher for the clayey grassland ecosystem compared to the sandy savanna ecosystem (Table 2). The highest CV value of 86% was for urease activity in the clayey grassland ecosystem. Nevertheless, different trends evolved for the different rangeland management systems within these ecosystems (Fig. 2). The activity of β-glucosidase enzymes showed opposite trends in the respective grazing gradients of the clayey grassland ecosystem: it decreased from poor to good grazing condition in rotational grazing but increased from poor to good grazing condition in continuous grazing (significant with Tukey-HSD = 2803 in both main effects as well as the interaction term between rangeland management systems and grazing conditions, [F(2,16) = 5.932, p = 0.011] (Fig. 2)). In the sandy savanna ecosystem β-glucosidase activity showed similar trends for the rotational grazing but varying values for the continuous grazing. In the continuous grazing system β-glucosidase activity even exceeded values of rotational grazing with the exception of poor grazing condition (Fig. 2). Interestingly, the rotational grazing systems exhibited similar trends in all enzyme activities along the grazing gradient: rangelands under poor grazing condition showed the highest enzyme activities, followed by moderate grazing condition, while lowest enzyme activities were found in good grazing condition.
A comparison of three soil microbial enzyme activities a β-glucosidase, b urease and, c dehydrogenase, in the clayey grassland and sandy savanna ecosystems, between the rotational and continuous grazing systems within poor, moderate and good grazing conditions. Standard deviation is indicted by error bars. Significant differences are present when error bars do not overlap
The activity of urease enzyme showed no significant interactions in the clayey grassland ecosystem [F(2,18) = 1.080, p = 0.360], as well as in the sandy savanna ecosystem [F(2,12) = 0.961, p = 0.410] (Fig. 2). For rotational grazing, urease behaved similarly along the grazing gradient as β-glucosidase activity (see above), with highest activity under poor grazing condition. However, for urease, the patterns in the grazing gradients were now also more or less similar in continuous grazing, with again highest urease activity in poor grazing, followed by moderate grazing and the lowest activities in good grazing. Moreover, urease activity of continuous grazing exceeded the activities of rotational grazing, with the exception of poor grazing condition (Fig. 2).
The activity of dehydrogenase enzyme again followed similar trends as β-glucosidase enzyme, with decreasing activities from poor to good grazing condition in both rotational and continuous grazing, respectively. An exception was found for the continuous grazing system in the sandy savanna ecosystem. Here, β-glucosidase activity did not differentiate between rotational and continuous grazing, and dehydrogenase activity declined from good to poor grazing condition in both rotational and continuous grazing (Fig. 2). Hence, dehydrogenase activity showed a significant interaction among rangeland management systems in the clayey grassland but not in the sandy savanna ecosystem (Fig. 2). In the latter system, however, dehydrogenase activity was significantly greater in continuous grazing than in rotational grazing [F(1,18) = 5.570, p = 0.03; Table 2].
For alkaline and acid phosphatase activities, no clear pattern were found between the different rangeland management systems, also interaction terms were insignificant (data not shown). However, in the clayey grassland ecosystem, higher activity were evident in rotational grazing (18907 μg ρNP g−1 soil h−1) compared to continuous grazing (4846 μg ρNP g−1 soil h−1), throughout the grazing gradients for acid-phosphatase [F(1,18) = 10.921, p = 0.004], and alkaline-phosphatase [F(1,18) = 9.572, p = 0.006], with rotational grazing having a higher activity of 1612 μg ρNP g−1 soil h−1 compared to continuous grazing with an activity of 540 μg ρNP g−1 soil h−1. The activity of alkaline-phosphatase even showed a trend among grazing conditions, with elevated values under poor grazing condition (2914–3100 μg ρNP g−1 soil h−1) compared to moderate grazing condition (552–566 μg ρNP g−1 soil h−1.
Phospholipid fatty acids
Total microbial lipid biomass (PLFA) was higher in the topsoils of the clayey grassland ecosystem than in those of the sandy savanna ecosystem (Table 2). A very high CV value of 92% for the sandy savanna ecosystem, compare to a CV value of 54% for the clayey grassland ecosystem was calculated. Within the clayey grassland ecosystem, the patterns followed those of β-glucosidase and dehydrogenase, which, similar to the amount of total PLFA, are also proxies for living microbial biomass. There was no significant interaction between rangeland management systems and grazing gradients, F(2,18) = 2.572, p = 0.104; yet, rotational grazing exhibited on average higher PLFA content than continuous grazing [F(1,18) = 11.318, p = 0.003]. In the sandy savanna ecosystem, similar trends between both rangeland management systems could be anticipated, except for the good grazing condition of rotational grazing, which showed exceptionally high PLFA contents while a similar peak in enzyme activities was lacking. On the other hand, the differences in PLFA between the two rangeland management systems were not significant (Fig. 3).
