Abstract
Conservation biologists often deal with species that have small, fragmented populations throughout their range, making it necessary to prioritize populations for management. Genetics provides tools to assist with prioritization according to the levels and distribution of genetic diversity and evolutionary distinctiveness. Many studies have used nuclear microsatellite loci to measure genetic diversity in disparate populations and mitochondrial DNA to assess genetic distinctiveness. However, comparing metrics based on microsatellite genotypes ascertained in different laboratories is complicated by the selection of different loci with distinct nucleotide repeat motifs. This issue may be resolved by comparing metrics to a well-characterized reference population with shared microsatellite markers. The Asian elephant, Elephas maximus, is an endangered species with 50–60% of populations in India and Sri Lanka, and small, fragmented populations throughout southeast and insular Asia. We assessed range-wide genetic diversity of the Asian elephant by directly comparing allelic diversity and heterozygosity estimates from 35 populations, overcoming marker selection bias by calibrating metrics to a large population on the Nakai Plateau, Lao PDR, genotyped at 25 loci. We coupled these results with mtDNA analysis to evaluate genetic distinctiveness and identify potential conservation management units. We found the greatest diversity in the populations of southeast Asia and the greatest genetic distinctiveness among the subspecies designations, particularly Borneo and Sumatra, and other southeast Asian populations. The populations of southeast Asia, albeit small, fragmented, and at high risk of extirpation, contain valuable diversity and distinctiveness and are thus of high priority for the preservation of the Asian elephant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Conservation biologists often deal with species characterized by small, fragmented populations throughout their range. Given limited resources, managers must prioritize populations and determine the appropriate actions required to reduce the rate of further declines and increase the probability of recovery of the species adaptive potential (Hoban 2018). Intraspecific genetic variation increases adaptive potential under changing environmental conditions (Reed and Frankham 2003) and can alleviate the deleterious effects of inbreeding depression (Frankham 2005). Quantifying genetic diversity and differentiation within and between declining populations is therefore a critical component of conservation management.
Here, we evaluate the levels of genetic diversity and genetic distinctiveness among populations of Asian elephants. Despite being icons among the charismatic megafauna, elephant populations continue to decline across their native range. Conservation efforts generally focus on the African savanna elephant (Loxodonta africana), although populations of the African forest elephant (L. cyclotis) and the Asian elephant (Elephas maximus) each comprise only 10% the size of the African savanna elephant population (Gobush et al. 2021; Williams et al. 2020). Asian elephant populations are declining at alarming rates in India, Bangladesh, Bhutan, Cambodia, Lao PDR, Thailand, Vietnam, China, Myanmar, Malaysia, Indonesia, and Nepal, largely due to habitat degradation, urbanization, and fragmentation (Williams et al. 2020). While 50–60% of the total Asian elephant population is contained in India and Sri Lanka, smaller populations throughout southeast Asia continue to decline from habitat loss, with some already extirpated and only 16% of suitable habitat under protection (Choudhury et al. 2008; Williams et al. 2020).
Until recently, studies of genetic diversity within populations have been heavily reliant on nuclear microsatellite loci. However, common metrics derived from microsatellites, such as allelic diversity, heterozygosity, and fixation indices (FIS, FST), are notoriously non-comparable among studies (Skrbinsek et al. 2012) due to non-repeatability of genotype scoring among researchers, inconsistent sample sizes, and differing properties of the microsatellite loci selected for the study. Discrepancies in genetic inference due to the selection of microsatellite loci (marker bias) were clearly demonstrated in studies of isolated Asian elephant (Elephas maximus) populations in Borneo, where Fernando et al. (2003) found extremely low levels of nuclear genetic diversity with an allelic diversity (A) of 1.4 and expected heterozygosity (HE) of 0.041, primarily using loci characterized by Fernando et al. (2001). In contrast, Goossens et al. (2016) found a two-fold increase in A to 2.8 and 30% greater HE (0.34) in the same populations using a panel of microsatellite loci developed by Kongrit et al. (2008). Although few studies have demonstrated such notable differences in genetic diversity within the same population, large differences in genetic diversity between populations are common. It is unknown whether these differences are biologically important or are artifacts of marker bias. To quantify the levels and distribution of genetic diversity within populations, managers will need methods for direct comparison across independent studies.
The most commonly used microsatellite loci in Asian elephant studies are those characterized by Fernando et al. (2001; the EMX loci) and those by Kongrit et al. (2008; the EMU loci). Diversity estimates differ widely across the species range (Table 1). However, it is unknown whether these differences are biologically important or are artifacts of marker bias as estimates within and between populations may be due to the structure of the microsatellites themselves. While the EMX loci contain complex motifs of tri- and tetra-nucleotide repeats, the EMU loci consist of di-nucleotide repeat regions. Di-nucleotide repeats have been shown to accumulate mutations at a rate 1.5–2.0 times greater than tetra-nucleotides (Chakraborty et al. 1997). Thus, the choice of loci could substantially affect the number of alleles detected per locus and result in substantially different estimates of diversity.
