Abstract
DNA methylation is an epigenetic regulator of gene expression, and this process has been shown to be disrupted by environmental contaminants. Di-2-(ethylhexyl) phthalate (DEHP) and related phthalate esters have been shown to affect development in early life stages of fish and can alter genomic methylation patterns in vertebrates. The objectives of this study were the following: (1) Describe the expression patterns of the DNA methyltransferase (dnmt) genes during early fathead minnow (FHM) development. These genes are critical for methylation and imprinting during development. (2) Determine the effects of DEHP on the development of FHM larvae [1 and 14 days post-hatch (dph)]. (3) Determine the effect of DEHP on dnmt expression and global methylation status in larval FHM. FHMs were first collected over a developmental time course [1, 3, 5, 6, and 14 days post-fertilization (dpf)] to investigate the expression patterns of five dnmt isoforms. The expression of dnmt1 and dnmt7 was relatively high in embryos at 1 dpf but was variable in expression, and these transcripts were later expressed at a lower level (>3 dpf); dnmt3 was significantly higher in embryos at 1 dpf compared to those at 3 dpf. Dnmt6 showed more of a constitutive pattern of expression during the first 2 weeks of development, and the mRNA levels of dnmt8 were higher in embryos at 5 and 6 dpf compared to those at 1 and 3 dpf, corresponding to the hatching period of the embryos. A waterborne exposure to three concentrations of DEHP (1, 10 and 100 µg/L) was conducted on 1-day FHM embryos for 24 h and on larval fish for 2 weeks, ending at 14 dpf. DEHP did not negatively affect survival, hatch rate, or the expression of dnmt isoforms in FHMs. There were no differences in global cytosine methylation following DEHP treatments in 14 dpf larvae, suggesting that environmentally relevant levels of DEHP may not affect global methylation at this stage of FHM development. However, additional targeted methylome studies are required to determine whether specific gene promoters are differently methylated following exposure to DEHP. This study offers new insight into the roles of the dnmt enzymes during FHM development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epigenetics refers to heritable changes in gene expression caused by molecular mechanisms that do not involve a change in the DNA sequence of the organism. Several processes involved in epigenetics are histone modification (e.g., acetylation and methylation), noncoding RNA interactions, and 5-cytosine methylation of DNA (Vandegehuchte and Janssen 2011). Methylation of 5-cytosine in DNA is involved in many aspects of gene regulation, including transcriptional silencing, embryonic development, genomic imprinting, and X-chromosome inactivation (Stein et al. 1982; Razin 1998; Licchesi et al. 2005; Wilkinson et al. 2007; Latham et al. 2008). It has also been hypothesized that DNA methylation increases genomic stability and protection for the cell from possible disruptions such as transposable elements (Yoder et al. 1997; Rizwana and Hahn 1999). It is therefore well documented that DNA methylation is a crucial regulatory mechanism for normal development and physiological function.
In eukaryotes, DNA methylation is a heritable regulatory biochemical process that involves the covalent transfer of a methyl group to the 5-position of cytosine from S-adenosyl-l-methionine to form 5-methylcytosine (Hermann et al. 2004). DNA methylation occurs in all known vertebrate genomes, and the primary site of action is at CpG dinucleotide sites (Tweedie et al. 1997; Jeltsch 2002). DNA methylation is primarily catalyzed by a group of enzymes known as DNA methyltransferases (DNMTs; Jeltsch 2002; Hermann et al. 2004). Currently, three classes of DNA methyltransferases have been characterized in mammals, DNMT1, DNMT2 (though DNMT2 does not appear to be involved in DNA methylation), and DNMT3; moreover, there are three different types of DNMT3 known as DNMT3a, DNMT3b, and DNMT3L (DNA methyltransferase-like protein; Jones and Takai 2001; Jeltsch 2002; Hermann et al. 2004). Homologous genes to the mammalian classes, as well as several other related genes (dnmt4, dnmt5, dnmt6, dnmt7, and dnmt8), have been identified in zebrafish (Shimoda et al. 2005; Smith et al. 2011). The dnmt1 gene in zebrafish appears to have a maintenance role similar to the mammalian homologue (Shimoda et al. 2005). The dnmt6 and dnmt8 genes appear to be structurally related to DNMT3a, while dnmt3, dnmt4, dnmt5, and dnmt7 are related to DNMT3b. Additional evidence for these relationships is based upon data demonstrating that dnmt6 and dnmt8 show early expression patterns similar to DNMT3a, while dnmt3, dnmt4, dnmt5, and dnmt7 show comparable expression patterns to DNMT3b (Shimoda et al. 2005; Smith et al. 2011; Campos et al. 2012). The various functions of these genes in fish are still under investigation.
Endocrine-disrupting compounds (EDCs) in the environment are ubiquitous chemicals that affect normal functioning of the hormone systems. Environmental contaminants have also been shown to induce changes in DNA methylation patterns, and exposure to several environmental contaminants including tributyltin, triphenyltin, benzo[a]pyrene, and nickel has been shown to affect DNA methylation across multiple taxa (Lee et al. 1998; Wang et al. 2009; Vandegehuchte and Janssen 2011). For research in fish toxicology and physiology, there has been a new focus on epigenetics mechanisms of gene regulation (Williams et al. 2014). A recent study by Olsvik et al. (2014) demonstrated that contaminants that included dioxin (TCDD) and methylmercury affected methylation patterns of ~200 genes in the F1 generation, and it is now recognized that this process (i.e., epigenetics) is a primary mechanism by which chemicals exert long-term effects in aquatic organisms (Mirbahai and Chipman 2014).