Total PLFA (nmol PLFA g−1 soil) measured in the clayey grassland and sandy savanna ecosystems, comparing rotational and continuous grazing systems within poor, moderate and good grazing conditions. Standard deviation is indicted by error bars. Significant differences are present when error bars do not overlap
Remarkably, the two ecosystems responded contrasting in terms of significant differences evident in the individual PLFAs. The clayey grassland ecosystem had the most significant differences when comparing rotational and continuous grazing. In addition, significant interactions were found between rangeland management systems and grazing conditions. On the other hand, the sandy savanna ecosystem had the most significant differences when comparing the grazing conditions (poor, moderate and good) within the rangeland management systems, and not when comparing rotational and continuous grazing. No significant interactions were found in this ecosystem. From a total of thirty-two different PLFAs identified, only twelve PLFAs (14:0, i15:0, a15:0, 15:0, 2OH14:0, i16:0, 16:0, i17:0, 17:0, 18:2ω9, 12, 18:1ω9c, 18:0) were consistently present in the samples. They were thus chosen for principal component analysis. This analysis confirmed substantial differences in soil microbial community composition among study sites (Fig. 4).
Principle component analysis (PCA) dividing the PLFA analyses for the clayey grassland (a, c) and sandy savanna (b, d) ecosystems. For the clayey grassland ecosystem PC1 (66.89%) and PC2 (19.75%) explained 86.64% of the total variance in the data. For the sandy savanna ecosystem PC1 (54.14%) and PC2 (30.52%) accounted for 84.66% of the total variance in the phospholipid fatty acids. In a, b rotational grazing systems are shown in orange and continuous grazing systems in blue. In c, d, the fungal fatty acid signatures (18:1ω9c and 18:2ω9,12) are indicated in green, Gram+ bacteria (i15:0, a15:0, i16:0, and i17:0) in orange, Gram− (14:0, 15:0, 2OH14:0 and 17:0) in blue, and common bacteria (16:00 and 18:00) in black. (Color figure online)
For the clayey grassland ecosystem the first principal component (PC1) explained 66.9 and the second (PC2) 19.8% of the total variance in the PLFAs (Fig. 4a, c). The PCA plot in Fig. 4a also showed that all three grazing conditions (poor, moderate, good) are grouped together for rotational grazing, as well as for continuous grazing, i.e. it appears possible to structure the data according to rangeland management system, though with an overlap for the good grazing condition. Also interesting to note is that for PC2 the poor grazing condition for both rotational and continuous grazing had positive loading scores, compared to the other grazing conditions, which had negative loading scores. The twelve fatty acids chosen for data analysis also showed clear groupings. For PC1, lipid signatures i15:0, a15:0, 15:0, 2OH14:0, i16:0, 17:0, 18:2ω9,12 and 18:1ω9c had higher positive loading scores while 16:0 and 18:0 had lower negative loading scores (Fig. 4c). Both fungal biomarkers 18:2ω9,12 and 18:1ω9c had positive loading scores for this ecosystem.
For the sandy savanna ecosystem PC1 explained 54.1 and PC2 30.5% of the total PLFA variance (Fig. 4b, d). The PC1 plot in Fig. 4b had a high positive loading score only for the poor grazing condition in continuous grazing, whereas all the other sites had lower to negative loading scores. For PC2 the poor grazing condition for rotational grazing showed a high positive loading score, whereas the moderate grazing condition for rotational grazing showed an opposite high negative loading score. Hence, the PCA separates the rangeland management systems less clearly than in the clayey grassland ecosystem, only the poor grazing condition has different factor loadings than the other two grazing conditions. The fatty acids in this sandy savanna ecosystem load the fungal biomarkers 18:2ω9,12 and 18:1ω9c, in the negative score for PC1 and in the positive score for PC2 (Fig. 4d). The common bacteria signatures 16:0 and 18:0 both had positive loading scores for PC1, and negative scores for PC2. On the other hand, the Gram+ bacteria (i15:0, a15:0, i16:0 and i17:0) do not show any clear groupings for the sandy savanna ecosystem in this PCA, while the Gram− bacteria (14:0, 15:0 and 17:0) grouped together in the top right hand quadrant of the PCA. Overall, and similar to the clayey grassland ecosystem, differences in PLFA patterns are less clearly disentangled among the different groups of organisms than among the farming system.
