Abstract
Bread-making quality is a core trait for wheat breeding programs. Recently, the expression of a novel gene named wheat bread making (wbm) gene has been associated with good bread-making quality. In this study, 54 historical and modern bread wheat genotypes from CIMMYT were screened by PCR marker for the presence of the allele associated with high wbm expression. Eight of the 54 wheat genotypes tested positive for the wbm allele and the genotype Waxwing was identified as the most likely donor of the allele in part of CIMMYT germplasm. The wbm allele had a significant effect on overall gluten quality, gluten strength, gluten extensibility and bread-making quality, although its effect was smaller than the effects of other quality related genes as Glu-D1, Glu-B1 or the negative effect of the 1BL.1RS translocation. The wbm allele was associated with higher values of the traits mentioned but not with higher protein content. The identification of this new wbm gene/protein is a step forward in understanding wheat quality genetic control. Implementation of marker assisted selection in breeding programs to detect the wbm allele is highly recommended.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Wheat is one of the most important crops in the world, occupying 219 million hectares and resulting in global production of about 715 million tonnes a year (FAOSTAT 2013). High adaptability to diverse environments is important but probably the key characteristic for understanding the success of this crop are the unique properties of the dough formed from wheat flours, which is processed into a range of products including cakes, biscuits, pasta, noodles, and other processed foods of which probably bread is the most important worldwide (Shewry 2009).
Bread-making quality is a core trait for wheat breeding programs. The International Maize and Wheat Improvement Center (CIMMYT) wheat breeding program aims to develop new wheat genotypes for release as varieties in developing countries that not only produce high yields for farmers but also satisfy subsequent actors in the value chain, including food manufacturers and consumers. Integrating bread-making quality in a breeding program is not simple, as quality analyses are expensive, time-consuming and require certain amounts of seed that are usually not available until late generations of the breeding process. Molecular markers can be useful for determining various wheat quality components and enhancing selection for bread-making quality (Kuchel et al. 2007).
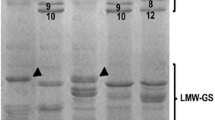
The viscoelastic properties of wheat dough occur due to the seed storage proteins, which form a complex responsible of the elasticity and extensibility of the dough called gluten when the flour is mixed with water. Gluten quality is important to understand and ensure good bread-making quality. Intensive research has been carried out to identify its components and understand their underlying genetics (see Wrigley et al. 2006 for a complete review). Glutenins (high and low molecular weight subunits, HMW and LMW, respectively) and gliadins have been identified as the main components of storage proteins. HMW glutenins have been the most studied, probably for their relatively simple pattern by SDS-PAGE electrophoresis and for their great impact in rheological and bread-making properties, in spite of the fact that they only represent 10 % of the total storage protein in the grain. The genes coding for these proteins (Glu-A1, Glu-B1 and Glu-D1, Payne and Lawrence 1983) and their alleles are well known, and molecular markers are available to detect them (D´Ovidio and Anderson 1994; Liu et al. 2008). The presence of 1BL.1RS translocation has also been associated to low quality (Liu et al. 2005; Peña et al. 1990), due to the loss of proteins from the Glu-B3 and Gli-B1 and incorporation of secalins from rye. However, observed polymorphisms of these and other grain proteins associated with additional quality traits (hardness, starch properties, etc.) still do not explain the genetic variation found in bread-making quality.
Recently Furtado et al. (2015) identified a new gene that is expressed in developing seeds, called the wheat bread making (wbm) gene. This gene codifies for a small sulphur-rich protein not previously associated with wheat quality. The wbm gene has shown highly differential expression in genotypes varying in bread-making quality: genotypes with high wbm expression all had good bread-making quality. Furtado et al. (2015) also identified the sequence variant in promoter region of the gene (GWseqVar3) associated with high expression of the gene, which presence can be determined by a simple molecular marker using PCR. Therefore, the objective of the current study was to screen CIMMYT germplasm for the presence of GWseqVar3 sequence variant associated to high wbm expression and to analyse the effect of this gene on processing and bread-making quality traits.