In addition to the aforementioned chemicals, phthalate esters have also been shown to have effects on DNA methylation. These chemicals are used in plasticizers in polyvinyl chloride (PVC) products such as toys and hospital supplies. Phthalate esters have been detected in the environment in water, sediments, and sewage sludge, on a global scale [Canadian Council of Ministers of the Environment (CCME) 1999; Beauchesne et al. 2008; Crisp et al. 2009; Keil et al. 2011]. The most commonly used phthalate ester is di-2-(ethylhexyl) phthalate (DEHP; Beauchesne et al. 2008). DEHP has been found in surface waters as high as 300 µg/L in Canadian freshwater systems (CCME 1999). Of interest, maternal DEHP exposure has been found to increase the global methylation level and DNMT expression in rat testes and causes testicular malformations that resemble human testicular dysgenesis syndrome (Wu et al. 2010). Another study found that maternal DEHP exposure caused changes in promoter methylation of adults rats in several genes, including mineralocorticoid receptor, estrogen receptors alpha and beta, androgen receptor, and peroxisome proliferated-activated receptor alpha (Martinez-Arguelles et al. 2009). There is also evidence that exposure to phthalate chemicals can alter insulin signaling in utero via epigenetic mechanisms (Rajesh and Balasubramanian 2014) and reprogram cardiomyocytes, changing the methylation status of specific CpGs in peroxisome proliferator-activated receptor subunits (Schaedlich et al. 2014). In teleost fishes, DEHP has been shown to increase mortality, delay growth, induce oxidative stress, and interfere with normal endocrine and reproductive function (Chikae et al. 2004a, b; Mankidy et al. 2013; Ye et al. 2014). Given the importance of DNA methylation in development, it is plausible that the disruption of methylation processes is one of the mechanisms through which DEHP exhibits adverse effects in aquatic organisms.
The objectives of this study were to first describe the expression patterns of the dnmt isoforms in the fathead minnow (FHM; Pimephales promelas) in early development. FHMs are a model freshwater toxicological species and are a cyprinid fish found ubiquitously in North America. Due to the large collection of knowledge about their biology, their ability to survive in a range of water types, and their general hardiness and tolerance of handling and experimentation, they have been used extensively in toxicological research for over 50 years (Ankley and Villeneuve 2006). While a number of studies on the FHM transcriptome have been conducted in recent years, there is a paucity of research on the mechanisms of methylation in this species. Therefore, we aimed to address this knowledge gap by first characterizing dnmt expression and then determining whether DEHP exposure affects FHM larvae development via epigenetic mechanisms by measuring the expression of the dnmt genes and in vivo DNA methylation after exposure.
Materials and methods
Development profiles of the dnmt genes
For generating the developmental profiles of the dnmt genes, adult FHMs were taken from a breeding stock at the University of New Brunswick (Saint John) and placed into eight aquariums in 1:2 male/female ratio per tank and a breeding tile was placed in each tank. All animal use protocols were carried out ethically in accordance with Animal Care Committee protocol from the University of New Brunswick, Saint John (approval #2013-3s-08). Each tank was aerated using an air stone, which maintained a mean oxygen content of 86.9 %. The pH was measured daily, and the mean pH was 7.1. Fish remained on a light/dark schedule of 16:8 h, respectively. The experimental room was maintained at approximately 20 °C, while the water was maintained at approximately 25.5 °C. These conditions were maintained for 2 days until breeding was completed (six of the eight aquariums had produced eggs, yielding a total of approximately 360 fertilized embryos) at which point all embryos were pooled and transplanted from the original breeding tiles into petri dishes. The pooled embryos were divided evenly across eight petri dishes, with each petri dish containing 35 embryos. Embryos were maintained at room temperature, also on a 16:8-h light/dark schedule, and water changes occurred every second day. Water changes were performed using a stock tank of water which was maintained at 98.4 % oxygen per liter of water, a pH of 7.4, and a mean room and water temperature both of 20 °C. Mortality rates for embryos were recorded individually as they occurred with mortality rates ranging from 6 to 26 %, with an overall mean mortality rate of 17.5 %.
Embryos were collected and stored at the following time points: 1 day post-fertilization (dpf), 3 dpf, 5 dpf, 1 day post-hatch (6 dpf), and 14 dpf. During the first 3 dpf, the following morphological events occur: completion of embryonic development, post-embryonic development as yolk-sac larvae (yolk-sacs still attached), the switch from endogenous feeding (yolk-sac) to exogenous feeding (food gained via eating), inflation of the swim bladder, development of the anterior dorsal section of the mouth, and development of the gut and intestinal tracts (Kimmel et al. 1995). The first three time points corresponded to the neurula stage, the commencing of pectoral fin blood flow and dorsal swim bladder pigmentation, and pre-hatch (based on work in FHMs by Devlin et al. 1996). The fourth time point (6 dpf) was immediately post-hatch, and the fifth time point was collected after 8 days of larval growth (14 dpf). Pre-hatching development of the FHM P. promelas Rafinesque is described in EPA/600/R-96/079 (1996). Micrographs of these stages have been published by us recently in Wood et al. (2015). The embryos were collected using a stainless steel spatula and placed into 2.0 ml vials, after which a sterile eyedropper was used to remove excess water from the vials. The embryos were placed into the vials in groups of two, while the larvae (6 and 14 dpf samples) were collected individually. Samples were then flash frozen on dry ice and later stored in 1.5 mL microcentrifuge tubes at −80 °C until used in RNA extraction.
DEHP exposures (embryo and larval)
In order to assess the effects of DEHP on pre-hatch embryos and dnmt expression, a 48-h exposure was performed. The first 24-h period is critical for methylation, and we aimed to determine whether DEHP affected dnmt expression during this sensitive period of development. FHMs were collected after fertilization and all embryos that had progressed past the late cleavage stage were removed. The embryos were evenly distributed into 12 glass petri dishes per treatment group containing 50 mL of water (water control), 0.001 % ethanol solution (carrier control), and nominal concentrations of 1 µg/L DEHP, 10 µg/L DEHP, and 100 µg/L DEHP-treated freshwater. The chorion was left on for all exposures to mimic a natural exposure in the environment. The exposure was carried out for 48 h at ambient temperature, and the chemicals were prepared fresh and renewed at 24 h with a 90 % water change. At 24 h, up to 3 individuals per replicate were collected, flash frozen, and stored at −80 °C for gene expression analysis (N = 12). At 48 h, the number of surviving embryos was assessed in each treatment.
