Abstract
The diverse timing of the heading stage, mainly determined by the VRN genes, contributes to the wide spread of bread wheat and the realization of adaptive and breeding potential. Wild wheat species are valuable sources for expanding the bread wheat genetic diversity by the introgression of new gene alleles, including VRN genes. In this study, a near-isogenic line of the winter wheat cultivar Bezostaya 1 with a VRN-A1 dominant allele was obtained with a Triticum aestivum ssp. petropavlovskyi accession as the donor. Using known PCR markers for the promoter and first intron sequences of the VRN-1 gene and subsequent sequencing of PCR fragments, the presence of the Langdon deletion was revealed in the first intron (Vrn-A1L allele), previously described only in tetraploid wheat. The allele composition of VRN genes was determined in T. aestivum ssp. petropavlovskyi accessions and the presence of the Vrn-A1L allele was established in all accessions. It was shown that the Vrn-A1L dominant allele increased the shoots-heading period under long- and short-day conditions, which is associated with a prolongation period before the first node formation. The comparative study of productivity characteristics of near-isogenic lines with Vrn-A1a and Vrn-A1L alleles on spike and plant productivity is presented.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For cereals, including bread wheat, the growth season duration and, in particular, the onset time and duration of certain stages of organogenesis, is an important trait that allows plants to adapt to certain natural and climatic conditions, being in close relationship with the resistance to adverse biotic and abiotic factors and, ultimately, productivity (Worland 1996; Snape et al. 2001; Cockram et al. 2007; Kamran et al. 2014). To date, data has been complied on the genetic control of growth season length and molecular mechanisms of flowering regulation in wheat and other cereal species (Distelfeld et al. 2009). It is known that the duration of the bread wheat growth season and the transition from the vegetative to the generative stage are controlled by complex interaction mechanisms between several genetic systems, among which the vernalization response genes (VRN) and photoperiod sensitivity genes (PPD) play the leading role (Snape et al. 2001; Kamran et al. 2014). One of the most effective mechanisms underlying the regulation of the wheat transition from the vegetative stage to the generative stage is the need for vernalization, which distinguishes spring wheat from winter wheat (Loukoianov et al. 2005). The need to vernalize winter crops is an important adaptive mechanism to prevent low temperature damage to the sensitive apical meristem of the growth apex (Yan et al. 2003; Trevaskis et al. 2007).
The main genes determining the wheat genetic diversity in terms of heading time were cloned: VRN-1 (Yan et al. 2003), VRN-2 (Yan et al. 2004a), VRN-3 (Yan et al. 2006), and VRN-D4 (Kippes et al. 2015). The VRN-1 locus encodes the MADS-box transcription factor, the homolog of the Arabidopsis meristem identity gene APETALA1 (Yan et al. 2003), which controls the transition of plants from the vegetative stage to the generative stage. For the beginning of wheat flowering, the VRN-1 transcription must exceed a certain threshold value (Loukoianov et al. 2005). The VRN-2 gene encodes two proteins: ZCCT1 and ZCCT2, the products of which contain the putative domains: zinc finger and CCT and acts as a dominant repressor of VRN-3 and flowering (Yan et al. 2004a). The VRN-3 (TaFT1) locus is a homolog of the Arabidopsis FT (FLOWERING LOCUS T) gene. It is an integrator of various pathways involved in the formation of heading time, up-regulated by a long day, which is a regulator of the VRN-1 expression. The bread wheat VRN-B3 gene is mapped in the short arm of the 7B chromosome (Yan et al. 2006). The VRN-4 gene is a VRN-A1 gene paralog resulting from the insertion of the 290 kb region of the 5A chromosome long arm, with the VRN-A1 gene, into the short arm of chromosome 5D (Kippes et al. 2015). Low positive temperatures activate the expression of VRN-1, which initiates the apical meristem transition to the generative stage of development, and suppresses the action of VRN-2, which is a TaFT1 repressor (Yan et al. 2004b; Trevaskis et al. 2007). The VRN-1 locus is represented by three homeologous genes: VRN-A1, VRN-B1, and VRN-D1 localized in the long arms of 5A, 5B, and 5D chromosomes, respectively, which mainly determine the need for vernalization in bread wheat (Snape et al. 2001; Yan et al. 2003). In wheat, winter, spring, and facultative types are known, which are distinguished according to allelic variations in the VRN-1 locus. The VRN-A1, VRN-B1, and VRN-D1 dominant alleles determine the spring type of development with a varying degree of genotype sensitivity to vernalization, and recessive ones determine the winter type. The lowest plant sensitivity to vernalization is determined by the VRN-A1 gene, while genotypes with VRN-B1 and VRN-D1 dominant genes are more sensitive to vernalization and determine later heading (Stelmakh 1992), correlating with the relative expression of these genes and the effect of transcripts on reducing the VRN-2 expression (Loukoianov et al. 2005).