As could be expected, the absolute abundance of specific microbial groups was highest in the clayey grassland ecosystem compared to the sandy savanna ecosystem, with higher CV values in the sandy savanna ecosystem (Table 2). When comparing rangeland management systems and grazing conditions within the clayey grassland ecosystem, all the bacterial, fungal, Gram+ and Gram− PLFAs showed similar changes along the grazing gradients in the clayey grassland ecosystem, with rotational grazing always having higher concentrations than continuous grazing (Fig. 5). These trends therefore, largely explained the changes of bulk PLFA contents as displayed in Fig. 3. Interestingly, rotational grazing always had the highest concentrations of these PFLAs in the poor grazing condition and the lowest in the good grazing condition of the clayey grassland ecosystem, whereas continuous grazing exhibited the opposite trend with the lowest concentrations in the poor grazing condition and the highest in the good rangeland condition. Due to the similarity of the behavior of individual PLFAs, the ratios of fungal/bacterial PLFA as well as G+/G− ratios showed similar trends for both rotational and continuous grazing, with not much variance between the ratios. Although not significant, the moderate grazing condition always tended to have the higher ratio for both these indices, irrespective of rangeland management system. All microbial groups and ratios did thus also not reflect any significant interactions between rangeland management systems and grazing conditions, except for Gram−, F(2,18) = 5.768, p = 0.012. There were, however, statistically significant main effects for rangeland management systems, with clear differences between rotational and continuous grazing for all the microbial groups, viz. bacteria: F(1,18) = 8.628, p = 0.009; fungi: F(1,18) = 12.176, p = 0.003; Gram+: F(1,18) = 8.621, p = 0.009; Gram−: F(1,18) = 18.573, p < 0.001; fungi/bacteria: F(1,18) = 10.219, p = 0.005.
Sums and ratios of phospholipid fatty acids (PLFAs) of microbial groups measured in the clayey grassland and sandy savanna ecosystems, comparing rotational and continuous grazing systems within poor, moderate and good grazing conditions. Standard deviation is indicted by error bars. Significant differences are present when error bars do not overlap
For the sandy savanna ecosystem the absolute abundance of the different microbial groups did not show clear trends or patterns, except that the moderate grazing condition for both rotational and continuous grazing seemed to have lower concentrations for bacterial, fungal and Gram+ PLFAs, when compared to the poor and good grazing conditions. These minima, however, were not significant due to the large standard deviation during analyses of these sandy savanna sites. The fungi/bacteria ratio for the sandy savanna ecosystem seemed to be influenced differently when comparing rotational and continuous grazing. This ratio for rotational grazing had the lowest value for the moderate grazing condition, whereas continuous grazing had the highest value for the moderate grazing condition (note that in continuous grazing bush encroachment was most pronounced in the moderate veld). The Gram+/Gram− ratio also tended to have the highest value for the moderate grazing condition for both rotational and continuous grazing. No significant interactions between rangeland management system and grazing condition existed in this ecosystem for any of the microbial groups or ratios. The only significant main effect manifested for Gram− bacteria in the grazing condition [F(2,12) = 7.261, p = 0.009], where the poor grazing condition in continuous grazing had the most Gram− bacteria.
In summary, the two ecosystems responded differently to rangeland management when considering individual PLFAs as indicators for microbial response reactions. In the clayey grassland ecosystem, rotational grazing preserved microbial properties better than continuous grazing. In addition, significant interactions were found between rangeland management systems and grazing conditions. The sandy savanna ecosystem, in contrast, exhibited the largest differences in microbial properties within the grazing gradient (i.e., among poor, moderate and good grazing conditions) but not between the rangeland management systems at rotational and continuous grazing. This is also supported by the relative high CV values indicated in Table 2 for most measured biological parameters.
Soil enzyme efficiency
To give an indication of how rangeland management affected soil enzyme efficiency, selected soil microbiological properties were normalized and expressed as enzyme or microbial activity per gram soil C or N (β-glucosidase, dehydrogenase, total-PLFA normalized to C; and urease normalized to N) (Table 4). There were significant differences between the two ecosystems, when comparing the normalized data, However, no significant effects or interactions were found within each ecosystem, when comparing rangeland management systems and grazing conditions. Contrasting to what was found with the total levels of enzyme and microbial activities, all normalized enzyme activities except for urease activity, were now highest in the sandy savanna ecosystem. In the clayey grassland ecosystem, the continuous and rotational grazing systems followed the same trends along the grazing gradients as described above for the bulk soil parameters: the highest C normalized enzyme activities were measured in the poor grazing condition and the lowest in the good grazing condition. In the sandy savanna ecosystem, however, no clear trend was visible. Intriguingly, the lowest C-normalized enzyme activities in the rotational grazing system were also found in the good grazing condition.
To further understand microbial physiology, enzyme activities were normalized and expressed as enzyme per μmol PLFA (Table 5). There were again significant differences between the two ecosystems, when comparing the PLFA normalized data. When comparing PLFA normalized enzyme activity within each ecosystem, there were only significant effects in the clayey grassland ecosystem, when comparing rangeland management systems. However, no clear trends were evident for the PLFA normalized data, in both ecosystems.