A second experiment was performed to assess the effects of DEHP on FHM development over a longer period. This experiment aimed to determine whether exposure to DEHP over the first 2 weeks of development affected global methylation patterns in larvae. The DEHP exposure was conducted in 40 glass petri dishes arranged into 4 groups of 10. The experiment was carried out in a room with a 16:8-h light/dark photoperiod. Four larval [0 days post-hatch (dph)] FHM were randomly placed in each petri plate. This life stage was chosen for the exposure because of research conducted in zebrafish showing that there is high dnmt expression in the post-hatch stage (Smith et al. 2011). The larvae were fed twice each day using 0.05 mL of Artemia culture. The 4 treatments were a carrier control (n = 10), 1 µg/L DEHP (n = 10), 10 µg/L DEHP (n = 10), and 100 µg/L DEHP (n = 10); these concentrations are all considered environmentally relevant (CCME 1999). Ethanol was used as a carrier solvent for DEHP. The control group contained only water and the solvent. Each dish contained 50 mL of freshwater at room temperature. The water in each dish was changed every 2 days at 1 PM using a reservoir of freshwater kept at ambient temperature (May 2013), a mean pH of 6.38, and dissolved oxygen (DO2) level of >95 %. Using stock water, three chemical solutions were prepared fresh every second morning; this included the control water (ethanol added), the D1 water (DEHP dissolved in ethanol added to make a 1.0-µg/L solution), the D2 water (DEHP dissolved in ethanol added to make a 10.0-µg/L solution), and the D3 water (DEHP dissolved in ethanol to make a 100.0-µg/L solution). A 100 % water change was performed for each dish using the reservoir, and the freshly prepared contaminant was added to yield the desired concentration for each treatment group. The ethanol concentration was <0.01 %. Each replicate was checked daily for mortalities (completely white body and/or lack of a heartbeat under a dissection microscope). Mortalities were recorded each morning before the water was changed and the exposure was carried out for fourteen days post-hatch (~19–20 days total post-fertilization).
RNA extraction and cDNA synthesis
RNA was extracted from whole FHM embryo/larval samples (from both the developmental profile and the DEHP exposures) using the Allprep DNA/RNA Mini Kit (Qiagen; Toronto, ON). Samples (whole FHM larvae or 2 embryos) were homogenized in 350 µL Buffer RLT Plus and processed according to the manufacturer’s instructions. During the extraction, the DNA spin column was collected and stored at 4 °C for later DNA extraction (Sect. 2.6). The eluted RNA concentration was quantified using the Nanodrop 2000 spectrophotometer (Thermo Scientific; Wilmington, DE). RNA samples were tested for RNA integrity with the Agilent RNA 6000 Nano Assay Kit (Agilent Technologies; Santa Clara, CA), and RNA Integrity Numbers (RIN) were generated using the Agilent 2100 Bioanalyzer as per manufacturers’ instructions. Samples collected from the DEHP exposures had a mean RIN of 8.8 (±1.9). Complementary DNA (cDNA) was synthesized using DNAse-treated RNA as a template following methods outlined in Chishti et al. (2014). The cDNA product was stored at −20 °C until used in real-time PCRs (Sect. 2.5).
Primer design
The DNA sequences for the DNA methyltransferase genes (dnmt1, dnmt3, dnmt4, dnmt5, dnmt6, dnmt7, and dnmt8) were obtained by searching for the nucleotide sequences in the National Center for Biotechnology Information (NCBI) database. As the FHM sequences were not available, the homologous sequences for Danio rerio were used. The sequences for the genes in D. rerio and at least one other species (e.g., Carassius auratus) were aligned using the Clustal Omega program (EMBL-EBI; Cambridge, UK) to identify conserved regions of DNA sequence. Using these conserved regions and regions specific to each isoform, the forward and reverse primers for the gene transcript were designed using Primer3 (Untergrasser et al. 2012). The aforementioned primers were first tested by amplifying the genes using polymerase chain reaction (PCR) and Taq polymerase (Life Technologies; Carlsbad, CA) as per manufacturer’s protocols. Following the PCR, a 1 % agarose gel was prepared with SYBR® Safe DNA gel stain to visualize amplicons.
Despite multiple attempts, we were unsuccessful at obtaining sequence information for FHM dnmt4 and dnmt5. However, these transcripts appear to be present in the FHM genome as a survey of a new 8 × 60 K microarray for FHMs (Garcia Reyero et al. personal communication) suggests that the following genes are present: DNMT1, DNMT3, DNMT3A, DNMT3B, DNMT3L, DNMT4, DNMT5, DNMT6, DNMT7, and DNMT8. The DNMT3A, DNMT3B, and DNMT3L are homologous to the mammalian DNA methyltransferases. Thus, based on zebrafish, dnmt3, 4, 5, and 7 are orthologs of mammalian 3B, and dnmt6 and 8 are orthologs of mammalian 3A. Interesting, DNMT3L also appears to be present in FHMs, and its function is thought to be the recruitment of DNMT3A and B to methylate cytosine residues. We point out that nomenclature for the dnmt isoforms conforms to that presented in Campos et al. (2012). All gene-specific primers are listed in Table 1.