It was shown that the known allelic variation in the VRN-1 locus associated with the need or lack of vernalization was mainly determined by mobile element insertions, deletions, and duplications in two regulatory regions: the promoter and the first intron (Distelfeld et al. 2009). Three Vrn-A1a alleles are described: Vrn-A1a.1, Vrn-A1a.3 (syn. Vrn-A1a, described for hexaploid and tetraploid wheat) and Vrn-A1a.2, which have MITE insertion and duplication in the promoter region. Vrn-A1a.2 contains a 16 bp deletion and four single nucleotide deletions within the MITE insertion compared to Vrn-A1a.1 (Yan et al. 2004b; Muterko et al. 2016a). Seven Vrn-A1b allele variants with 20 bp deletion, with different polymorphisms in A-tract and C-rich fragment within the VRN-box were detected (Konopatskaia et al. 2016; Muterko et al. 2016a). Vrn-A1d, Vrn-A1e, Vrn-A1f, Vrn-A1g, Vrn-A1h, and Vrn-A1k are characterized by small deletions in the promoter region in diploids and tetraploids (Yan et al. 2004b; Golovnina et al. 2010; Muterko and Salina 2017). Vrn-A1i, described in T. turgidum and T. durum, has a point mutation in the VRN-box (Muterko et al. 2016a). In the T. compactum hexaploid wheat, the Vrn-A1j allele with a deletion in the promoter region was detected (Muterko et al. 2016b). The vrn-A1u and Vrn-A1ins alleles characterized by a 1.4 kb deletion or 0.5 kb insertion in the first intron were described for diploid species (Dubcovsky et al. 2006; Shcherban et al. 2015). The Vrn-A1c and Vrn-A1L alleles found in tetraploids (Vrn-A1c and Vrn-A1L) and bread wheat (Vrn-A1c) are characterized by extended deletions of 5.5 kb and 7.2 kb in the first intron, respectively (Fu et al. 2005; Shcherban et al. 2015). Sehgal et al. (2015) identified the allele with 6 kb deletion the first intron marked as Vrn-A1f in T. aestivum landraces. T. araraticum and T. timopheevii are characterized by Vrn-A1f-del and Vrn-A1f-ins alleles containing the Vrn-A1f promoter (50 bp deletion) with 2.7 kb deletion and 0.4 kb insertion in the first intron (Shcherban et al. 2016), and Vrn-A1f-like with additional 8 bp deletion, 0.4 kb insertion, 2.7 kb deletion in the first intron and SNP in the coding region (Ivaničová et al. 2016).
The dominant alleles of the VRN-B1 and VRN-D1 loci differ from recessive alleles mainly by the presence of insertions/deletions in the first intron (Fu et al. 2005; Shcherban and Salina 2017; Shi et al. 2019). Tetra- and hexaploid wheat species are characterized by allelic variants with mutations in the first intron: Vrn-B1a (6.8 kb deletion) (Fu et al. 2005), Vrn-B1b (6.8 kb and 36 bp deletions) (Santra et al. 2009), Vrn-B1c (6.8 kb and 0.8 kb deletions and 0.4 kb duplication) (Milec et al. 2012; Shcherban et al. 2012) and Vrn-B1d (6.8 kb and 187 bp deletions, one SNP and one 4 bp mutation) (Zhang et al. 2018). Mutant alleles with changes in the promoter region are represented by the Vrn-B1a allele with 127 bp insertion (Golovnina et al. 2010), Vrn-B1ins (5.4 kb insertion) (Chu et al. 2011), Vrn-B1dic (11% dissimilarity vs. vrn-B1) (Konopatskaia et al. 2016). The Vrn-D1a and Vrn-D1b dominant alleles of spring bread wheat have a 4.2 kb deletion in the first intron differing from each other by a single nucleotide mutation (Fu et al. 2005; Zhang et al. 2012). Vrn-D1s (0.8 kb insertion) and Vrn-D(t)1 (5.4 kb deletion) alleles are described in T. spelta and Ae. tauschii (Takumi et al. 2011; Muterko et al. 2015). The Vrn-D1c allele in bread wheat is associated with a 174 bp insertion in the promoter region (Zhang et al. 2015). Of the described allelic diversity of the VRN-1 locus in bread wheat, the following Vrn-A1a, Vrn-A1b, Vrn-A1c, Vrn-A1f (6 kb deletion in the first intron), Vrn-B1a, Vrn-B1b, Vrn-B1c, Vrn-D1a, Vrn-D1b, Vrn-D1c, and Vrn-D1s alleles were founded.
The VRN-B3 locus is represented by five dominant alleles found in bread wheat and tetraploid species associated with structural changes in the promoter. Vrn-B3a in the Chinese Spring/Hope 7B substitution line contains a retrotransposon insertion of 5.3 kb (Yan et al. 2006). The Vrn-B3c allele differs from Vrn-B3a one by two deletions (4 bp and 20 bp) within the retrotransposon region (Chen et al. 2013). The Vrn-B3b allele has 890 bp insertion (Chen et al. 2013). The Vrn-B3d and Vrn-B3e alleles have 1615 bp and 160 bp insertions, respectively (Berezhnaya et al. 2021).
It is important to study the effect of different alleles associated with structural changes in the regulatory regions of the VRN-1 genes on the beginning of heading and flowering of bread wheat. Near-isogenic lines (NILs) of bread wheat are the most suitable object for studying the VRN-1 allele polymorphism in heading time control. It was found that genotypes with the Vrn-A1a dominant allele were the fastest growing. Cultivars with the dominant Vrn-A1b allele have a few days later heading than ones with the dominant Vrn-A1a allele (Koval and Goncharov 1998). The Vrn-B1a dominant allele increases the duration of the shoots-heading period relative to the Vrn-B1c allele (Efremova et al. 2011).
Since, as mentioned above, the currently known genetic diversity of bread wheat is described by a small number of VRN gene alleles found in diploid and tetraploid cereals. Therefore, the transfer of VRN gene alleles from wild and cultivated wheat species will make it possible to study the effect of alleles on heading time and obtain new genotypes for breeding. In this study, the research team obtained and studied a NIL of the Bezostaya 1 (Bez1) winter cultivar with a Vrn-A1pet dominant allele whose donor was a T. aestivum ssp. petropavlovskyi (Udacz. et Migusch.) N.P. Gontsch., hexaploid wheat (2n = 6x = 42, BBAuAuDD) (endemic to Xinjiang and Tibet) accession that has a species-specific P1 gene determining the elongated external glumes (Xiao et al. 2021). Previous studies contain limited information on the genetic control of the growth season duration of T. aestivum ssp. petropavlovskyi (Efremova et al. 2000; Goncharov 2012; Dragovich et al. 2021). Therefore, the purpose of this work was to identify the allelic variants of the VRN genes in NIL and T. aestivum ssp. petropavlovskyi accessions and to evaluate the influence of the VRN-A1 gene structural features on the developmental phase duration and productivity traits.