Discussion
Response of ecosystems on rangeland management
Substantial differences existed in all the measured microbial enzyme activities, biomass and soil microbial community composition when comparing the clayey grassland ecosystem with the sandy savanna ecosystem. This was also evident in the high CVs calculated, which is common for soil biological parameters (Baldrian 2014). These differences could be attributed to the large variation between these two ecosystems in soil texture and climate, which influenced soil water content and subsequently the grass species growing in that specific ecosystem. Furthermore, clayey soils promote soil organisms directly by providing larger surface areas for growth, and indirectly by improved moisture status, elevated organic matter contents and enhanced aggregation, when compared with sandy soils, (Prieto et al. 2011). Both Sessitsch et al. (2001), Xu et al. (2014) confirmed in their studies that soil texture has a regulatory role in soil biological processes and thus affected the soil microbial community structure.
Intriguingly, the C and N normalized data indicated that when comparing the two ecosystems, the organic matter in the sandy savanna ecosystem supported significant larger enzyme and microbial activities than in the clayey grassland ecosystem, just opposite to the non-normalized levels of enzyme activities and biomass contents. Apparently, there was a more intense cycling of nutrients in the sandy savanna than in the clayey grassland ecosystem. In this regard, the soils of the sandy savanna ecosystem might also respond faster to land-use change than in the clayey grassland ecosystem. Alternatively, soil microbes in the clayey grassland ecosystem may be less physiologically restrained and respond more to changes in soil and N.
When enzyme activities were expressed per unit microbial biomass (PLFA normalized enzyme activity), the sandy savanna ecosystem only supported significant larger β-glucosidase enzyme activity compared to the clayey grassland ecosystem. Shifts in mass-specific enzyme activity indicate that microbial communities allocated a greater amount of energy to enzyme production rather than growth and is indicative of lower C use efficiency. All other normalized enzyme activities remained consistent across ecosystems, rangeland management systems and grazing gradients. When assuming enzyme activity was representative of recently active members of the microbial community, results indicate that changes in physiological capacity of the microbial community explained patterns in enzyme production, rather than microbial biomass.
Response of grazing gradients on rangeland management
In any rangeland management system, the composition and structure of grass cover and biomass are ultimately influenced by the animals grazing on the land. In both ecosystems the rotational grazing system had both greater grass cover and biomass than the continuous grazing system, indicating that sufficient resting time is essential for grass to recover from grazing. This was also confirmed by Linstädter et al. (2014) who evaluated the development of plant functional types in the same study area. Tesseman et al. (2011), Banerjee et al. (2000) also found reduced vegetation growth, diversity, abundance and biomass in continuous grazed pastures than in rotationally grazed ones. As could be expected, all grazing gradients in both ecosystems, irrespective of rangeland management system, also showed the highest values for grass cover as well as biomass under good grazing condition, where grazing pressure was low (Rutherford and Powrie 2011). In the sandy savanna ecosystem the difference between poor and good grazing conditions were two-fold for grass cover and four times larger for biomass, showing that this environment was more sensitive to vegetation degradation.
Changes in the contents of soil C and N in the studied grazing gradients have been described in detail by Kotzé et al. (2013), Sandhage-Hofmann et al. (2015). The results are shortly summarized here again since they are relevant for understanding soil biological performance. In the clayey grassland ecosystem, both soil C and N contents had been negatively affected by high grazing pressure, accompanied by a depletion of plant cover and litter input, additional trampling and resulting mixing of surface soil with subsoil, or erosion. Hence, the contents declined from good to poor grazing condition in continuous grazing, whereas in the poor grazing condition of rotational grazing some enrichment due to urine and dung of the animals was observed (Perkins and Thomas 1993; Kotzé et al. 2013). The effects were paralleled by changes in the C/N ratio within the grazing gradients for both rangeland management systems. The lower C/N ratio in the poor grazing condition indicated a higher soil N content, which could be attributed to the concentrated urine and dung in this area due to the water or entry point of the camp. Because the soil C content increased as grass condition improved towards the good grazing condition, the C/N ratio also increased.
The sandy savanna ecosystem had less rainfall, less grass cover and therefore lower C contents than the clayey grassland ecosystem. The trend within a rangeland management system was also dissimilar, because invasion of bush at heavy grazing may enhance C contents, particularly under moderate grazing condition of the continuous grazing system (Sandhage-Hofmann et al. 2015). Lower C/N ratios in the continuous grazing system compared to the rotational grazing system could also be due to the presence of more Acacia species, which contributed to higher N fixation in soil (Cramer et al. 2007; Isaac et al. 2011).
The above mentioned changes went along with aggregate breakdown with trampling and soil organic matter loss (clayey grassland ecosystem; Kotzé et al. 2013), as well as by accrual of particulate organic matter with enhanced input of plant debris (sandy savanna ecosystem; Sandhage-Hofmann et al. 2015). All these parameters are well known to co-affect soil microbial performance. Soil microbial diversity, for instance, varies significantly among soil aggregate classes and particle-size fractions, as well as associated enzyme activities (e.g., Stemmer et al. 1998; Kandeler et al. 2000; Kihara et al. 2012). This must be considered, therefore, when elucidating changes of these parameters with grazing pressure and rangeland management system.