Skrbinsek et al. (2012) developed a computational method to overcome differences in microsatellite marker selection and sample size among studies by calibrating independent estimates of allelic diversity and heterozygosity against a reference “yardstick” population. To utilize their model, the researcher first generates the yardstick population by genotyping a large number of samples on a broad array of loci, ideally representing all loci that have been previously used. For each independent study, the model subsets only loci shared with the yardstick population and randomly resamples corresponding yardstick genotypes to match the study sample size, and this procedure is repeated over a pre-determined number of iterations (Leberg 2002). Allelic diversity and heterozygosity values, along with their standard errors, are calculated for each iteration. Finally, values are averaged over all iterations and the results are compared to the original study estimates using simple ratios. The Skrbinsek et al. (2012) model was validated by the authors with microsatellite data from 10 studies of brown bears that cover 30 populations of varying sizes from across the species range. It was also used to aid in the analysis of genetic diversity in water buffalo (Bubalis bubalis and B. carabanensis) based on SNP data from 31 populations from across the worldwide range of these species (Colli et al. 2018). It has been used to study diversity patterns in several other mammals, including European brown bears (Ursus arctos; Karamanlidis et al. 2018) and has been proposed as a potential tool for conserving black bears (Ursus americanus; Murphy et al. 2018) and grey wolves (Canis lupus; de Groot et al. 2016). Hindrikson et al. (2016) found that results were in accordance with the recorded demography and population history of 10 wolf (Canis lupus) populations of various sizes across Europe.
In addition to within-population genetic diversity, it is important to consider the presence of genetically distinct lineages to identify populations of high conservation priority. Genetically differentiated, locally adapted groups may reflect evolutionary potential and warrant designation as evolutionarily significant units or management units (Moritiz 1994). Although genetic differentiation among populations can be estimated based on microsatellite data, the complexities of microsatellite evolution make interpretation of those measures difficult over any but the most recent timeframes (Ellegren 2004; Eggert et al. 2009). Instead, mitochondrial DNA (mtDNA) control region sequences have been used as they are directly comparable and can reflect ancestral maternal lineages and recent patterns of population divergence.
Nearly every population genetic study on the mtDNA of Asian elephants has used an approximately 630 bp fragment containing a portion of the C terminal of cytochrome b, the threonine and proline tRNAs, and the 5’ end of the noncoding control region (d-loop), described in Fernando et al. (2000). Using data from this fragment, studies have found deep-rooted mitochondrial clades in Asian elephants, which were first suggested by Fernando et al. (2000) and Fleischer et al. (2001) and further validated across the species’ range by Vidya et al. (2009). These mtDNA clades are described as the α-clade, persisting primarily in the north-east areas of the range, and the β-clade, primarily found in the south and south-west (including the Indonesian and Malaysian islands) with the two clades overlapping in Myanmar and Thailand.
Our primary objective was to ascertain populations for conservation priority in the Asian elephant by identifying lineages with high genetic diversity and genetic distinctiveness. We calibrated microsatellite data from previous Asian elephant population genetic studies by analyzing shared microsatellite markers in a large “yardstick” reference population, allowing direct comparison of allelic diversity and heterozygosity despite differing marker selection. Using a GIS framework, we then applied Bayesian kriging prediction to our calibrated allelic diversity and heterozygosity metrics to infer continuous patterns of diversity across the species distribution, including unstudied regions. Finally, because the yardstick model considers only population level genetic diversity estimates based on microsatellite loci, we did not use those data to compute genetic distances among populations. Instead, we evaluated populations using evolutionary distinctiveness (ED, Isaac et al. 2007), an index that measures the relative contribution of a taxon (i.e. population) to the phylogenetic diversity of the species. We calculated scores of compiled mtDNA sequences spanning the species distribution to assess genetic differentiation and subspecies designations utilizing evolutionary significant unit and management unit definitions (Moritiz 1994).
Materials and methods
Yardstick reference population
To generate a reference (i.e., “yardstick”) population, we used DNA samples (N = 91) that were confirmed to be unique Asian elephants in a previous survey of the Nakai Plateau in Lao PDR (Budd 2021). For all individuals, we amplified 25 microsatellite loci including EMX and EMU loci and four African elephant loci (LA4, Eggert et al. 2000; FH94, Comstock et al. 2000; LafMS02, LafMS03, Nyakaana and Arctander 1998) that have been most often used in previous studies. DNA samples were arranged into 6 multiplexed panels (Supplementary Table 1). We included negative (no DNA) and positive controls in each panel for contamination detection and genotype standardization.
Polymerase chain reactions (PCRs) were performed in 8 µl volumes comprised of 2µM primer mix, 0.8mM BSA, and Platinum® Multiplex PCR Master Mix (2X Master Mix, GC Enhancer; Applied Biosystems, Foster City, CA). Thermocycler conditions were as follows: 95 °C for 2-min; 40 cycles of 94 °C for 30-sec, multiplex-specific annealing temperature for 90-sec (Supplementary Table 1), 72 °C for 1-min; followed by a final extension at 60 °C for 30-min. Preparations for all PCRs were conducted in a UV–Sterilized hood.