Materials and methods
Plant material
The material for the study was the NIL of the winter cultivar Bez1 with the Vrn-A1pet dominant allele, whose donor was an accession of T. aestivum ssp. petropavlovskyi (Udacz. et Migusch.) N.P. Gontsch (catalog number unknown), designated “KIZ”, from the Federal Research Center N.I. Vavilov Institute of Plant Genetic Resources (VIR, Russian Federation). i:Bez1Vrn-A1pet was obtained by crossing of spring accession T. aestivum ssp. petropavlovskyi (KIZ) with recurrent winter cultivar Bez1. In eight backcrosses, spring heterozygotes tagged with the Vrn-A1pet dominant allele were used as fathers. After selfing BC8F1 plants, homozygotes were isolated in generation BC8F2. In addition, the i:Bez1Vrn-A1a, obtained earlier by the authors, as well as seven accessions of T. aestivum ssp. petropavlovskyi from the VIR collection, were used in the work.
DNA extraction, PCR amplification and sequencing
Total DNA was isolated from 100 mg of leaves using the DNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s protocol. Polymerase chain reactions (PCR) were performed in a 20 μl volume with 1 × reaction buffer (10 mM TrisHCl, pH 8.8; 4 mM MgCl2; 1 mM (NH4)2SO4), 200 μM of each dNTP, 0.5 μM of each primer and 1 unit of Taq DNA polymerase and 0.1 μg of genomic DNA. The structure of the used primers for amplifying the promoter and first intron sequences of VRN-A1, VRN-B1, VRN-D1, and VRN-B3 genes and PCR conditions were consistent with the published protocols (Table 1). The reaction was run on a Bio Rad T100 Thermal Cycler (USA). The amplification products were separated by electrophoresis on a 1.5% agarose gel in 1 × TAE buffer with the addition of ethidium bromide. After electrophoresis, the gel was photographed in ultraviolet light using the Doc-Print II gel documentation system (Vilber Lourmat, France).
PCR products were separated by agarose gel electrophoresis and purified using a QIAquick Gel Extraction Kit (QIAGEN, Germany) according to the manufacturer’s protocol. Sequencing reactions were performed with the ABI BigDye Terminator Kit and corresponding specific primers on an ABI 3130XL Genetic Analyser (Applied Biosystems) in the SB RAS Genomics Core Facility (http://www.niboch.nsc.ru/doku.php/corefacility) following the manufacturer’s protocol. The search for nucleotide sequence homology was performed using the FASTA and BLAST algorithms in the NCBI databases (www.ncbi.nlm.nih.gov/Database/).
Analysis of the heading time, the developmental phases duration and the productivity traits
NILs with dominant alleles of VRN-A1 loci and accessions of T. aestivum ssp. petropavlovskyi were grown in the experimental field of the Breeding and Genetics Laboratory of the Institute of Cytology and Genetics (ICG SB RAS) under a natural long photoperiod (55° 2,49’ N, 82° 56,80’ E; day length for the May–August period, 17 h) in 2017–2019 and 2021. Seed sowing was on May 15 in 2017, May 22 in 2018, May 13 in 2019 and May 17 in 2021, respectively. Also, NILs and T. aestivum ssp. petropavlovskyi (KIZ) were grown in the greenhouse of the Laboratory of Artificial Plant Growth at ICG SB RAS at 20–25 °C under a long (16–18 h light) and short (12–14 h light) photoperiod, with or without vernalization treatment. The following developmental phases were studied: shoots, third leaf, first node, flag leaf, heading, and ripening. The first node phase was recorded when the first node appeared on the main shoot at a height of 1 cm above the soil surface. The heading was recorded when the head had fully emerged from the flag leaf. A total of 20–30 plants of each line and accessions of T. aestivum ssp. petropavlovskyi were examined. Also, the duration of the shoots-heading phase of F1 and F2 hybrids of NILs Bez1Vrn-A1a and Bez1Vrn-A1pet was studied.
For a comparative productivity study, the best 25–30 samples from each NIL were selected. The productivity components of the main spike (spike length, number of spikelets, grain number and weight) and plant (spike number, grain number and weight), the weight of 1000 grains, and plant height were examined.
Statistical treatment of the data
Statistical treatment of the obtained data was carried out using Microsoft Excel 2013. To assess the statistical significance of the differences between the mean values, Student’s test was used (t-test). A two-factor ANOVA was used to investigate the effect of VRN-A1 dominant alleles on spike and plant productivity.
Meteorological conditions
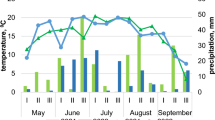
The four field experiments performed in the 2017, 2018, 2019 and 2021 seasons had different climatic conditions (Fig. S1). In our studies, groundbased observations of the local weather station were used. June and July 2017 were characterized by higher temperatures compared to the annual average (deviation was 2.4 °C). In the remaining months the values corresponded to the annual average. Precipitation in May was 73% of the normal, while June, July and September were, on the contrary, wetter (precipitation was 30–63% higher than the annual average). In 2018, the average temperature in May was 4 °C below annual average. June and September, on the contrary, were on average warmer by 2 °C. May precipitation was 200% and June 130% of annual average; August and September precipitation totals were only 49% and 11% of normal, respectively. In 2019, August was warmer by 2.6 °C. Temperatures in the other months were no different than annual average. In 2019, there was a significant deficit in precipitation in all summer months, amounting to only 17% and 11% of normal in June and August, and not exceeding 60% in May, July, and September. In 2021, temperatures were 3.3 °C higher in May and 1.9 °C higher in August. May and July were more dry (68% and 37% of annual average precipitation, respectively), in contrast, in June, the amount of precipitation was 30% higher than annual average.