For this purpose 54 CIMMYT bread wheat lines (ESM 1), including historical and modern varieties and advanced lines, released from 1960 to 2014, were sown in an alpha lattice design with three replicates in 2013 and 2014 crop seasons in Ciudad Obregon in the state of Sonora, Mexico, under six different field management conditions, including drought and heat stress. Grain from two field replicates was analysed for seven quality traits, including grain protein content (%) (GPC), SDS-sedimentation volume (ml) (SDSS), mixograph optimum dough development time (min) (DDT) and torque (%) (TQ), alveograph gluten strength (ALVW) and tenacity/extensibility ratio (ALVP/L), and bread loaf volume (LV), using the methodologies of the American Association of Cereal Chemist (AACC 2000), with some modifications (Guzman et al. 2015). In addition, the HMW glutenins composition and the presence of the 1BL.1RS translocation were determined by SDS-PAGE in polyacrylamide gels, according to the methodology of Peña et al. (2004). To detect the presence of the wbm allele associated to high expression, PCR screening was carried out using genomic DNA extracted from young leaves, with primers NWPFor and NWPRev and PCR conditions described by Furtado et al. (2015). Two more bread wheat lines, Kiritati and Waxwing, were tested for the wbm allele.
Eight of the 54 wheat genotypes tested positive for the wbm allele associated to high expression of the gene, including some popular varieties released in different countries, such as Seher 06 (Pakistan) and Baj #1 (India). Two of the advanced lines that were positive for the wbm allele had the line Waxwing in their pedigree as well as Baj #1, Grackle #1 and Munal #1. Therefore it seemed that Waxwing was probably the source of the wbm allele in part of CIMMYT germplasm, which was confirmed with the PCR marker. Kiritati, which is the other parental of Munal #1, tested also positive for the wbm allele. The origin of the wbm allele in two other genotypes (Kanchan and Babax/Lr42//Babax*2/4/Sni/Trap#1/3/Kauz*2/Trap//Kauz) that tested positive for the marker is not clear and further screenings in more lines will be required to elucidate it.
The effect of the presence of the wbm allele on the seven measured quality traits was analysed by a combined variance analysis across the two years and six field managements, together with the composition of HMW glutenins (Glu-A1, Glu-B1 and Glu-D1) and 1BL.1RS translocation data (data not shown) (Table 1). The analysis revealed that wbm allele had a significant effect on all of the quality traits analysed, including those related to overall gluten quality (SDSS), gluten strength (TQ and ALVW), gluten extensibility (ALVP/L) and bread-making quality (LV). The same results were revealed for the additional genes included in the study. For most of the traits, the locus Glu-D1 showed the greatest effect, followed by the Glu-B1 locus and the 1B/1R translocation. The Glu-A1 effect was lower, but also more significant than the effect of the wbm allele for TQ and ALVW. Overall, the effect of wbm was less significant than that of the Glu-1 genes in traits related to gluten strength, but in traits related to gluten extensibility (ALVP/L and to a minor extent SDSS) its effect was more prominent and comparable to that of the other genes but far from Glu-D1. On bread-making, the wbm effect was small, particularly if compared with the effect of Glu-D1. The two major alleles at the Glu-D1 locus, 5 + 10 and 2 + 12, have repeatedly shown a contrasting effect on quality traits (Gupta et al. 1994; He et al. 2005), so it was not surprising to find a strong Glu-D1 effect in this study. The interaction of the wbm gene and other genes, except Glu-D1, with the environment was not significant.
The results shown in Table 2 reveal that the presence of the wbm allele was associated with higher quality for all the traits. This higher quality is not associated with higher protein content in the genotypes carrying the wbm allele, which agrees with Furtado et al. (2015). The association of the wbm allele with higher gluten strength and extensibility (lower ALVP/L value) is what probably leads to higher loaf volumes values. The deduced protein sequence of the wbm gene showed the presence of four cysteine residues which could participate in intra- or inter-molecular disulphide bonds with other gluten proteins enhancing the visco-elastic properties of the dough (Furtado et al. 2015). However, based on current results, we cannot state that the presence of the allele associated to high expression of the gene automatically leads to good bread-making quality. In the current study, lines with low loaf volume values carrying the wbm allele were found, and vice versa (data not shown). Furtado et al. (2015) made the same discovery in the cv. Bobwhite, which carries the 1BL.1RS translocation.
In summary, the identification of the wbm allele in several CIMMYT genotypes is important, as CIMMYT develops spring wheat lines for all major wheat growing areas in the developing world (Rajaram and van Ginkel 2001). Advanced breeding lines are distributed annually to around 250 partners in more than 60 countries, where they are released directly as varieties or are used as parents for crossing (Singh et al. 2007). Therefore CIMMYT lines have served as donors of the highly expressed wbm allele in many other breeding programs, helping to enhance bread-making quality worldwide. The frequency of the wbm gene (14 %) is still relatively low in CIMMYT germplasm and can be increased. Dough systems are a complex mixture of diverse proteins, carbohydrates and lipids, each of them contributing to the dough and products characteristics. The identification of this new wbm gene/protein is a step forward in understanding wheat quality genetic control. Further studies are necessary to analyse in detail the functional role of this protein in combination with other grain factors. Implementation of marker assisted selection in breeding programs to detect the wbm allele is highly recommended.