Real-time PCR
Primer sets that yielded a single amplicon were tested on a standard curve plate using real-time PCR. Primers that yielded a single melt curve, a R 2 > 0.97, and efficiency (E) between 90 and 110 % were deemed successful. Real-time PCR (RT-PCR) was performed to measure gene expression of 4 reference genes for normalization and the 5 dnmt genes tested. Sample sizes for the developmental profile were the following: 1 dpf = 6, 3 dpf = 8, 5 dpf = 8, 6 dpf = 5, 14 dpf = 7 using Ssofast EvaGreen® RT-PCR mix (Bio-Rad; Hercules, CA) and ~300 nM primer. A seven-point standard curve was generated by diluting 1 µL of pooled cDNA in 9 µL of ultrapure water and then performing a 1:5 dilution series. An eighth point in the curve was a no-template control (NTC) using water. The sample cDNA was diluted 1:20 in ultrapure water prior to real-time PCR using a CFX96 instrument (Bio-Rad) as per details in Chishti et al. (2014). The NRTs and NTCs were evaluated on each plate to rule out genomic DNA contamination. The RT-PCR data were analyzed using the gene study option in the CFX96 software (Bio-Rad). The reference genes were loaded into the gene study in different combinations in order to find the most stable combination; ef1 alpha and actb yielded the most stable arrangement with M = 1.10 and CV = 0.43 for the developmental profile and ef1 alpha, rps12, and rps18 were most stable for the DEHP exposure experiments (M = 0.67; CV = 0.26).
Here we point out that many genes commonly used as reference genes in real-time PCR experiments are present in very low amounts before the midblastula transition, and these genes may not be suitable for studies at early time points. This is a limitation for developmental studies in gene expression. Here, ef1 alpha did not statistically vary across the five time points; however, actb did vary significantly and was lower in expression in the first 24 h compared to all other groups (p < 0.01). Differences in Cq values were even more pronounced for rps12 and rps18, and these were not used to normalize data. Noteworthy was that no time point after 24 h differed in terms of the expression level (based on RNA input) for all four control genes tested (i.e., no difference in Cq values), and the differences were confined to comparisons with embryos collected 1 dpf. Ef1 alpha has been reported to be one of the most stable housekeeping genes during teleost development (Fernandes et al. 2008; McCurley and Callard 2008; Øvergård et al. 2010). However, actb can vary from species to species in terms of stability. Fernandes et al. (2008) showed in halibut that actb was one of the most stable across genes across developmental stages, while Overgard et al. (Øvergård et al. 2010) reported that actb varied significantly over time. Similarly, McCurley found ef1 alpha to be stable during zebrafish development, while actb was not very stable. Thus, species differences can exist in the stability of common reference genes over development, and there is a balance between using a sufficient number of genes to achieve a stable baseline for relative comparisons of genes of interest and variation in their expression from stage to stage. In our study, the combination of ef1 alpha and actb provided the most stable M value (or baseline) than other combinations of normalizer genes (ef1 alpha, actb, rps12, and rps18). Thus, we believe that we achieved a reasonable stable baseline from which to draw our relative comparisons. We used ef1 alpha expression as this gene is considered to be one of the most stable across developmental stages and species compared to several other reference genes. Our CV = 0.43 is actually quite good for control genes based on our experience in calculating M values for multiple normalizers (Chishti et al. 2014). However, we caution that the early time points (i.e., <24 h) may show low expression for reference genes in real-time PCR experiments.
The normalized unscaled expression of the genes of interest was analyzed to determine differences in expression between treatment groups. Normalized gene expression was extracted using CFX Manager™ software and gene expression differences determined using the relative ΔΔCq method. All amplicons were verified as correct target genes by Sanger Sequencing at the McGill University and Génome Québec Innovation Centre (Montréal, QB, Canada) (Supplemental Table S1).
DNA extraction and global DNA methylation
DNA purification was conducted using the Allprep DNA/RNA Mini Kit (Qiagen; Toronto, ON) as per manufacturer’s protocol. Final DNA concentration was then quantified using the Nanodrop 2000 spectrophotometer (Thermo Scientific; Wilmington, DE). Global methylation of the genomic DNA was measured using the MethylFlash™ Methylated DNA Quantification Kit (Epigentek; Farmingdale, NY) without modification. The colorimetric assay plate was read on an iMark Microplate Absorbance Reader (Bio-Rad) at 450 nm. The relative percentages of cytosine methylation as well as the absolute quantity (ng) of methylated DNA were calculated using formulas described by the manufacturer (Epigentek).
Statistics
All statistical analyses were performed using GraphPad Prism 5.0 (Graphpad Software, Inc., La Jolla, CA). Differences in survival and hatch were tested using a Mantel–Cox log-rank test. The methylation absorbance data (relative quantity as well as absolute), morphometric data, and gene expression data were tested for normality using the Shapiro–Wilks test and for homogeneity of variance using a Levene’s test. If the assumptions of normality and homogeneity were met, the data were analyzed using a one-way ANOVA test. If differences were detected, a post hoc analysis using Tukey’s HSD test was used to determine which groups differed from the control. If the assumptions of normality and homogeneity were not met, the nonparametric Kruskal–Wallis test (denoted as an H-test statistic) was used and any post hoc differences were tested for using a Dunn’s test. The Spearman rank correlation was used to determine whether dnmt isoforms were significantly associated with each other in expression during development. A p < 0.05 was considered significant.