Results
Determination of allelic variants of VRN genes in NIL by the Bez1 cultivar
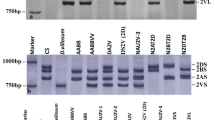
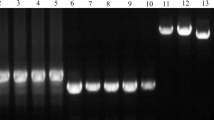
The goal was to determine the allelic variants of VRN genes in NIL by the Bez1 cultivar with an unidentified allele from T. aestivum ssp. petropavlovskyi Udacz. et Migusch. (Vrn-A1pet), which is non-allelic to the known Vrn-A1a and Vrn-A1b loci. Using a pair of VRN1AF and VRN1-INT1R primers to determine allelic variants of the VRN-A1 promoter in i:Bez1Vrn-A1pet and T. aestivum ssp. petropavlovskyi (KIZ), a PCR product of about 700 bp was amplified and then sequenced (Fig. 1a). The sequenced regions were found to be completely identical to the sequences of the intact promoter vrn-A1 T. aestivum NIL Triple Dirk C. The first intron of the VRN-A1 gene was analysed using three primer pairs. When Intr1/C/F and Intr1/AB/R primers were used, which amplify the intact sequences of the first intron, there was no PCR product of 1068 bp. No PCR products of 5.5 kb deletion IL369 identified in hexaploid Afghanistan landrace IL369 were detected. Using the Ex1/C/F and Intr1/A/R3 primer pair in i:Bez1Vrn-A1pet and T. aestivum ssp. petropavlovskyi (KIZ), a 522 bp product was amplified (Fig. 1b), indicating the 7.2 kb deletion (391 bp to 7612 bp) in the first intron. Such deletion was first described for the tetraploid cultivar Langdon. The recessive alleles vrn-B1 and vrn-D1 were also detected (Fig. 1c, d).
Amplification products with primers: VRN1AF and VRN1-INT1R (a), Ex1/C/F and Intr1/A/R3 (b), Ex1/B/F3, Intr1/B/F, Intr1/B/R3, and Intr1/B/R4 (c), Intr1/D/F and Intr1/D/R4 (d) for the regions of the promoter and the first intron of VRN-A1, VRN-B1, and VRN-D1: 1—i:Bez1Vrn-A1L, 2—T. aestivum ssp. petropavlovskyi (KIZ). M—100 bp Ladder
Thus, the isogenic line identified is designated as i:Bez1Vrn-A1L. Since the Vrn-A1L dominant allele has not been previously detected in bread wheat accessions, it is of interest to establish its effect on the growth season duration.
The analysis of heading time and the developmental phase duration of NILs with VRN-1 gene dominant alleles
During the experimental years, the duration of the period from shoots to heading in i:Bez1Vrn-A1a under field conditions varied within 42–50 days. NILs with the Vrn-A1L dominant allele, as well as the parent of this allele, T. aestivum ssp. petropavlovskyi (KIZ), were later mature than the i:Bez1Vrn-A1a line. The duration of the period from shoots to heading of i:Bez1Vrn-A1L was 49 to 61 days, 6–17 days later than that of i:Bez1Vrn-A1a) (P < 0.001) (Table 2).
In the analysis of Table 3, we can note a response to the day length of the i:Bez1Vrn-A1L. Thus, the increase in the average number of days before heading under short-day (12–14 h) relative to long-day (16–18 h) conditions was about 5 days. In i:Bez1Vrn-A1L compared with i:Bez1Vrn-A1a, heading came later by 18–29 and 17–19 days under short- and long-day conditions, respectively (P < 0.001). A similar trend was showed in T. aestivum ssp. petropavlovskyi (KIZ).
When analyzing the duration of individual phases of development (Figs. 2 and 3), we can note that the increasing shoots – heading period in i:Bez1Vrn-A1L versus i:Bez1Vrn-A1a is associated with an increasing period to form the first node. This was established in the experiments under both field and greenhouse conditions. The duration of the first node – flag leaf and flag leaf – heading periods varied insignificantly. Under field conditions, i:Bez1Vrn-A1L versus i:Bez1Vrn-A1a was characterized by an acceleration of the period from heading to ripening, while under greenhouse conditions, this trend was not confirmed. The specifics of individual phases of T. aestivum ssp. petropavlovskyi development were similar to i:Bez1Vrn-A1L. In short-day conditions, an increase in the total duration of the growing season due to the vegetative phase lengthening was also confirmed. The increase in the duration of the studied phases of development in NILs with a decrease in day length is explained by the Bez1 cultivar characteristic response to day length.
Thus, it was found that the Vrn-A1L dominant allele caused later heading compared to the Vrn-A1a allele, which is mainly due to the prolongation of the period before the first node formation.
The effect of vernalization duration on the heading time of the i:Bez1Vrn-A1L and T. aestivum ssp. petropavlovskyi (KIZ)
The i:Bez1Vrn-A1a is known to be insensitive to vernalization. Without vernalization, the duration of the period from shoots to heading in the i:Bez1Vrn-A1L and T. aestivum ssp. petropavlovskyi (KIZ) was 63 days under 16 h photoperoid (Fig. 4), while when vernalized from 14 to 49 days, both genotypes responded sensitively by accelerating heading. The duration of the period before heading in i:Bez1Vrn-A1L plants at a vernalization duration of 14, 35 and 49 days decreased by 5, 16 and 22 days, respectively compared to the non-vernalized plants. After 49 days of vernalization, the plants formed ears in 42 days, which coincides with the time of heading of non-vernalized i:Bez1Vrn-A1a plants.
The duration of the shoots-heading period in F1 and F2 hybrids obtained by crossing NILs carrying the dominant alleles Vrn-A1a and Vrn-A1L
The duration of the shoots-heading period in the NILs Bez1Vrn-A1a and Bez1Vrn-A1L was 39 and 62 days, respectively. In the F1 hybrids between these lines, the value was intermediate but closer to the early-ripening parental form and was 48 days. Among the F2 hybrids, not a single winter plant was segregated, indicating the allelism of the Vrn-A1a and Vrn-A1L loci. F2 population segregation shows that the plants are divided into early heading (37–45 days as in i:Bez1Vrn-A1a) and late heading (58–72 days as in i:Bez1Vrn-A1L), as well as an intermediate class (46–57 days), corresponding in heading time to F1 hybrids (Fig. 5).