Amplified products were submitted for fragment analysis in an ABI 3730xl DNA analyzer (Thermo Fisher Scientific, Waltham, MA) at the University of Missouri DNA Core facility with added 600LIZ size standard. Chromatograms were visualized and individuals were genotyped using GeneMarker v.1.97 (SoftGenetics, State College, PA; Holland and Parson 2010).
Yardstick calibration model
We used the yardstick genotypes as a reference population for comparison of genetic diversity metrics ascertained from multiple Asian elephant populations across independent studies. We compiled data from previous Asian elephant studies (test populations) that shared loci with our yardstick population (avg shared loci 8.5 ± 4.0 SD (range 4–16; Table 1.). From each test population, we used sample size (Ntest), expected heterozygosity (Htest), allelic diversity (Atest) and corresponding standard errors (HSEtest; ASEtest) for shared loci as inputs for our yardstick calibration model.
We developed a yardstick calibration model following Skrbinsek et al. (2012) in R v.4.0.3 (R Core Team 2018). For each test population, we first subset the yardstick population to retain only shared loci using the R package genepopedit v.1.0.0.6 (Stanley et al. 2017). Then, for each of 1,000 iterations, we randomly sampled yardstick genotypes, without replacement, from individuals matching the number of samples in the test population (Ntest) and calculated mean and standard error values for expected heterozygosity (Hyardstick, HSEyardstick) and allelic diversity (Ayardstick,ASEyardstick) using the R package adegenet v.2.1.5 (Jombart 2008; Jombart and Ahmend 2011).
We then calculated (1) pairwise heterozygosity ratios (HR) of Htest and the corresponding yardstick population mean (Hyardstick) and (2) pairwise allelic diversity ratios (AR) of Atest and the corresponding yardstick population mean (Ayardstick) to assess the relative diversity of each test population following Skrbinsek et al. (2012):
Diversity ratios (HR, AR) were therefore relative to the yardstick population, where values of 1.000 indicated equal diversity between the test and yardstick population, and deviations greater or lower than 1.000 indicated higher or lower diversity, respectively. We also calculated the standard error for 3) heterozygosity ratios (HRSE) and 4) allelic diversity ratios (ARSE) as in Skrbinsek et al. (2012):
To assess differences in inference that were made possible by using the yardstick method, we ranked the populations according to the values reported in the original studies (Htest and ARtest) and the diversity ratios (HR and AR) and compared the rankings in a Spearman’s rank order correlation test (Zar 2005) in Statziki (https://www.statziki.com/Spearman).
Spatial patterns of genetic diversity
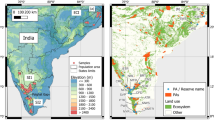
We applied empirical Bayesian kriging prediction in ArcGIS Pro (ESRI) to HRSE and ARSE values to predict spatial patterns of genetic diversity across Asian elephant populations. For the genetic input, we applied a log-empirical transformation with a K-Bessel semi-variogram model and used standard circular neighborhood parameters. For the geographic input, we generated the centroid of each population based on the source literature indications. The predictive raster was then masked using the IUCN Asian elephant shapefile (Williams et al. 2020) to the current extent of the Asian elephant distribution. We repeated empirical Bayesian kriging using the test population reported values and standardized the scale of the test population and the predicted diversity ratios to 100 to allow for spatial comparisons.
Evaluation of distinct lineages
We compiled mtDNA sequences from published articles, reports, and GENBANK accessions. We continued the naming convention set forth by Vidya et al. (2005a) to the haplotypes described in these additional studies for clarity and to avoid name duplicates (such as “New Haplotype”) that were frequently encountered in the literature; original haplotype names and citations are provided in Supplementary Table 2. We manually trimmed and aligned sequences using ClustalW (Thompson et al. 1994) in Geneious v.8.0.5 (Kearse et al. 2012). We then collapsed sequences into haplotypes using FaBox v.1.5 (Villesen 2007) and removed duplicate haplotypes. We conducted maximum likelihood analyses in PAUP* v.4.0a169 (Swofford 2002) with a heuristic search and 1000 bootstrap replicates to assess support. The DNA substitution model was selected using the automated model selection tool and determined using the optimal AICc.
For each haplotype, we calculated evolutionary distinctiveness scores using fair proportions (Isaac et al. 2007) using Picante (Kembel et al. 2010) in R. Visualization into a median joining network (Bandelt et al. 1999) was generated in POPART (Leigh and Bryant 2015) with individual haplotypes colored to signify source country.
Results
Evaluation of the yardstick model
To validate the model, we compared the results to several studies that were done in our lab by different researchers. Genotyping results for these studies were scored both manually and electronically and different sequencing platforms and sizing standards were used over the 10 year period at the University of Missouri DNA Core Facility. The similarities and differences were as expected: the yardstick population at Nakai in 2018/2019 was compared to the Nakai 2011 population (Ahlering et al. 2011) and found to be quite similar (Table 1); a genetic MRC survey (Eggert and Ruiz-Lopez 2012a) found that the Sepon, Lao PDR, population was smaller and the yardstick calibration found lower allelic diversity but similar heterozygosity; a genetic MRC survey (Eggert and Ruiz-Lopez 2011) found that the Seima, Cambodia, population was also smaller and the yardstick model found it to have less allelic diversity but similar heterozygosity; the Bago Yoma, Myanmar, population, while similar in size was more isolated geographically and had much lower allelic diversity but similar heterozygosity (Budd et al. 2021). All these findings were in accordance with expectations based on the demographic and geographic history of the populations.