Within the clayey grassland ecosystem, all enzyme activities, total microbial biomass as well as all the various microbial PLFA groups had elevated values in the poor grazing condition of the rotational grazing system, whereas the opposite was true for the continuous grazing system. Xu et al. (2014) found similar results with a study in China, where soil microbial communities also showed a positive relationship with enzyme activities. This finding can be attributed to the urine and dung of the animals deposited nearer to the centre of the piosphere (Banerjee et al. 2000). Furthermore, under rotational management, there was a resting time of the soil, i.e., the adding of urine and dung nearby the water point went along with a maintenance in vegetation, and as such, also of microbial activity: high enzyme activities were recorded. In contrast, under continuous grazing, the intensive trampling destroyed aggregates (Kotzé et al. 2013), vegetation, and soil organic matter was lost, likely also by erosion. Hence, even despite enhanced input of urine and dung, microorganisms lacked additional C sources and nutrients, and we found lower enzyme activities than under the other grazing conditions.
Due to the lack of aggregates in the sandy savanna ecosystem, the effects of trampling on aggregated disruption was absent, when compared to the clayey grassland ecosystem. Both rangeland management systems had a similar effect on the enzyme activities measured, with the highest values in the poor grazing condition and comparable values in the moderate and good grazing conditions. These data can again be explained by higher inputs of dung, which also went along with an enrichment of particulate organic matter (POM) (Sandhage-Hofmann et al. 2015). POM fractions are known to host substrates for enlarged microbial and enzyme activities (e.g. Kandeler et al. 2000; Moore-Kucera et al. 2008; Yan et al. 2009), so changes in enzyme activity with grazing coincide with other changes in soil properties. Similar to what Watts et al. (2010) found, the dehydrogenase activity, which is a measure of microbial respiration and a reliable index of microbial activity in soil, responded well with the individual microbial groups (fungi, bacteria, Gram+, Gram−) in both ecosystems. Interestingly, the rotational grazing system had the lowest PLFA values in the moderate grazing condition. This finding corresponded to the reaction of organic C and POM contents, which were also lowest in moderate grazing condition at rotational grazing (Sandhage-Hofmann et al. 2015). We interpret this loss of microbial biomass as an indicator that the formerly good grazing condition had degraded to moderate condition; whereas further degradation to poor grazing condition did not continue to impair microorganisms due to the immense additional input of dung excrements, as explained above. PLFA analysis exposed the structural characteristics of the living microbial community at the time of sampling and is therefore suitable for sensing rapid changes in microbial communities due to rangeland management.
In the sandy savanna ecosystem, moderate grazing condition in the continuous grazing system went along with bush encroachment (Sandhage-Hofmann et al. 2015). This process did not have any effect on total microbial activity, however it influenced fungi/bacteria and G+/G− bacteria ratios. The higher ratios in moderate grazing condition indicated that this zone promotes selected soil organisms, likely adapted to the encroaching bushes. Total soil organic matter stocks and C/N ratios were also elevated for moderate grazing condition: the encroaching bushes deteriorate the rangeland but prevent the site from further soil degradation (Sandhage-Hofmann et al. 2015). Fungi are known to grow on C/N wide substrates (Rousk and Baath 2007; Strickland and Rousk 2010), i.e., the elevated C/N ratios in the continuous grazing system support their accrual. Vice versa, the abundance of bacteria in all sites was favoured at subsites with lower C/N ratio, which supports studies by Bossuyt et al. (2001), Hiltbrunner et al. (2012), for example.
The elevated portions of Gram- bacteria in poor grazing condition were likely again associated with the better supply of dung-derived organic matter and high nutrient availability from the urine. Hiltbrunner et al. (2012) found similar results in a Swiss sub-alpine pasture. Our findings can also be reconciled with Hamer et al. (2009) who reported lower proportions of Gram- bacteria in bare soil than under grass cover, as is evident in the poor grazing condition of particularly the continuous grazing system studied.
Nevertheless, and despite the differences in ecosystems, soil microbial communities were clearly grouped according to the rangeland management system as well as grazing condition, suggesting that rotational and continuous grazing systems generated different and unique ecological niches in soil microbial communities (Jangrid et al. 2011; Vallejo et al. 2012), especially in the clayey grassland ecosystem. Although the two ecosystems differed significantly, microbial efficiency remained consistent across rangeland management system and grazing gradient, within both the ecosystems. Grazing-induced shifts in microbial activity may be responding to shifts in soil C and N pools. The higher C normalized enzyme and microbial activities that was evident in poor grazing condition, corresponded well with the urine and dung deposits in this areas. More in depth research along clear time-series would now be needed to finally investigate and explain soil vulnerability and resilience in both ecosystems.