A comparative analysis of spike and plant productivity of the NILs Bez1Vrn-A1a and Bez1Vrn-A1L
A comparative analysis of the spike and plant productivity of the NILs Bez1Vrn-A1a and Bez1Vrn-A1L was carried out under field conditions for 4 years (Table 4). The influence of different alleles on production performance was found to be unequal in different years. Thus, i:Bez1Vrn-A1L was more productive compared to i:Bez1Vrn-A1a in 2017–2018, while the opposite was observed in 2019 and 2021. In i:Bez1Vrn-A1L, the number of spikelets in the spike was higher than in i:Bez1Vrn-A1a in all experiments. i:Bez1VRN-A1L had a slightly long stem than i:Bez1Vrn-A1a.
Two-factor ANOVA revealed a significant effect of the Vrn-A1a and Vrn-A1L alleles on the number of spikelets per spike, plant productivity traits (the number of spikes, grains, and grain weight), and plant height. The vegetation conditions had an effect on all studied traits. The genotype × environment interaction influenced the variability of spike length, the weight of grains per spike, the weight of 1000 grains, and plant productivity traits (Table S1).
Allelic diversity of VRN genes in T. aestivum ssp. petropavlovskyi accessions from the VIR collection
Since the Vrn-A1L dominant allele was detected in T. aestivum ssp. petropavlovskyi (KIZ) accession, it's necessary to determine the allelic diversity of the VRN genes in other T. aestivum ssp. petropavlovskyi accessions from the VIR collection (Table 5, Fig. S1). These T. aestivum ssp. petropavlovskyi accessions have a spring type of development; the shoots-heading period in the field conditions varied from 46 to 59 days. All the accessions studied using the VRN1AF and VRN1-INT1R primers amplified a 713 bp PCR product characteristic of the vrn-A1 allele, which characterizes the intact promoter, while the Intr1/A/F2 and Intr1/A/R3 primers detected a 522 bp fragment of the Vrn-A1L allele. Four T. aestivum ssp. petropavlovskyi accessions had the Vrn-B1a dominant allele, and three had the vrn-B1 recessive allele. The Vrn-D1a dominant allele was detected in five samples when amplified with Intr1/D/F and Intr1/D/R3 primers (1671 bp PCR product); the remaining accessions showed a vrn-D1 recessive allele with Intr1/D/F and Intr1/D/R4 primers (PCR product of about 1000 bp). A recessive allele vrn-B3 was detected in the accessions studied. Thus, differences were revealed in the allelic state of the VRN-B1 and VRN-D1 loci.
The spring type of development of K-51763, K-43351, and K-43376 T. aestivum ssp. petropavlovskyi accessions is determined by three dominant alleles: Vrn-A1L, Vrn-B1a, and Vrn-D1a. K-44126 and K-51766 accessions carry two dominant alleles: Vrn-A1L Vrn-B1a and Vrn-A1L VRN-D1a. K-51764 and KIZ are monogenic carriers; their spring type of development is due to the Vrn-A1L allele. This may explain the longer shoots-heading period of these accessions compared to accessions with two or three dominant genes.
Discussion
In terms of expanding the breeding and adaptive potential of bread wheat, wild and cultivated wheat species are of special interest because they carry valuable genetic variants not represented in the bread wheat genome. Examples of introgression of biotic resistance genes are the most common. However, the interspecific or intergeneric introgression of gene alleles, which determine different heading initiation times, provide an important opportunity to control the duration of the growing season, resulting in increased adaptive capacity of bread wheat, especially in changing climate conditions. However, there are only a few known examples of such introgressions. For example, Ivaničová et al. (2016) introgressed the first discovered Vrn-A1f-like allele from T. militinae into the bread wheat genome. In the research by Takumi et al. (2020), segments of Ae. tauschii chromosomes with early heading date genes were introgressed into Japanese elite cultivars through synthetic hexaploid wheat lines. In our present work, in winter cultivars Filatovka and Ulʼynovka, the recessive vrn-A1 allele of wheat was replaced with the dominant rye VRN-R1 gene, which led to a change in the winter type of development to the spring type (Efremova et al. 2022). In this study, T. aestivum ssp. petropavlovskyi hexaploid wheat was used for this purpose to increase the bread wheat genetic diversity by allelic variants of the VRN-A1 gene. According to the latest data, T. aestivum ssp. petropavlovskyi, also known as "Daosuimai" or rice-head wheat endemic, is a subspecies of bread wheat, which has a limited range of growing in the wild in Xinjiang and Tibet.
In this work, we studied the genetic control of heading time of T. aestivum ssp. petropavlovskyi accessions. It was shown that the spring type of development in T. aestivum ssp. petropavlovskyi accessions is determined by one to three dominant alleles: Vrn-A1L, Vrn-B1a, and Vrn-D1a, associated with deletions in the first intron. The Vrn-B1a and Vrn-D1a alleles are common among bread wheat cultivars, while Vrn-A1L has not been found in wheat hexaploid forms including T. aestivum. Previously, the Vrn-A1L was described only in tetraploid wheat belonging to the Dicoccoides section: T. aethiopicum, T. carthlicum, T. dicoccoides, T. dicoccum, T. durum, T. polonicum, and T. turgidum (Fu et al. 2005; Oliveira et al. 2012; Shcherban et al. 2015; Konopatskaia et al. 2016; Muterko et al. 2016a). Phylogenetic analysis indicates that T. aestivum ssp. petropavlovskyi originated in Xinjiang as a result of divergence between T. polonicum tetraploid wheat and unknown hexaploid landrace either by spontaneous introgression or by a breeding effort (Liu et al. 2022). It can be assumed that the introgression of Vrn-A1L into the genome of T. aestivum ssp. petropavlovskyi from T. polonicum which participated in the origin as one of the presumed ancestors (Liu et al. 2022). The detection of the Vrn-A1L dominant allele in all analyzed T. aestivum ssp. petropavlovskyi accessions in the present research suggests a wide distribution of this allele in T. aestivum ssp. petropavlovskyi along with tetraploid species. The presence of the Vrn-D1a dominant allele in T. aestivum ssp. petropavlovskyi is explained by the distribution of the VRN-D1 gene among local and commercial Chinese bread wheat cultivars (Zhang et al. 2010; Goncharov 2012; Chen et al. 2013; Guo et al. 2015).