Results
Developmental expression of dnmt genes
The developmental expression profiles for dnmt1, dnmt3, dnmt6, dnmt7 and dnmt8 are shown in Fig. 1. Dnmt3 transcript abundance differed between 1 and 3 days post-fertilization (H = 10.9; p = 0.028; Fig. 1b), and there was a significant reduction in mRNA levels between these time points. Dnmt8 transcripts differed significantly (H = 18.0; p = 0.0012) in abundance between time points, with transcripts being more abundant on days 5 (p < 0.05) and 6 (p < 0.01) compared to levels at day 1 (Fig. 1e). There were no significant differences in transcript abundance between the 5 time points for dnmt1 (H = 7.3; p = 0.12), dnmt6 (H = 3.4; p = 0.49), or dnmt7 (H = 5.69; p = 0.22) (Fig. 1a, c, d). The Spearman correlation revealed significant (p < 0.05) correlations for expression patterns between dnmt1 and dnmt3 as well as between dnmt1 and dnmt7 (Fig. 1f).
Normalized gene expression for the dnmt isoforms (A–E) across five time points in development. Data are the mean-normalized expression ± standard error of the mean. Time points marked with the same letter do not significantly differ from one another. The figure also shows the correlation coefficient for each transcript (F)
DEHP exposure did not affect survival in embryos or larvae
In the first experiment, a 48-h exposure to one of three concentrations of phthalates did not result in any significant mortality compared to the controls (H = 6.16; p = 0.10). In the second experiment, larvae survival ranged from 80 to 85 % among the four groups over the 14 days (Fig. 2). Survival of the FHM larvae was not different across the experimental groups (H = 0.51; p = 0.92).
DEHP exposure did not alter the expression of dnmt or the global methylation
Following a 24-h exposure, no differences were detected among groups for mRNA levels of dnmt1 (df = 3; H = 1.5; p = 0.68), dnmt3 (H = 3.9; p = 0.28), dnmt6 (H = 3.5; p = 0.33), dnmt7 (H = 3.5; p = 0.34), or dnmt8 (H = 0.51; p = 0.92) .
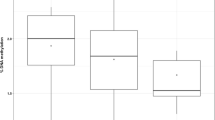
Genomic methylation levels ranged from ~1.4 to 1.5 % among the four groups in larvae. There were no significant differences in global cytosine methylation levels between the four groups (H = 1.061; p = 0.7865; Fig. 3).
Discussion
Developmental expression of the dnmt isoforms in fathead minnows
Similar to other studies in fish, there was differential expression of the dnmt isoforms during FHM development. Cytosine methylation is associated with a number of biochemical and physiological processes. Two notable examples of these are gene silencing and cell differentiation during development (Reik 2007). The zebrafish dnmt3 (and its paralogues dnmt4, 5, 7, and 8) is similar to the mammalian DNMT3 genes that are required for de novo cytosine methylation, while dnmt1 is a homologue of the mammalian DNMT1 which plays a role in maintenance methylation as well as tissue-specific terminal differentiation (Shimoda et al. 2005; Campos et al. 2012).
We determined that at least 5 dnmt isoforms are expressed in FHM embryos; however, it looks as though there are more based upon recent sequencing of the FHM genome and production of a new high-density (8 × 60 K) microarray platform. The expression levels of dnmt1, dnmt3, and dnmt7 were highest at 1 dpf and then decreased by day 3. Dnmt1 expression was significantly correlated with both dnmt4 and dnmt7 expressions. These results are consistent with data generated by Smith et al. (2011). These authors examined the expression of dnmt7 (among others) during zebrafish development and found that dnmt7 mRNA was most abundant during the first 24 h, but by 72 h post-fertilization, mRNA levels had dropped drastically below the detection limit of the bioassay. To better determine the developmental patterns of expression for these genes, it will be necessary to sample at various time points within the first 24 h of development. DNMT7 is known to specifically methylate the zebrafish ntl gene which is crucial to notochord development in fish (Shimoda et al. 2005) and appears to be fish specific (Seritrakul and Gross 2013). Dnmt3 is highly expressed in developing zebrafish up until 72 h, when it falls below the limit of detection of the assay (Smith et al. 2011) and this is a pattern that we also observed in FHMs. As previously stated, dnmt3 is related to mammalian Dnmt3b which is involved in de novo methylation during development (Campos et al. 2012). During zebrafish development, there is a complete demethylation and remethylation of the genome within 6 hpf (MacKay et al. 2007). If FHM development follows a temporal pattern similar to that of zebrafish, the higher levels of dnmt3 and other genes in the mammalian dnmt3b family (such as dnmt4) observed at 1 dpf may be involved in this reprogramming of the FHM genome. In addition to its role in maintaining methylation patterns, DNMT1 plays a critical role in organ formation in zebrafish development by directing histone methylation by SUV39H1 (Rai et al. 2006) which may explain its higher activity in early embryonic development observed in this study. The correlation between dnmt1 and dnmt3 and dnmt1 and dnmt7 also suggests that these genes all play a role in early methylation events in FHM within the first 24 h of development.
Dnmt6 and dnmt8 showed a different pattern of expression compared to dnmt1, 3, and 7, consistent with data collected in zebrafish. The mRNA levels of dnmt8 increased at days 5 and 6 relative to day 1 before dropping again to near-day 1 levels by day 14. There was no difference detected in the expression of dnmt6 over time points. These genes are phylogenetically related to mammalian DNMT3A and are highly similar to each other in nucleic acid sequence (Campos et al. 2012). The study by Smith et al. (2011) found that dnmt6 and dnmt8 mRNA were expressed up until at least 72 h in zebrafish and were ubiquitously expressed throughout different embryo tissues. According to previous research, these genes are important to neural development in fish, being highly expressed in retinal ganglion cells and adult brain tissue (Smith et al. 2011; Seritrakul and Gross 2013). The timeline from fertilization to hatch in this study was ~24 h longer than that reported for FHMs by the Environmental Protection Agency (EPA 1996). The beginning of the rise in dnmt6 and dnmt8 mRNA at approximately 3 days in this study coincides with several aspects of brain and optic development reported from day 2 to hatch (EPA 1996).