Population genetic and spatial diversity
We compiled data from 35 test populations from India, Lao PDR, Vietnam, Thailand, China, Myanmar, Cambodia, Sumatra, Borneo, peninsular Malaysia and US Zoos (Table 1). Among the reference populations, AR ranged from 0.241 (ARSE ± 0.032) to 1.715 (± 0.183) and HR ranged from 0.325 (HRSE ± 0.082) to 1.178 (± 0.083). Rankings of the values from the original studies and the ratios of diversity generated under the yardstick model differed significantly (Spearman’s rank correlation Atest vs. AR = 0.991774, p < 0.05; Htest vs. HR = 0.91911, p < 0.05). For both allelic diversity and heterozygosity, we found the highest ratios of diversity in populations in Thailand, Lao PDR, Cambodia, and Myanmar while populations from Borneo and India contained the lowest diversity (Table 1). For populations in China, rankings were mixed, with Mengyang ranking 10th in Atest and 6th in AR but 30th in both Htest and HR, Shangyong ranking 24th in Atest and 11th in AR but 35th in Htest and 34th in HR, and Mengla ranking 16th in Atest and 15th in AR but 32nd in Htest and 31st in HR. All other Chinese populations had diversity ratios in the lowest third of the rankings for allelic diversity (Atest and AR) and heterozygosity (Htest and HR).
Analysis of genetic diversity based on geographic patterns showed hotspots for expected heterozygosity in southeast Asia (Myanmar, Thailand, Cambodia, Lao PDR) and southern India (Fig. 1A). Based on allelic diversity, mainland southeast Asia was the primary diversity hotspot (Fig. 1B). The lowest relative diversity using each metric was seen in the small populations of Borneo and China (Fig. 1A, B).
Evolutionary distinctiveness
We compiled mtDNA sequences from populations in 12 home range countries and North American zoos (Supplemental Table 2). We found 69 unique Asian elephant haplotypes; 31 from the α-clade and 38 from the β-clade. Of these, only 19 were shared between two or more range countries, allowing for 50 private haplotypes. Additionally, two haplotypes (AO, AP) were found only in zoo populations (Lei et al. 2011).
The optimal substitution model for the maximum likelihood analysis of evolutionary distinctiveness was determined to be HKY + I + G. MtDNA haplotypes with the highest evolutionary distinctiveness scores were found to be AI (0.054; Myanmar), BBG (0.046; peninsular Malaysia), BD (0.036; Borneo), BY (0.031; Lao PDR), BR (0.027; Sumatra), and AP (0.027; North American Zoos), while the lowest scores were AH (0.000; multiple countries), AB (0.001; multiple countries), AD (0.001; multiple countries), AR (0.002; Lao PDR), and BF (0.002; India, North American Zoos; Fig. 2). The most widely distributed haplotypes were BO, found in 5 range countries and North American Zoos, and AD and BP, each of which was found in four range countries and North American Zoos.
Discussion
The Asian elephant is experiencing alarming decreases in the number and sizes of populations across its range (Blake and Hedges 2004; Hedges 2006). Evaluating the levels and distribution of genetic diversity is vital to conservation, as it allows managers to infer migratory pathways across fragmented landscapes, estimate census and effective population sizes and demographic parameters, and infer the proximate and ultimate causes of human-elephant conflict. As populations decline in size, a reduction in allelic diversity due to genetic drift is predicted to occur before the loss of heterozygosity (Nei et al. 1975; Cornuet and Luikart 1996). Thus, monitoring levels of both measures of genetic diversity is essential to allow managers to detect genetic erosion and, to the extent possible, avoid the deleterious effects of inbreeding depression (Hoban et al. 2021).
In their study of isolated populations of elephants on Goossens et al. (2016) suggested that using highly polymorphic markers, such as the microsatellite loci developed by Kongrit et al. (2008), rather than randomly selected markers to estimate genetic diversity likely produced overestimated values. Specifically, they suggested that although both their Bornean values and the values reported by Ahlering et al. (2011) for the Nakai population were similar to values reported for populations of other rare or endangered species, they were likely overestimates. Their study did not suggest a remedy for such over- or underestimation, presumably caused by the use of different loci. The yardstick method we used in this study allows for direct comparison of studies after calibration. While it does not produce “corrected” values for allelic diversity and heterozygosity, it does produce ratios that managers can use to compare populations.
Although the largest Asian elephant populations are found in India, we found that the Southeast Asian populations of Thailand, Lao PDR, Cambodia and Myanmar harbored higher levels of allelic diversity and heterozygosity. The higher levels of diversity are in accordance with the results of Ahlering et al. (2011), who suggested that the high levels of diversity found in the Nakai Plateau, Lao PDR population underscored its importance for the conservation of the species. As has been shown previously, we found that studies of Chinese and Bornean populations consistently reveal low diversity, which is especially notable after the data are corrected for marker bias.