It is well known that the differences between spring and winter forms of wheat are associated with mutations in the regulatory regions of the VRN-1 gene, which are the promoter and the first intron. Therefore, the analysis of the structure of these gene regions is extremely important for understanding the mechanisms and features underlying the regulation of heading time. An important task is to investigate the role of first intron structural features in the regulation of plant transition from the vegetative stage to the generative stage. Earlier results confirm the importance of first intron in the sensitivity of plants to vernalization and in determining the heading time. Mutations described above typically affect a putative 2.8 kb vernalization critical region located near the left side of first intron (Fu et al. 2005), where insertions or deletions (including the Langdon mutation characterized by 7222 bp deletion from 391 to 7612 bp) affect changes in VRN-1 expression (Fu et al. 2005; Shcherban et al. 2013).
It was shown that the Vrn-A1L dominant allele in the NIL caused significantly later heading versus the Vrn-A1a allele. The main differences in the duration of individual phases, which ultimately determine the duration of the growth season, were observed in the period before the formation of the first node, which reached 20–22 days in a greenhouse. The range of differences in the field under the possible influence of environmental factors decreased slightly, but this trend persisted. The results confirm the data that the differences in the duration of the shoots-heading period are largely due to changes in the length of the tillering – first node period, which is the key stage determining the differences in the duration of the growing season among the genotypes (Pánková and Košner 2004; Emtseva et al. 2013; Chumanova et al. 2020).
In addition, in the discussed experiments, i:Bez1Vrn-A1L was late ripening compared to NILs carrying alleles of the VRN-B1 locus, so, the longer shoots-heading period of two T. aestivum ssp. petropavlovskyi accessions compared to other studied accessions is associated with the one Vrn-A1L dominant allele in these genotypes. However, the four accessions with different combinations of Vrn-B1a/vrn-B1 and Vrn-D1a/vrn-D1 were characterized by almost the same duration of time to heading. This may be due to the different allelic states of other loci involved in the formation of differences in the time of heading in these accessions, as well as differences in the time of heading in monogenic carriers of the Vrn-A1L dominant allele. The possible allelic differences in the PPD genes confirm the presence of a response to the length of day in the T. aestivum ssp. petropavlovskyi (KIZ) accession, which requires additional research.
It is known that the variability of wheat flowering timing, determined to a greater extent by VRN genes, is of great practical importance, since, being in close relationship with resistance to biotic and abiotic factors, it affects yield components (Worland 1996; Snape et al. 2001; Cockram et al. 2007; Kamran et al. 2014). On the one hand, under the Western Siberia conditions, fast-growing and midseason-ripening cultivars of spring wheat are more productive due to the faster initial organogenesis when the laying of spike productivity traits takes place, which allows them to avoid damage by possible late spring frosts and to form a productive spike with limited precipitation. On the other hand, the longer tillering – first node phase in late maturing cultivars increases the productive tilling capacity and the number of grains under favorable conditions. In different years the influence of different alleles on production performance was found to be unequal. Thus, in 2017–2018, i:Bez1Vrn-A1L was more productive compared with i:Bez1Vrn-A1a, while in 2019 and 2021, the opposite was observed, which is probably due to less favorable conditions for the vegetative phase of plants, in particular, the drier period in early spring and May–June when the elements of yield structure were laid. However, in all years of observations, i:Bez1Vrn-A1L was characterized by a large number of spikelets per spike.
Conclusions
The authors introgressed the Vrn-A1L dominant allele from T. aestivum ssp. petropavlovskyi, characterized by the presence of the Langdon deletion in the first intron, into the bread wheat genome by producing a NIL of the Bez1 winter cultivar. An analysis of the allele composition of VRN genes in T. aestivum ssp. petropavlovskyi accessions revealed that the spring type of development was determined by one to three dominant alleles: Vrn-A1L, Vrn-B1a, and Vrn-D1a, suggesting a wide distribution of the Vrn-A1L allele in T. aestivum ssp. petropavlovskyi along with tetraploid species. It was shown that the Vrn-A1L allele, compared with the Vrn-A1a allele, increases the heading time of NIL plants, with the maximum differences associated with the time from seedlings to the first node formation. No unambiguous effect of VRN-A1 alleles on spike and plant production performance could be detected in a comparative study of NILs. In summary, the new scientific information can be valuable for breeding and further research the mechanisms of regulation of heading time.