A consideration for future studies with FHMs is the timing used in sampling. The vertebrate genome is thought to be demethylated and progressively methylated to adult levels again following fertilization. Research on zebrafish has demonstrated this, with low levels of genome methylation being detected 1–2 h post-fertilization and subsequently reaching near-adult levels by 6 h post-fertilization (Mhanni and McGowan 2004). Assuming that FHM have a similar rate of post-fertilization methylation, then the 1 dpf measurement of dnmt genes offered only a glimpse into an ongoing process which may have ended by the second time point (72 h). Zebrafish also have a faster rate of development and shorter hatching time than FHM which may factor into determining the appropriate window for detection of dynamic changes in dnmt expression.
DEHP exposure and survival
There were no effects in embryo or larval survival following DEHP treatment, which is consistent with several studies in teleost fishes. A 21-day exposure by Chikae et al. (2004b) using medaka (Oryzias latipes) fry did not detect differences in survival between waterborne DEHP-treated (0.01–10 µg/L) and control groups, nor did a study by DeFoe et al. (1990) which examined juvenile FHM, rainbow trout (Onchorynchus mykis), Japanese medaka, and Daphnia magna using DEHP concentrations at or above its water solubility. A recent study exposing adult Chinese rare minnow (Gobiocypris rarus) to environmentally relevant concentrations (0–117.6 µg/L) of DEHP for 21 days found that there were no mortalities following DEHP exposure (Wang et al. 2013). Similarly, a feeding study with Atlantic salmon (Salmo salar) detected no significant differences in mortality between control and treated fish (Norman et al. 2007). Conversely, there are studies that have demonstrated significant mortality due to DEHP exposure. A second study by Chikae et al. (2004a) on Japanese medaka embryos found that low-dose (0.01–1 µg/L) waterborne DEHP exposure caused significantly higher mortalities than the control group. Guppy fry (Poecilia reticulata) exposed continuously to a waterborne exposure of 10 µg/L for ~3 months exhibited higher mortalities than control animals (Zanotelli et al. 2010). These animals also showed reduced growth and condition factor, suggesting that there may be long-term consequences in fish following exposure to DEHP. Conflicting data between this study and those above may be due to the concentrations of DEHP used, as levels in the µg range may not be acutely toxic to FHM at this life stage. Mankidy et al. (2013) observed 30 % mortality when FHM eggs at 1 hpf were exposed to 1 mg DEHP/L until 96 hpf, and this was associated with increased lipid peroxidation. Thus, higher concentration of DEHP may be needed to exert lethal effects in FHM embryos.
Environmental exposures to chemicals have been recently shown to alter the expression of dnmt isoforms in fish, such as the case with dioxins (Aluru et al. 2015). However, in this study, DEHP exposure had no effect on dnmt expression in the FHM embryos and larvae. Murine Dnmt1, Dnmt3a and Dnmt3b transcription has been shown to increase in response to prenatal DEHP exposure in mouse testes with concurrent hypermethylation in the genome of these cells (Wu et al. 2010). Another phthalate, diethyl phthalate, caused altered expression in DNMT3A and DNMT3B (DNMT3A was decreased by serum deprivation, but both genes were increased during serum-deprivation-induced apoptosis) in human PC12 cells (Sun et al. 2013). The absence of expression changes in dnmt transcripts in this study may be due to the differences in the dose for the exposed animals (feeding for mammals and direct cellular exposure for PC12 cells as opposed to waterborne exposure at environmentally relevant levels). The regulation and expression of these genes may also simply not be sensitive to the concentrations of DEHP used in this study.
DEHP exposure did not affect global methylation patterns or the expression of dnmt isoforms at environmentally relevant concentrations in larvae
Exposure to DEHP did not affect the abundance of 5-methylcytosine in the genomes of FHM larvae. DEHP and other phthalates have previously been found to alter genomic methylation patterns in mammalian studies, causing heritable changes that can be detected in offspring of the exposed animals (Kang and Lee 2005; Wu et al. 2010). However, a limitation is that the DNA methylation assay only tested for a global shift in methylcytosine content and does not detect changes at specific loci. Thus, epigenetic changes may still occur without affecting the overall level of methylcytosine (i.e., increase in one promoter with a concurrent decrease in another). This is important as studies show that the estrogen receptor alpha gene in cancer cells shows decreased methylation in the promoter of the gene following dibutyl phthalate and butyl benzyl phthalate treatments (Kang and Lee 2005). Methylation changes in promoter regions of several developmentally important genes (e.g., steroid hormone receptors and IGF1; Martinez-Arguelles et al. 2009; Rajesh and Balasubramanian 2014) may still be a significant mechanism underlying adverse effects of DEHP to aquatic organisms. As there were no detectable changes in 5-methylcytosine levels in the FHM genome, global methylation status in FHM larvae may not be a sensitive approach and a more precise assay (e.g., bisulfite sequencing) will better determine whether DEHP affects specific loci in FHMs.
The global level of 5-methylcytosine observed in FHM was between 1.4 and 1.5 %, which is less at this stage of larval development than that observed in the close relative D. rerio at a comparable stage (~2 %; Fang et al. 2013). Other fish have been shown to have higher levels of cytosine methylation, for example, adult three-spined stickleback (Gasterosteus aculeatus) have 6–8 % of cytosine residues methylated (Aniagu et al. 2008), while adult rare minnow (G. rarus) range from 1 to 3 % (Liu et al. 2014). It is possible that differences in expression patterns between orthologous genes of related species may be regulated in part by this epigenetic variation.
Conclusions
In conclusion, this study is the first to describe the expression patterns of the DNA methyltransferase genes, as well as to measure cytosine methylation in vivo in FHM. This study provides new insight into the roles of the dnmt isoforms during FHM development (e.g., those perhaps more involved in de novo methylation versus maintenance methylation). Further studies should examine the expression of the dnmt genes in more detail prior to 24 hpf, to better define their role in genomic methylation reprogramming. In addition, these data suggest that DEHP at environmentally relevant concentrations may not have significant effects on developing FHM in the short term (i.e., not acutely toxic). However, as other studies in fish suggest, chronic exposures to DEHP may still adversely affect growth and survival.