In addition to monitoring levels of diversity, managers must also consider genetic differentiation among populations. Many studies of Asian elephants have used mtDNA to estimate levels of differentiation due the relative ease of amplifying it from non-invasively collected dung samples, its higher mutation rate than nuclear DNA which enables studies within and among populations, and its maternal mode of inheritance in a largely matrilineally structured species. Ballard and Whitlock (2004) argued that mtDNA should not be used for this purpose as it may be subject to natural selection, Galtier et al. (2009) argued that mtDNA mutation rates differ among species and do not exhibit “molecular clock-like” behavior, and Toews and Brelsford (2012) cite instances of mito-nuclear discordance that could be problematic in biogeographic studies. While we do not disagree with these limitations, our study used mtDNA to document and compare levels of genetic differentiation. The action of natural selection on mtDNA would enhance differentiation, which would suggest that managers should use caution when managing highly divergent populations as a single unit. We do not attempt to date population divergences, thus no molecular clock was applied. While mito-nuclear discordance has been documented in African elephants (Roca et al. 2005), we are unaware of such findings in Asian elephants. In any case, our study did not reveal surprising asssociations among populations that might suggest the presence of discordance.
He et al. (2020) suggested that inbreeding in Chinese populations might be alleviated by establishing connectivity among remnant Chinese populations and with regions such as Lao PDR or by initiating further actions such as translocations to facilitate genetic rescue. However, prior to taking such actions managers should evaluate genetic differentiation between potential donor and recipient populations. For example, despite their findings of low diversity in Bornean elephants, Goossens et al. (2016) expressed concerns with the potential for outbreeding depression if gene flow was established between Bornean and mainland elephant populations.
In Asian elephants, three subspecies are currently recognized: the Asian mainland elephant or Indian elephant, E. m. indicus, the Sri Lankan elephant, E. m. maximus, and the Sumatran elephant, E. m. sumatranus. Additionally, the Bornean elephant, E. m. borneensis, has been supported by genetic studies (Fernando et al. 2003; Sharma et al. 2018) but awaits further range-wide assessment prior to designation (Williams et al. 2020). Our mtDNA results support the evolutionary distinctiveness of the Sumatran elephant subspecies (E. m. sumatranus; 5 haplotypes BP, BR, BS, BT, BU with evolutionary distinctiveness values from 0.009 to 0.027) and the proposed Bornean elephant subspecies (E. m. borneensis; single haplotype BD with high evolutionary distinctiveness 0.036). In our haplotype network, the Sumatran haplotypes group together with multiple steps between them and other β clade haplotypes. Thus, they meet the criteria of evolutionary significant units, or lineages of populations that maintain their identity from other such lineages and demonstrate independent historical fates (Wiley 1978); management units that demonstrate reciprocal monophyly at mtDNA (Mortiz 1994); and/or phylogenetic species that possess a combination of derived traits and a unique evolutionary history (Cracraft 1982). Regardless of taxonomic classification, the present study underscores Goossens et al. (2016) concerns for the potential of outbreeding depression if translocations are used to enhance genetic variability, especially for insular populations.
We also found evolutionary distinctiveness among Indian elephant (aka mainland elephant, E. m. indicus) populations, particularly among peninsular Malaysia and Myanmar populations. Although the largest populations of Asian elephants are found in India, we found lower regions of diversity and distinction in these populations than in the small, fragmented populations of southeast and insular Asia. The largest Asian elephant population, Nilgiris, is characterized by a single mtDNA haplotype (BN; Vidya et al. 2005a) that is of low evolutionary distinctiveness. Nilgiris is also among the populations with levels of nuclear diversity lower than those in Thailand, Lao PDR, Cambodia, and Myanmar. The lower diversity found in this large population underscores the importance of conservation efforts focusing not only on the large populations of India, but also on the smaller populations of southeast and insular Asia for the preservation of diversity and distinctive lineages within the species.
Across southeast Asia, surveys have produced varying estimates of population sizes, especially in Lao PDR (ranging from 500 to 1000; Duckworth and Hedges 1998; Choudhury et al. 2008; Khounboline 2011), Vietnam (100–130 elephants; Williams et al. 2020) and Cambodia (250 to 2000 elephants; Kemf and Jackson 1995; Osborn and Vinton 1999; Murdoch 2008). In Myanmar, where more potential elephant habitat exists than in any other country in the species’ range, the total population is estimated at approximately 2,000 individuals, but the country’s recent and extreme poaching climate may drastically reduce those numbers in coming years (Leimgruber et al. 2011; Sampson et al. 2018). The small populations, such as those in southeast Asia that harbor the highest diversity, are also among those with the highest rates of habitat loss and fragmentation (Williams et al. 2020) and are therefore at high risk for local extirpation. The loss of diversity in southeast Asia is likely to be directly detrimental to the long-term preservation of the species.