References
AACC International (2000) Approved methods of the American Association of Cereal Chemists, 10th edn. AACC International, St. Paul
D´Ovidio R, Anderson OD (1994) PCR analysis to distinguish between alleles of a member of a multigene family correlated with bread-making quality. Theor Appl Genet 88:759–763
FAOSTAT (2013) Rome, Italy. http://faostat.fao.org/
Furtado A, Bundock PC, Banks PM, Fox G, Yin X, Henry RJ (2015) A novel highly differentially expressed gene in wheat endosperm associated with bread quality. Sci Rep 5:10446
Gupta RB, Paul JG, Cornish GB, Palmer GA, Bekes F, Rathjen AJ (1994) Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1, of common wheats. I. Its additive and interaction effects on dough properties. J Cereal Sci 19:9–17
Guzman C, Posadas-Romano G, Hernandez-Espinosa N, Morales-Dorantes A, Peña RJ (2015) A new standard water absorption criteria based on solvent retention capacity (SRC) to determine dough mixing properties, viscoelasticity, and bread-making quality. J Cereal Sci 66:59–65
He Z, Liu L, Xia XC, Liu JJ, Peña RJ (2005) Composition of HMW and LMW glutenin subunits and their effects on dough properties, pan bread, and noodle quality of Chinese bread wheats. Cereal Chem 82:345–350
Kuchel H, Fox R, Reinheimer J, Mosionek L, Willey N, Bariana H, Jefferies S (2007) The successful application of a marker-assisted wheat breeding strategy. Mol Breed 20:295–308
Liu L, He Z, Yan J, Zhang Y, Xia X, Peña RJ (2005) Allelic variation at the Glu-1 and Glu-3 loci, presence of the 1B. 1R translocation, and their effects on mixographic properties in Chinese bread wheats. Euphytica 142:197–204
Liu S, Chao S, Anderson JA (2008) New DNA markers for high molecular weight glutenin subunits in wheat. Theor Appl Genet 118:177–183
Payne PI, Lawrence GJ (1983) Catalogue of alleles for the complex gene loci, Glu-A1, Glu-B1 and Glu-D1 which code for high-molecular weight subunits of glutenin in hexaploid wheat. Cereal Res Commun 11:29–35
Peña RJ, Amaya A, Rajaram S, Mujeeb-Kazi A (1990) Variation in quality characteristics associated with some spring 1B/1R translocation wheats. J Cereal Sci 12:105–112
Peña RJ, Gonzalez-Santoyo J, Cervantes F (2004). Relationship between Glu-D1/Glu-B3 allelic combinations and breadmaking quality-related parameters commonly used in wheat breeding. In: Masci S, Lafiandra D (eds) Proceedings 8th gluten workshop. RACI, Australia, pp 156–157
Rajaram S, van Ginkel M (2001) Mexico: 50 years of international wheat breeding. In: Bonjean AP, Angus WJ (eds) The World wheat book. A history of wheat breeding. Lavoisier publishing, Paris, pp 579–608
Shewry PR (2009) Wheat. J Exp Bot 60:1537–1553
Singh RP, Huerta-Espino J, Sharma R, Joshi AK, Trethowan R (2007) High yielding spring bread wheat germplasm for global irrigated and rainfed production systems. Euphytica 157:351–363
Wrigley C, Bekes F, Bushuk W (eds) (2006) Gliadin and glutenin: the unique balance of wheat quality. AACC International Press, St Paul
Acknowledgments
We greatly appreciate financial support from the CGIAR Research Program-Wheat of CGIAR consortium and from Fondo Sectorial SAGARPA-CONACYT (No. 146788—Sistema de mejoramiento genético para generar variedades resistentes a royas, de alto rendimiento y alta calidad para una producción sustentable en México de trigo) of the Mexican government. We are also very grateful for the field work of CIMMYT scientists Suchismita Mondal and Enrique Autrique.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guzmán, C., Xiao, Y., Crossa, J. et al. Sources of the highly expressed wheat bread making (wbm) gene in CIMMYT spring wheat germplasm and its effect on processing and bread-making quality. Euphytica 209, 689–692 (2016). https://doi.org/10.1007/s10681-016-1659-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-016-1659-5