References
Berezhnaya A, Kiseleva A, Leonova I, Salina E (2021) Allelic variation analysis at the vernalization response and photoperiod genes in Russian wheat varieties identified two novel alleles of Vrn-B3. Biomolecules 11(12):1897. https://doi.org/10.3390/biom11121897
Chen F, Gao M, Zhang J, Zuo A, Shang X, Cui D (2013) Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China. BMC Plant Biol 13:199. https://doi.org/10.1186/1471-2229-13-199
Chu CG, Tan CT, Yu GT, Zhong S, Xu SS, Yan L (2011) A novel retrotransposon inserted in the dominant Vrn-B1 allele confers spring growth habit in tetraploid wheat (Triticum turgidum L.). G3 Genes Genomes Genet 1:637–645. https://doi.org/10.1534/g3.111.001131
Chumanova EV, Efremova TT, Kruchinina YV (2020) The effect of different dominant VRN alleles and their combinations on the duration of developmental phases and productivity in common wheat lines. Russ J Genet 56:822–834. https://doi.org/10.1134/S1022795420070029
Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA, Greenland AJ (2007) Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J Exp Bot 58:1231–1244. https://doi.org/10.1093/jxb/erm042
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184. https://doi.org/10.1016/j.pbi.2008.12.010
Dragovich AY, Fisenko AV, Yankovskaya AA (2021) Vernalization (VRN) and photoperiod (PPD) genes in spring hexaploid wheat landraces. Russ J Genet 57:329–340. https://doi.org/10.1134/S1022795421030066
Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L (2006) Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol 60:469–480. https://doi.org/10.1007/s11103-005-4814-2
Efremova TT, Maystrenko OI, Laikova LI, Arbuzova VS, Popova OM (2000) Comparative genetic analysis of hexaploid wheats Triticum petropavlovskyi Udacz. et Migusch. and Triticum aestivum L. Genetika 36:1362–1369
Efremova T, Arbuzova V, Leonova I, Makhmudova K (2011) Multiple allelism in the Vrn-B1 locus of common wheat. Cereal Res Commun 39:12–21. https://doi.org/10.1556/CRC.39.2011.1.2
Efremova TT, Chumanova EV, Zhukova IM (2022) Winter hardiness analysis of wheat-rye 5R(5A)-substituted lines in Western Siberia. Cereal Res Commun 50:25–35. https://doi.org/10.1007/s42976-021-00147-z
Emtseva MV, Efremova TT, Arbuzova VS (2013) The influence of Vrn-B1a and Vrn-B1c alleles on the length of developmental phases of substitution and near-isogenic lines of common wheat. Russ J Genet 49:545–552. https://doi.org/10.1134/S1022795413050050
Fu D, Szűcs P, Yan L, Helguera M, Skinner JS, Von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65. https://doi.org/10.1007/s00438-004-1095-4
Golovnina KA, Kondratenko EY, Blinov AG, Goncharov NP (2010) Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats. BMC Plant Biol 10:168. https://doi.org/10.1186/1471-2229-10-168
Goncharov NP (2012) Comparative genetics of wheat and its relatives. Geo, Novosibirsk (in Russian)
Guo X, Wang Y, Meng L, Liu H, Yang L, Zhou Y, Zhang H (2015) Distribution of the Vrn-D1b allele associated with facultative growth habit in Chinese wheat accessions. Euphytica 206:1–10. https://doi.org/10.1007/s10681-015-1440-1
Ivaničová Z, Jakobson I, Reis D, Šafář J, Milec Z, Abrouk M, Doležel J, Järve K, Valárik M (2016) Characterization of new allele influencing flowering time in bread wheat introgressed from Triticum militinae. New Biotechnol 33:718–727. https://doi.org/10.1016/j.nbt.2016.01.008
Kamran A, Iqbal M, Spaner D (2014) Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica 197:1–26. https://doi.org/10.1007/s10681-014-1075-7
Kippes N, Debernardi JM, Vasquez-Gross HA, Akpinar BA, Budak H, Kato K, Chao S, Akhunov E, Dubcovsky J (2015) Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc Natl Acad Sci USA 112:E5401–E5410. https://doi.org/10.1073/pnas.1514883112
Konopatskaia I, Vavilova V, Kondratenko EY, Blinov A, Goncharov NP (2016) VRN1 genes variability in tetraploid wheat species with a spring growth habit. BMC Plant Biol 16:244. https://doi.org/10.1186/s12870-016-0924-z
Koval SF, Goncharov NP (1998) Multiple allelism at the VRN1 locus of common wheat. Acta Agron Hung 46:113–119
Liu J, Yao Y, Xin M, Peng H, Ni Z, Sun Q (2022) Shaping polyploid wheat for success: origins, domestication, and the genetic improvement of agronomic traits. J Integr Plant Biol 64:536–563. https://doi.org/10.1111/jipb.13210
Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol 138:2364–2373. https://doi.org/10.1104/pp.105.064287
Milec Z, Tomková L, Sumíková T, Pánková K (2012) A new multiplex PCR test for the determination of Vrn-B1 alleles in bread wheat (Triticum aestivum L.). Mol Breed 30:317–323. https://doi.org/10.1007/s11032-011-9621-7
Muterko AF, Salina EA (2017) Analysis of the VERNALIZATION-A1 exon-4 polymorphism in polyploid wheat. Vavilovskii Zhurnal Genet Sel 21:323–333. https://doi.org/10.18699/VJ16.19-o. (in Russian)
Muterko A, Balashova I, Cockram J, Kalendar R, Sivolap Y (2015) The new wheat vernalization response allele Vrn-D1s is caused by DNA transposon insertion in the first intron. Plant Mol Biol Rep 33:294–303. https://doi.org/10.1007/s11105-014-0750-0
Muterko A, Kalendar R, Salina E (2016a) Novel alleles of the VERNALIZATION1 genes in wheat are associated with modulation of DNA curvature and flexibility in the promoter region. BMC Plant Biol 16:9. https://doi.org/10.1186/s12870-015-0691-2
Muterko A, Kalendar R, Salina E (2016b) Allelic variation at the VERNALIZATION-A1, VRN-B1, VRN-B3, and PHOTOPERIOD-A1 genes in cultivars of Triticum durum Desf. Planta 244:1253–1263. https://doi.org/10.1007/s00425-016-2584-5
Oliveira HR, Campana MG, Jones H, Hunt HV, Leigh F, Redhouse DI, Lister DL, Jones MK (2012) Tetraploid wheat landraces in the Mediterranean basin: taxonomy, evolution and genetic diversity. PLoS ONE 7:e37063. https://doi.org/10.1371/journal.pone.0037063
Pánková K, Košner J (2004) Chromosome substitutions with dominant loci Vrn-1 and their effect on developmental stages of wheat. Czech J Genet Plant Breed 40:37–44. https://doi.org/10.17221/3698-CJGPB