Conservation management aims to reduce the rate of population declines and increase the probability of recovery of a species adaptive potential under changing environmental conditions; thus, methods for quantifying genetic diversity and differentiation within and between declining populations are essential tools. The issue of marker selection bias in the comparison of genetic diversity across a species range is not unique to elephants as yardstick calibration models have also been used in studies of brown bears (Skrbinsek et al. 2012) and water buffalo (Colli et al. 2018) and European wolves (Hindrikson et al. 2016). Despite the overwhelming evidence of marker selection bias using microsatellites, few studies address it. One important limitation of yardstick calibration modeling is the need to create a large, well-studied population that is amplified at all microsatellites in the comparison, which may be costly and time-consuming. However, for study species such as Asian elephants, where separate incomparable panels have dominated the literature, yardstick calibration modeling can be an important tool for identifying patterns of diversity to aid in conservation management.
Data availability
Data from the yardstick population (Nakai Plateau, 25 microsatellite loci) and the R code used to calculate the diversity ratios have been deposited in DRYAD: https://doi.org/10.5061/dryad.vdncjsz11. Genetic diversity data from populations used for comparison to the yardstick population are provided in Table 1. Accession numbers for all mtDNA sequences used in the evolutionary distinctiveness analyses are shown in Supplementary Table 2.
References
Ahlering MA, Hedges S, Johnson A, Tyson M, Schuttler SG, Eggert LS (2011) Genetic diversity, social structure and conservation value of the elephants of the Nakai Plateau, Lao PDR, based on non-invasive sampling. Conserv Genet 12:413–422
Ballard JWO, Whitlock MC (2004) The incomplete natural history of mitochondria. Mol Ecol 13:729–744
Bandelt H, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Blake S, Hedges S (2004) Sinking the flagship: the case of forest elephants in Asia and Africa. Conserv Biol 18:1191–1202
Budd K (2021) Conservation genetics of conflict in the asian elephant, Elephas maximus. University of Missouri–Columbia (PhD dissertation). https://doi.org/10.32469/10355/88043
Chakraborty R, Kimmel M, Stivers DN, Davison LJ, Deka R (1997) Relative mutation rates at di-, tri-, and tetranucleotide loci. Proc Nat Acad of Sci USA 94(3):1041–1046
Chakraborty S, Boominathan D, Desai AA, Vidya TNC (2014) Using genetic analyses to estimate population size, sex ratio, and social organization in an asian elephant population in conflict with humans in Alur, southern India. Conserv Genet 15:897–907
Choudhury A, Lahiri Choudhury DK, Desai A, Duckworth JW, Easa PS, Johnsingh AJT, Fernando P, Hedges S, Gunawardena M, Kurt F, Karanth U, Lister A, Menon V, Riddle H, Rübel A, Wikramanayake E, IUCN SSC Asian Elephant Specialist Group (2008) Elephas maximus. IUCN Red List Threat Species. https://doi.org/10.2305/IUCN.UK.2008.RLTS.T7140A12828813.en
Colli L, Milanesi M, Vajana E, Iamartino D, Bomba L, Puglisi F, Del Corvo M, Nicolazzi EL, Ahmed SSE, Herrera JRV, Cruz L, Zhang S, Liang A, Hua G, Yang L, Hao X, Zuo F, Lai S-J, Wang S, Liu R, Gong Y, Mokhber M, Ym Mao, Guan F, Vlaic A, Vlaic B, Ramunno L, Cosenza G, Ahmad A, Soysal I, Ünal E, Ketudat-Cairns M, Garcia JF, Utsunomiya YT, Baruselli PS, Amaral MEJ, Parnpai R, Drummond MG, Galbusera P, Burton J, Hoal E, Yusnizar Y, Sumantri C, Moioli B, Valentini A, Stella A, Williams JL, Ajmone-Marsan P (2018) New insights on water buffalo genomic diversity and post-domestication migration routes from medium density SNP chip data. Front Genet. https://doi.org/10.3389/fgene.2018.00053
Comstock KE, Wasser SK, Ostrander EA (2000) Polymorphic microsatellite DNA loci identified in the african elephant (Loxodonta africana). Mol Ecol 9:1004–1006
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cracraft J (1982) Geographic differentiation, cladistics, and vicariance biogeography: reconstructing the tempo and mode of evolution. Am Zool 22:411–424. https://doi.org/10.1093/icb/22.2.411
de Groot GA, Nowak C, Skrbinsek T, Andersen LW, Aspi J, Fumagalli L, Godinho R, Harms V, Jansman HAH, Liberg O, Marucco F, Myslajek RW, Nowak S, Pilot M, Randi E, Reinhardt I, Smietana W, Szewczyk M, Taberlet P, Vila C, Munoz-Fuentes V (2016) Decades of population genetic research reveal the need for harmonization of molecular markers: the grey wolf Canis lupus as a case study. Mamm Rev 46:44–59
Duckworth W, Hedges S (1998). Tracking Tigers: a review of the status of tiger, asian elephant, gaur, and banteng in Vietnam, Laos, Cambodia, and Yunnan (China), with recommendations for future conservation action, 1st edn. WWF Indochina Programme, Hanoi