Conclusions
In this study two distinctive ecosystems were compared regarding the influence of rangeland management systems on soil microbiological and associated properties. The data showed that microbial communities and activities responded strongly to differences in soil texture and ecosystem type in grazed management systems. Our data further demonstrated that a decrease in grazing pressure on a rangeland, such as by commercial farmers practicing rotational grazing, could stimulate microbial activity, especially in a clayey grassland ecosystem. Likely there was a positive feedback between microbial mediated nutrient mineralization and plant growth, as all microbial biomass and activity as well as grass cover and biomass were elevated when grazing pressure changed. The differences are seen under poor overall grazing conditions: while intensive, continuous grazing degraded soils and reduced vegetation growth by animal trampling, urine and dung additions sustained microbial performance and vegetation growth after adequate resting times under rotational grazing. Greater enzyme activity per unit C, N and PLFA, indicated that microbes were more efficient in C and N cycling in the sandy savanna ecosystem than in the clayey grassland ecosystem. This finding might ultimately change our perception on how microbes function in sandy compared to clayey soils.
References
Baldrian P (2014) Distribution of extracellular enzymes in soils: spatial heterogeneity and determining factors at various scales. Soil Sci Soc Am J 78:11–18
Banerjee MR, Burton DL, Mccaughey WP, Grant CA (2000) Influence of pasture management on soil biological quality. J Range Manage 53(1):127–133
Bardgett RD, Leemans DK, Cook R, Hobbs PJ (1997) Seasonality of the soil biota of grazed and ungrazed hill grasslands. Soil Biol Biochem 29(8):1285–1294
Basson MS (1997) Overview of water resources availability and utilisation in South Africa. Department of Water Affairs and Forestry, Pretoria
Bossuyt H, Denef K, Six J, Frey SD, Merckx R, Paustian K (2001) Influence of microbial populations and residue quality on aggregate stability. Appl Soil Ecol 16(3):195–208
Bredenkamp G, Granger JE, Van Rooyen N (1996) Moist Sandy Highveld Grassland. In: Low AB, Rebelo AG (eds) Vegetation of South Africa, Lesotho and Swaziland. Departement of Environmental Affairs and Tourism, Pretoria
Cramer MD, Chimpango SBM, Van Cauter A, Waldram MS, Bond WJ (2007) Grass competition induces N2 fixation in some species of African Acacia. J Ecol 95:1123–1133
DEAT (2002) National core set of environmental indicators, Phase 3: selection of indicators: integrated and environmental management. Specialist Report 8 of 8. Pretoria
Degens BP, Schippers LA, Sparling GP, Vojvodic-Vukovic M (2000) Decreases in inorganic C reserves in soils can reduce the catabolic diversity of soil microbial communities. Soil Biol Biochem 32:189–196
Denef K, Roobroeck D, Manimel Wadu MCW, Lootens P, Boeckx P (2009) Microbial community composition and rhizodeposit-carbon assimilation in differently managed temperate grassland soils. Soil Biol Biochem 41:144–153
Department of Agriculture and Rural Development (2003) Free State Province. Map compiled by Farming Information Section
Dick RP, Breakwell DP, Turco RF (1996) Soil enzyme activities and biodiversity measurements as integrative microbiological indicators. In: Doran JW, Jones AJ (eds) Methods for assessing soil quality. Soil Science Society of America, Madison
Djukic I, Zehetner F, Mentler A, Gerzabek MH (2010) Microbial community composition and activity in different Alpine vegetation zones. Soil Biol Biochem 42:155–161
Frostegard A, Baath E (1996) The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fert Soils 22:59–65
Griffiths BSK, Ritz K, Bardgett RD, Cook R, Christensen S, Ekelund F, Sorensen SJ, Baath E, Bloem J, De Ruiter PC, Dolfing J, Nicolardat B (2000) Ecosystem response of pasture soil communities to fumigation-induced microbial diversity reductions: an examination of the biodiversity-ecosystem function relationship. Oikos 90:279–294
Hamer U, Makeschin F, An S, Zheng FI (2009) Microbial activity and community structure in degraded soils on the Loess Plateau of China. J Plant Nutr Soil Sc 172(1):118–126
Hargreaves SK, Hofmockel KS (2014) Physiological shifts in the microbial community drive changes in enzyme activity in a perennial agroecosystem. Biogeochem 117:67–79
Hiltbrunner D, Schulze S, Hagedorn F, Schmidt MWI, Zimmermann S (2012) Cattle trampling alters soil properties and changes soil microbial communities in a Swiss sub-alpine pasture. Geoderma 170:369–377