References
Aluru N, Kuo E, Helfrich LW, Karchner SI, Linney EA, Pais JE, Franks DG (2015) Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio). Toxicol Appl Pharmacol 284:142–151
Aniagu SO, Williams TD, Allen Y, Katsiadaki I, Chipman JK (2008) Global genomic methylation levels in the liver and gonads of the three-spine stickleback (Gasterosteus aculeatus) after exposure to hexabromocyclododecane and 17-β oestradiol. Environ Int 34(3):310–317
Ankley GT, Villeneuve DL (2006) The fathead minnow in aquatic toxicology: past, present and future. Aquat Toxicol 78:91–102
Beauchesne I, Barnabe S, Cooper DG, Nicell JA (2008) Plasticizers and related toxic degradation products in wastewater sludges. Water Sci Technol 57:367–374
Campos C, Valente LMP, Fernandes JMO (2012) Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene 500:93–100
Canadian Council of Ministers of the Environment (1999) Canadian water quality guidelines for the protection of aquatic life: Phthalate esters—DEHP, DBP and DOP Canadian environmental quality guidelines, 1999, Canadian Council of Ministers of the Environment, Winnipeg. http://ceqg-rcqe.ccme.ca/download/en/206
Chikae M, Hatano Y, Ikeda R, Morita Y, Hasan Q, Tamiya E (2004a) Effects of bis(2-ethylhexyl) phthalate and benzo[a]pyrene on the embryos of Japanese medaka (Oryzias latipes). Environ Toxicol Pharmacol 16:141–145
Chikae M, Ikeda R, Hatano Y, Hasan Q, Morita Y, Tamiya E (2004b) Effects of bis(2-ethylhexyl) phthalate, gamma-hexachlorocyclohexane, and 17-beta-estradiol on the fry stage of medaka (Oryzias latipes). Environ Toxicol Pharmacol 18:9–12
Chishti YZ, Feswick A, Martyniuk CJ (2014) Progesterone increases ex vivo testosterone production and decreases the expression of progestin receptors and steroidogenic enzymes in the fathead minnow (Pimephales promelas) ovary. Gen Comp Endocrinol 199:16–25
Crisp TM, Clegg ED, Cooper RL, Wood WP, Anderson DG, Baetke KP, Hoffman JL, Morrow MS, Rodier DJ, Schaeffer JE, Touart LW, Zeeman MG, Dargnat C, Blanchard M, Chevreuil M, Teil MJ (2009) Occurrence of phthalate ester in the Seine River estuary (France). Hydrol Process 23:1192–1201
DeFoe DL, Holcombe GW, Hammermeister DE, Biesinger KE (1990) Solubility and toxicity of eight phthalate esters to four aquatic organisms. Environ Toxicol Chem 9:623–636
Devlin EW, Brammer JD, Puycar RL, McKim, JM (1996) Prehatching development of the fathead minnow Pimephales promelas Rafinesque. EPA/600/R-96/079
Fang X, Corrales J, Thornton C, Scheffler BE, Willett KL (2013) Global and gene specific DNA methylation changes during zebrafish development. Comp Biochem Physiol B: Biochem Mol Biol 166(1):99–108
Fernandes JMO, Mommens M, Hagen Ø, Babiak I, Solberg C (2008) Selection of suitable reference genes for real-time PCR studies of Atlantic halibut development. Comp Biochem Physiol B 150(1):23–32
Hermann A, Gowher H, Jeltsch A (2004) Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci 61:2571–2587
Jeltsch A (2002) Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chem Bio Chem 3:274–293
Jones PA, Takai D (2001) The role of DNA methylation in mammalian epigenetics. Science 293:1068–1070
Kang SC, Lee BM (2005) DNA methylation of estrogen receptor alpha gene by phthalates. J Toxicol Environ Health A 68:1995–2003
Keil R, Salemme K, Forrest B, Neibauer J, Logsdon M (2011) Differential presence of anthropogenic compounds dissolved in the marine waters of Puget Sound, WA and Barkley Sound, BC. Mar Pollut Bull 62:2404–2411
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310
Latham T, Gilbert N, Ramsahoye B (2008) DNA methylation in mouse embryonic stem cells and development. Cell Tissue Res 331:31–55
Lee YW, Broday L, Costa M (1998) Effects of nickel on DNA methyltransferase activity and genomic DNA methylation levels. Mutat Res 415(3):213–218
Licchesi JC, Kelly WG, Panning B (2005) Chromatin remodeling in dosage compensation. Annu Rev Genet 39:615–651
Liu Y, Yuan C, Chen S, Zheng Y, Zhang Y, Goa J, Wang Z (2014) Global and cyp19a1a gene specific DNA methylation in gonads of adult rare minnow Gobiocypris rarus under bisphenol A exposure. Aquat Toxicol 156:10–16
MacKay AB, Aizeddin AM, McGowan RA, Krone PH (2007) Immunological detection of changes in genomic DNA methylation during early zebrafish development. Genome 50:778–785
Mankidy R, Wiseman S, Ma H, Giesy JP (2013) Biological impact of phthalates. Toxicol Lett 217:50–58
Martinez-Arguelles DB, Culty M, Zirkin BR, Papadopolous V (2009) In utero exposure to di-2-(ethylhexyl) phthalate decreases mineralocorticoid receptor expression in the adult testis. Endocrinology 150:5575–5585
McCurley AT, Callard GV (2008) Characterization of housekeeping genes in zebrafish: male–female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Bio 9:102
Mhanni AA, McGowan RA (2004) Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev Genes Evol 214:412–417
Mirbahai L, Chipman JK (2014) Epigenetic memory of environmental organisms: a reflection of lifetime stressor exposure. Mutat Res Genet Toxicol Environ Mutagen 764–765:10–17