Eggert LS, Ramakrishnan U, Mundy NI, Woodruff DS (2000) Polymorphic microsatellite DNA markers in the african elephant (Loxodonta africana) and their use in the asian elephant (Elephas maximus). Mol Ecol 9:2223–2225
Eggert LS, Beadell JS, McClung A, McIntosh CE, Fleischer RC (2009) Evolution of microsatellite loci in the adaptive radiation of hawaiian honeycreepers. J Hered 100:137–147. https://doi.org/10.1093/jhered/esn111
Eggert LS, Ruiz-Lopez M (2011) Analysis of fecal DNA samples to estimate size and sex ratio of the elephant population at Seima biodiversity conservation area in Cambodia using capture-recapture methods. Report to the Wildlife Conservation Society
Eggert LS, Ruiz-Lopez M (2012a) Analysis of fecal DNA samples to estimate the sex ratio and size of the sepon asian elephant population in the Lao PDR using capture-recapture methods. Report to the Wildlife Conservation Society
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5:435–445. https://doi.org/10.1038/nrg1348
Fernando P, Lande R (2000) Molecular genetics and behavioral analysis of social organization in the asian elephant (Elephas maximus). Behav Ecol Sociobiol 48(84):84–91
Fernando P, Pfrender ME, Encalada SE, Lande R (2000) Mitochondrial DNA variation, phylogeography and population structure of the asian elephant. Heredity 84:362–372
Fernando P, Vidya TNC, Melnick DJ (2001) Isolation and characterization of tri- and tetranucleotide microsatellite loci in the asian elephant, Elephas maximus. Mol Ecol Notes 1:232–233
Fernando P, Vidya TNC, Payne J, Stuewe M, Davison G, Alfred RJ, Andau P, Bosi E, Kilbourn A, Melnick DJ (2003) DNA analysis indicates that asian elephants are native to Borneo and are therefore a high priority for conservation. PLoS Biol 1(1):e6
Fleischer RC, Perry EA, Muralidharan K, Stevens EE, Wemmer CM (2001) Phylogeography of the asian elephant (Elephas maximus) based on mitochondrial DNA. Evolution 55(9):1882–1892
Frankham R (2005) Genetics and extinction. Biol Conserv 126(2):131–140
Galtier N, Nabholz B, Glémin S, Hurst GDD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18:4541–4550
Gobush KS, Edwards CTT, Balfour D, Wittemyer G, Maisels F, Taylor RD (2021) Loxodonta africana. IUCN Red List Threat Species 2021:e.T181008073A181022663
Goossens B, Sharma R, Othman N, Kun-Rodrigues C, Sakong R, Ancrenaz M, Ambu LN, Jue NK, O’Neill RJ, Bruford MW, Chikhi L (2016) Habitat fragmentation and genetic diversity in natural population of the Bornean elephant: implications for conservation. Biol Conserv 196:80–92
Gray TNE, Vidya YNC, Potdar S, Bharti DK, Sovanna P (2014) Population size estimation of an asian elephant population in eastern Cambodia through non-invasive mark-recapture sampling. Conserv Genet 15:803–810
He C, Du J, Zhu D, Zhang L (2020) Population viability analysis of small population: a case study for asian elephant in China. Integr Zool 15:350–362
Hedges S (2006) Conservation. In: Fowler ME, Mikota SK (eds) Biology, Medicine and surgery of elephants. Blackwell Publishing, Oxford, pp 475–490
Hindrikson M, Remm J, Pilot M, Godinho R, Stronen AV, Baltrūnaité L, Cazanomska SD, Leonard JA, Randi E, Nowak C, Åkesson M, López-Bao JV, Álvares F, Llaneza L, Echegaray J, Vilà C, Ozolins J, Rungis D, Aspi J, Paule L, Skrbinšek T, Saarma U (2016) Wolf population genetics in Europe: a systematic review, meta-analysis and suggestions for conservation and management. Biol Rev 92:1601–1629. https://doi.org/10.1111/brv.12298
Hoban S (2018) Integrative conservation genetics: prioritizing populations using climate predictions, adaptive potential and habitat connectivity. Mol Ecol Res 18:14–17. https://doi.org/10.1111/1755-0998.12752
Hoban S, Bruford MW, Funk WC, Galbusera P, Griffith MP, Grueber CE, Heuertz M, Hunter ME, Hvilsom C, Stroil BK, Kershaw F, Khoury CK, Laikre L, Lopes-Fernandes M, MacDonald AJ, Mergeay J, Meek M, Mittan C, Mukassabi TA, O’Brien D, Ogden R, Palma-Silva C, Ramakrishnan U, Segelbacher G, Shaw RE, Sjögren-Gulve P, Veličković N, Vernesi C (2021) Global commitments to conserving and monitoring genetic diversity are now necessary and feasible. Bioscience 71:964–976. https://doi.org/10.1093/biosci/biab054
Holland MM, Parson W (2010) GeneMarker HID: a reliable software tool for the analysis of forensic STR data. J Forensic Sci 56(1):29–35
Isaac NJB, Turvey ST, Collen B, Waterman, Baillie JEM (2007) Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2(3):e296
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27(21):3070–3071
Karamanlidis AA, Skrbinsek T, de Gabriel Hernando M, Krambokoukis L, Munoz-Fuentes V, Bailey Z, Nowak C, Stonen AV (2018) History-driven population structure and asymmetric gene flow in a recovering large carnivore at the rear-edge of its european range. Heredity 120:168–182