Hoffman MT, Cowling RM (1990) Vegetation change in the semi-arid Eastern Karoo over the last 200 years: an expanding Karoo—Fact or Fiction? S Afr J Sci 86:86–294
Isaac ME, Harmand J-M, Lesueur D, Lelon J (2011) Tree age and soil phosphorus conditions influence N2-fixation rates and soil N dynamics in natural populations of Acacia senegal. For Ecol Manage 261:582–588
Jacobs N (2003) Environment, power, and injustice: A South African history. Cambridge University, Cambridge
Jangrid K, Williams MA, Franzluebbers AJ, Schmidt TM, Coleman DC, Whitman WB (2011) Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biol Biochem 43:2184–2193
Kandeler E (2007) Physiological and biochemical methods for studying soil biota and their function. In: Paul EA (ed) Soil microbiology, ecology and biochemistry. Academic, Amsterdam
Kandeler E, Gerber H (1988) Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol Fert Soils 6:68–72
Kandeler E, Tscherko D, Bruce KD, Stemmer M, Hobbs PJ, Bardgett RD, Amelung W (2000) Structure and function of the soil microbial community in microhabitats of a heavy metal polluted soil. Biol Fert Soils 32:390–400
Kihara J, Martius C, Bationo A, Thuita M, Lesueur D, Herrmann L, Amelung W, Vlek PLG (2012) Soil aggregation and total diversity of bacteria and fungi in various tillage systems of sub-humid and semi-arid Kenya. Appl Soil Ecol 58:12–20
Kotzé E, Sandhage-Hofmann A, Meinel J-A, Du Preez CC, Amelung W (2013) Rangegeland management impacts on the properties of clayey soils along grazing gradients in the semi-arid grassland biome of South Africa. J Arid Environ 97:220–229
Linstädter A, Schellberg J, Brüser K, Moreno Garcia CA, Oomen RJ, Du Preez CC, Ruppert JC, Ewert F (2014) Are there consistent grazing indicators in drylands? Testing plant functional types of various complexity in South Africa’s grassland and savanna biomes. PLoS ONE 9(8):e104672. doi:10.1371/journal.pone.0104672
Marschner P (2007) Soil microbial community structure and function assessed by FAME, PLFA and DGGE—Advantages and limitations. In: Varma A, Oelmüller R (eds) Advanced techniques in soil microbiology. Springer, Berlin
Mofidi M, Rashtbari M, Abbaspour H, Ebadi A, Sheidai E, Motamedi J (2012) Impact of grazing on chemical, physical and biological properties of soils in the mountain rangelands of Sahand, Iran. Rangeland J 34:297–303
Moore-Kucera J, Azarenko AN, Brutcher L, Chozinski A, Myrold DD, Ingham R (2008) In search of key soil functions to assess soil community management for sustainable sweet cherry orchards. Hortic Sci 43(1):38–44
Mucina L, Rutherford MC (2006) The vegetation of South Africa. Lesotho and Swaziland, Pretoria
Muller T, Hoper H (2004) Soil organic matter turnover as a function of the soil clay content: consequences for model applications. Soil Biol Biochem 36(6):877–888
Nannipieri P, Giagnoni L, Landi L, Renella G (2011) Role of phosphatase enzymes in soil. In: Bunemann EK, Obreson A, Frossard E (eds) Phosphorus in action. Springer, Berlin
Oates LG, Balser TC, Jackson RD (2012) Subhumid pasture soil microbial communities affected by presence of grazing, but not grazing management. Appl Soil Ecol 59:20–28
Patra AK, Abbadie L, Clays-Josserand A, Degrange V, Grayston SJ, Loiseau P, Loualt F, Mahmood S, Nazaret S, Philippot L, Poly F, Prosser JI, Richaume A, Le Roux X (2005) Effects of grazing on microbial functional groups involved in soil N dynamics. Ecol Monogr 75(1):65–80
Patra AK, Le Roux X, Abbadie L, Clays A, Ply F, Loiseau P, Loualt F (2007) Effect of microbial activity and nitrogen mineralization on free-living nitrogen fixation in permanent grassland soil. J Agron Crop Sci 193:153–156
Perkins JS, Thomas DSG (1993) Spreading deserts or spatially confined environmental impacts? Land degradation and cattle ranching in the Kalahari Desert of Botswana. Land Degrad Rehabil 4:179–194
Prieto LH, Bertileer MB, Carrera AL, Olivera NL (2011) Soil enzyme and microbial activities in a grazing ecosystem of Patagonian Monte, Argentina. Geoderma 162:281–287
Reichel R, Rosendahl I, Peeters ETHM, Focks A, Groeneweg J, Bierl R, Schlichting A, Amelung W, Thiele-Bruhn S (2013) Effects of slurry from sulfadiazine-(SDZ) and difloxacin-(DIF) medicated pigs on the structural diversity of microorganisms in bulk and rhizosphere soil. Soil Biol Biochem 62:82–91
Rousk J, Baath E (2007) Fungal and bacterial growth in soil with plant materials of different C/N ratios. FEMS Microbiol Ecol 62(3):258–267
Rutherford MC, Powrie LW (2011) Can heavy grazing on communal land elevate plant species richness levels on the Grassland biome of South Africa? Plant Ecol 212:407–1418