Santra DK, Santra M, Allan RE, Campbell KG, Kidwell KK (2009) Genetic and molecular characterization of vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 in spring wheat germplasm from the Pacific Northwest region of the U.S.A. Plant Breed 128:576–584. https://doi.org/10.1111/j.1439-0523.2009.01681.x
Sehgal D, Vikram P, Sansaloni CP, Ortiz C, Pierre CS, Payne T, Ellis M, Amri A, Petroli CD, Wenzl P, Singh S (2015) Exploring and mobilizing the gene bank biodiversity for wheat improvement. PLoS ONE 10:e0132112. https://doi.org/10.1371/journal.pone.0132112
Shcherban AB, Salina EA (2017) Evolution of VRN-1 homoeologous loci in allopolyploids of Triticum and their diploid precursors. BMC Plant Biol 17:188. https://doi.org/10.1186/s12870-017-1129-9
Shcherban AB, Efremova TT, Salina EA (2012) Identification of a new Vrn-B1 allele using two near-isogenic wheat lines with difference in heading time. Mol Breed 29:675–685. https://doi.org/10.1007/s11032-011-9581-y
Shcherban AB, Khlestkina EK, Efremova TT, Salina EA (2013) The effect of two differentially expressed wheat VRN-B1 alleles on the heading time is associated with structural variation in the first intron. Genetica 141:133–141. https://doi.org/10.1007/s10709-013-9712-y
Shcherban AB, Strygina KV, Salina EA (2015) VRN-1 gene-associated prerequisites of spring growth habit in wild tetraploid wheat T. dicoccoides and the diploid A genome species. BMC Plant Biol 15:94. https://doi.org/10.1186/s12870-015-0473-x
Shcherban AB, Schichkina AA, Salina EA (2016) The occurrence of spring forms in tetraploid Timopheevi wheat is associated with variation in the first intron of the VRN-A1 gene. BMC Plant Biol 16:236. https://doi.org/10.1186/s12870-016-0925-y
Shi C, Zhao L, Zhang X, Lv G, Pan Y, Chen F (2019) Gene regulatory network and abundant genetic variation play critical roles in heading stage of polyploidy wheat. BMC Plant Biol 19:6. https://doi.org/10.1186/s12870-018-1591-z
Snape JW, Butterworth K, Whitechurch E, Worland AJ (2001) Waiting for fine times: genetics of flowering time in wheat. Euphytica 119:185–190. https://doi.org/10.1023/A:1017594422176
Stelmakh AF (1992) Genetic effects of Vrn genes on heading date and agronomic traits in bread wheat. Euphytica 65:53–60. https://doi.org/10.1007/BF00022199
Takumi S, Koyama K, Fujiwara K, Kobayashi F (2011) Identification of a large deletion in the first intron of the Vrn-D1 locus, associated with loss of vernalization requirement in wild wheat progenitor Aegilops tauschii Coss. Genes Genet Syst 86:183–195. https://doi.org/10.1266/ggs.86.183
Takumi S, Mitta S, Komura S, Ikeda TM, Matsunaka H, Sato K, Yoshida K, Murai K (2020) Introgression of chromosomal segments conferring early heading date from wheat diploid progenitor, Aegilops tauschii Coss. into Japanese elite wheat cultivars. PLoS ONE 15:e0228397. https://doi.org/10.1371/journal.pone.0228397
Trevaskis B, Hemming MN, Dennis ES, Peacock WJ (2007) The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci 12:352–357. https://doi.org/10.1016/j.tplants.2007.06.010
Worland AJ (1996) The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89:49–57. https://doi.org/10.1007/BF00015718
Xiao J, Chen Y, Lu Y, Liu Z, Si D, Xu T, Sun L, Wang Z, Yuan C, Sun H, Zhang X, Wen M, Wei L, Zhang W, Wang H, Wang X (2021) A natural variation of an SVP MADS-box transcription factor in Triticum petropavlovskyi leads to its ectopic expression and contributes to elongated glume. Mol Plant 14:1408–1411. https://doi.org/10.1016/j.molp.2021.05.022
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268. https://doi.org/10.1073/pnas.0937399100
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004a) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109:1677–1686. https://doi.org/10.1007/s00122-004-1796-4
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004b) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644. https://doi.org/10.1126/science.1094305
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586. https://doi.org/10.1073/pnas.0607142103
Zhang Y, Liu WC, Li J, Wei HT, Hu XR, Li YJ, Lu BR, Yang WY (2010) Distribution and selective effects of Vrn-A1, Vrn-B1, and Vrn-D1 genes in derivative varieties from four cornerstone breeding parents of wheat in China. Agric Sci China 9:1389–1399. https://doi.org/10.1016/S1671-2927(09)60230-3
Zhang J, Wang Y, Wu S, Yang J, Liu H, Zhou Y (2012) A single nucleotide polymorphism at the Vrn-D1 promoter region in common wheat is associated with vernalization response. Theor Appl Genet 125:1697–1704. https://doi.org/10.1007/s00122-012-1946-z
Zhang X, Gao M, Wang S, Chen F, Cui D (2015) Allelic variation at the vernalization and photoperiod sensitivity loci in Chinese winter wheat cultivars (Triticum aestivum L.). Front Plant Sci 6:470. https://doi.org/10.3389/fpls.2015.00470
Zhang B, Wang X, Wang X, Ma L, Wang Z, Zhang X (2018) Molecular characterization of a novel vernalization allele Vrn-B1d and its effect on heading time in Chinese wheat (Triticum aestivum L.) landrace Hongchunmai. Mol Breed 38:127. https://doi.org/10.1007/s11032-018-0870-6
Acknowledgements
Reproduction of the plant material was carried out on the basis of the Laboratory of Artificial Plant Growth of ICG SB RAS within the budgetary project FWNR-2022-0017.
Funding
This study was supported by the Russian Science Foundation (Grant No. 22-26-00085).
Author information
Authors and Affiliations
Contributions
EC: wrote and edited the article, performed PCR, field and greenhouse experiments, and data analysis. TE: conceived and designed research, edited the article, performed field and greenhouse experiments. VV: performed PCR, sequencing samples.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chumanova, E., Efremova, T. & Vavilova, V. Characterization of the VRN-A1 allele introgressed from T. aestivum ssp. petropavlovskyi that influences the heading time in bread wheat. Euphytica 219, 53 (2023). https://doi.org/10.1007/s10681-023-03178-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03178-1