Norman A, Borjeson H, David F, Tiempont B, Norrgren L (2007) Studies of uptake, elimination, and late effects in Atlantic salmon (Salmo salar) dietary exposed to di-2-ethylhexyl phthalate (DEHP) during early life. Arch Environ Contam Toxicol 52:235–242
Olsvik PA, Williams TD, Tung HS, Mirbahai L, Sanden M, Skjaerven KH, Ellingsen S (2014) Impacts of TCDD and MeHg on DNA methylation in zebrafish (Danio rerio) across two generations. Comp Biochem Physiol C: Toxicol Pharmacol 165:17–27
Øvergård AC, Nerland AH, Patel S (2010) Evaluation of potential reference genes for real time RT-PCR studies in Atlantic halibut (Hippoglossus Hippoglossus L.); during development, in tissues of healthy and NNV-injected fish, and in anterior kidney leucocytes. BMC Mol Biol 11:36
Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairs BR, Jones DA (2006) Zebra fish dnmt1 and suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol 26:7077–7085
Rajesh P, Balasubramanian K (2014) Phthalate exposure in utero causes epigenetic changes and impairs insulin signalling. J Endocrinol 223:47–66
Razin A (1998) CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J 17:4905–4908
Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425–432
Rizwana R, Hahn PJ (1999) CpG methylation reduces genomic instability. J Cell Sci 112:4513–4519
Schaedlich K, Schmidt JS, Kwong WY, Sinclair KD, Kurz R, Jahnke HG, Fischer B (2015) Impact of di-ethylhexylphthalate exposure on metabolic programming in P19 ECC-derived cardiomyocytes. J Appl Toxicol 35:861–869. doi:10.1002/jat.3085
Seritrakul P, Gross JM (2013) Expression of the de novo DNA methyltransferases (dnmt3–dnmt8) during zebrafish lens development. Dev Dyn 243:350–356
Shimoda N, Yamakoshi K, Miyake A, Takeda H (2005) Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev Dyn 233:1509–1516
Smith THL, Collins TM, McGowan RA (2011) Expression of the dnmt3 genes in zebrafish development: similarity to Dnmt3a and Dnmt3b. Dev Genes Evol 220:347–353
Stein R, Razin A, Cedar H (1982) In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci USA 79:3418–3422
Sun Y, Guo Z, Iku S, Saito T, Kurasaki M (2013) Diethyl phthalate enhances expression of SIRT1 and DNMT3a during apoptosis in PC12 cells. J Appl Toxicol 33:1484–1492
Tweedie S, Charlton J, Clark V, Bird A (1997) Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol 17:1469–1475
United States Environmental Protection Agency (1996) Prehatching development of the Fathead Minnow Rafinesque Office Res Dev Washington, DC
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40(15):e115
Vandegehuchte MB, Janssen CR (2011) Epigenetics and its implications for ecotoxicology. Ecotoxicology 20:607–624
Wang Y, Wang C, Zhang J, Chen Y, Zuo Z (2009) DNA hypomethylation induced by tributyltin, triphenyltin, and a mixture of these in Sebastiscus marmoratus liver. Aquat Toxicol 95(2):93–98
Wang X, Yang Y, Zhang L, Ma Y, Han J, Yang L, Zhou B (2013) Endocrine disruption by di-(2-ethylhexyl) phthalate in Chinese rare minnow. Environ Toxicol Chem 32:1846–1854
Wilkinson LS, Davies W, Isles AR (2007) Genomic imprinting effects on brain development and function. Nat Rev Neurosci 8:832–843
Williams TD, Mirbahai L, Chipman JK (2014) The toxicological application of transcriptomics and epigenomics in zebrafish and other teleosts. Brief Funct Genomics 13:157–171
Wood RK, Seidel JS, Martyniuk CJ (2015) Transcripts involved in steroid biosynthesis and steroid receptor signaling are expressed early in development in the fathead minnow (Pimephales promelas). Comp Biochem Physiol B: Biochem Mol Biol 182:64–72
Wu S, Zhu J, Li Y, Lin T, Gan L, Yuan X, Xiong J, Liu X, Xu M, Zhao D, Ma C, Li X, Wei G (2010) Dynamic epigenetic changes involved in testicular toxicity induced by di-2-(ethylhexyl) phthalate in mice. Basic Clin Pharmacol Toxicol 106:118–123
Ye T, Kang M, Huang Q, Fang C, Chen Y, Shen H, Dong S (2014) Exposure to DEHP and MEHP from hatching to adulthood causes reproductive dysfunction and endocrine disruption in marine medaka (Oryzias melastigma). Aquat Toxicol 146:115–126
Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13:335–340
Zanotelli VR, Neuhauss SC, Ehrengruber MU (2010) Long-term exposure to bis(2-ethylhexyl)phthalate (DEHP) inhibits growth of guppy fish (Poecilia reticulata). J Appl Toxicol 30:29–33
Acknowledgments
The authors have no conflict of interest to declare. This research was funded by a Natural Science and Engineering Research Council (NSERC) Discovery Grant to CJM (386275-2010) and an NSERC USRA to RKW. We thank Rosalinda Knight, Jennifer Loughery, and Kathleena Sarty for their assistance with FHM rearing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wood, R.K., Crowley, E. & Martyniuk, C.J. Developmental profiles and expression of the DNA methyltransferase genes in the fathead minnow (Pimephales promelas) following exposure to di-2-ethylhexyl phthalate. Fish Physiol Biochem 42, 7–18 (2016). https://doi.org/10.1007/s10695-015-0112-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-015-0112-3