Sandhage-Hofmann A, Kotzé E, Van Delden L, Dominiak M, Fouché HJ, Van der Westhuizen HC, Oomen RJ, Du Preez CC, Amelung W (2015) Rangeland management effects on soil properties in the savanna biome, South Africa: a case study along grazing gradients in communal and commercial farms. J Arid Environ 120:14–25
Sessitsch A, Weilhalter A, Gerzabek MH, Kirchmann H, Kandeler E (2001) Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microbiol 67:4215–4224
Six J, Frey SD, Thiet RK, Batten KM (2006) Bacterial and fungal contributions to carbon sequestration in agroeco-systems. Soil Sci Soc Am J 70:555
Smet M, Ward D (2006) Soil quality gradients around water points under different management systems in a semi-arid savanna, South Africa. J Arid Environ 64:251–269
Stemmer M, Gerzabek MH, Kandeler E (1998) Organic matter and enzyme activity in particle-size fractions of soils obtained after low-enery sonication. Soil Biol Biochem 30(1):9–17
Strickland MS, Rousk J (2010) Considering fungal:bacterial dominance in soils: methods, controls, and ecosystem implications. Soil Biol Biochem 42:1385–1395
Su YZ, Zhao HL, Zhang TH, Zhao XY (2004) Soil properties following cultivation and non-grazing of a semi-arid sandy grassland in northern China. Soil Till Res 75(1):27–36
Tabatabai MA (1994) Soil Enzymes. In: Weaver RW, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, Wollum A (eds) Method of soil analyses—Part two: microbiological and biochemical properties. Soil Science Society of America, Madison
Tainton NM (1999) The ecology of the main grazing lands of South Africa. In: Tainton NM (ed) Veld management in South Africa. University of Natal Press, Pietermaritzburg
Tefera S, Dlamini BJ, Dlamini AM (2010) Changes in soil characteristics and grass layer condition in relation to land management systems in the semi-arid savannas of Swaziland. J Arid Environ 74:675–684
Tesseman ZK, De Boer WF, Baars RMT, Prins HHT (2011) Changes in soil nutrients, vegetation structure and herbaceous biomass in response to grazing in a semi-arid savanna of Ethiopia. J Arid Environ 75:662–670
Vallejo VE, Arbeli Z, Teran W, Lorenz N, Dick RP, Roldan F (2012) Effect of land management and Prosopis juliflora (Sw.) DC trees on soil microbial community and enzymatic activities in intensive silvopastoral systems of Colombia. Agr Ecosyst Environ 150:139–148
Van der Westhuizen HC, Snyman HA, Fouché HJ (2005) A degradation gradient for the assessment of rangeland condition of a semi-arid sourveld in southern Africa. Afr J Range Forage Sci 22:47–58
Von Mersi W, Schinner F (1991) An improved and accurate method for determining the dehydrogenase activity of soils with iodonitrotetrazolium chloride. Biol Fert Soils 11:216–220
Watts DB, Torbert HA, Feng Y, Prior SA (2010) Soil microbial community dynamics as influenced by composted diary manure, soil properties, and landscape position. Soil Sci 175:474–486
WRB (2007) World Reference Base for Soil Resources 2006: a framework for international classification, correlation and communication. World Soil Resources Reports No. 103. FAO, Rome
Xu Z, Yu G, Zhang X, Ge J, He N, Wang Q, Wang D (2014) The variation in soil microbial communities, enzyme activities and their relationships with soil organic matter decomposition along the northern slope of Changbai Montain. Appl Soil Ecol 86:19–29
Xue D, Yao H, Ge D, Huang C (2008) Soil microbial structure in deverse land use systems: a comparative study using Biolog, DGGE, and PLFA analyses. Pedosphere 18(5):653–663
Yan J, Pan G, Li L, Quan G (2009) Cellulose activity in physically isolated fractions of a paddy soil. Int Conf Environ Inf Appl Technol ESIAT 3:613–616
Acknowledgements
The authors would like to express our appreciation to all staff members of the University of Bonn and the University of the Free State in Bloemfontein for their assistance in field and labor. We wish to thank the community members and farmers for allowing us to do our research on their land. The authors are grateful to the German Research Foundation for funding this project (DFG FOR 1501). We would also like to thank the anonymous reviewers for their valuable and insightful comments to improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kotzé, E., Sandhage-Hofmann, A., Amelung, W. et al. Soil microbial communities in different rangeland management systems of a sandy savanna and clayey grassland ecosystem, South Africa. Nutr Cycl Agroecosyst 107, 227–245 (2017). https://doi.org/10.1007/s10705-017-9832-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10705-017-9832-3