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Thierer T (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26(11):1463–1464
Kemf E, Jackson P (1995) Asian elephants in the wild. World Wide Fund for Nature, Gland
Khounboline K (2011) Current status of asian elephants in Lao PDR. Gajah 35:4
Kongrit C, Siripunkaw C, Brockelman WY, Akkarapatumwong V, Wright TF, Eggert LS (2008) Isolation and characterization of dinucleotide microsatellite loci in the asian elephant (Elephas maximus). Mol Ecol Res 8:175–177
Kusza S, Suchentrunk F, Pucher H, Mar KU, Zachos FE (2018) High levels of mitochondrial genetic diversity in asian elephants (Elephas maximus) from Myanmar. Hystrix 29:152–154
Leberg PL (2002) Estimating allelic richness: effects of sample size and bottlenecks. Mol Ecol 11:2445–2449
Lei R, Brenneman RA, Schmitt DL, Louis EE Jr (2011) Genetic diversity in north american captive asian elephants. J Zool 286:38–47
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116
Leimgruber P, Oo ZM, Myint Aung, Kelly DS, Wemmer C, Senior B, Songer M (2011) Current status of elephants in Myanmar. Gajah 35:76–86
Moritz C (1994) Defining evolutionary significant units for conservation. Trends Ecol Evol 9(10):373–375
Moßbrucker AM, Apriyana I, Fickel J, Imron MA, Pudyatmoko S, Sumardi, Suryadi H (2015) Non-invasive genotyping of Sumatran elephants: implications for conservation. Trop Conserv Sci 8:745–759
Murdoch G (2008) Factbox – Threats Facing Asia’s Endangered Wild Elephants. Reuters, Toronto
Murphy SM, Laufenberg JS, Clark JD, Davidson M, Belant JL, Garshelis DL (2018) Genetic diversity, effective population size, and structure among black bear populations in the Lower Mississippi Alluvial Valley, USA. Conserv Genet 19:1055–1067
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nyakaana S, Arctander P (1998) Isolation and characterization of microsatellite loci in the african elephant, Loxodonta africana. Mol Ecol 7:1436–1437
Osborn FV, Vinton MD (eds) (1999) Proceedings of the Conference: Conservation of the Asian Elephant in Indochina. Hanoi, Vietnam, 24–27 November 1999. FFI-Indochina. Asian Elephant Conservation Programme
Zar JH (2005) Spearman rank correlation. In: Armitage P, Colton T (eds) Encyclopedia of Biostatistics. Wiley, New York. https://doi.org/10.1002/0470011815.b2a15150
Parida J, Sharma R, De R, Kalam T, Sedhupathy A, Digal DK, Reddy PA, Goyal SP, Puyravaud J-P, Davidar P (2022) Genetic characterisation of fragmented asian elephant populations with one recent extinction in its eastern-central indian range. Ecol Genet Genom 24:100132
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. Available at https://www.R-project.org
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17(1):230–237
Roca AL, Georgiadis N, O’Brien SJ (2005) Cytonuclear genomic dissociation in african elephant species. Nat Genet 37:96–100
Sampson C, McEvoy J, Oo ZM, Chit AM, Chan AN, Tonkyn D, Leimgruber P (2018) New elephant crisis in Asia—early warning signs from Myanmar. PLoS ONE 13(3):e0194113
Sharma R, Goossens B, Heller R, Rasterio R, Othman N, Bruford MW, Chikhi L (2018) Genetic analyses favour an ancient and natural origin of elephants on Borneo. Sci Rep 8:880
Skrbinsek T, Jelencic M, Waits LP, Potocnik H, Kos I, Trontelj P (2012) Using a reference population yardstick to calibrate and compare genetic diversity reported in different studies: an example from the brown bear. Heredity 109:299–305
Stanley RRE, Jeffrey NW, Wringe BF, Dibacco C, Bradbury IR (2017) Genepopedit: a simple and flexible tool for manipulating multilocus molecular data in R. Mol Ecol Res 17:12–18
Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland
Thitaram C, Somgird C, Mahasawangkul S, Angkavanich T, Roongsri R, Thongtip N, Colenbrander B, van Steenbeek F, Lenstra J J (2010) Genetic assessment of captive elephant (Elephas maximus) populations in Thailand. Conserv Genet 11:325–330
Thitaram T, Dejchaisri S, Somgrid C, Angkawanish T, Brown J, Phumphuay R, Chomdech S, Kangwanpong D (2015) Social group formation and genetic relatedness in reintroduced asian elephants (Elephas maximus) in Thailand. Appl Anim Behav Sci 172:52–57
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Toews DPL, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21:3907–3930
Vidya TNC, Fernando P, Melnick DJ, Sukumar R (2005a) Population differentiation within and among asian elephant (Elephas maximus) population in southern India. Heredity 94:71–80
Vidya TNC, Fernando P, Melnick DJ, Sukumar R (2005b) Population genetic structure and conservation of asian elephants (Elephas maximus) across India. Anim Conserv 8:377–388
Vidya TNC, Varma S, Dang NX, Van Thanh T, Sukumar R (2007) Minimum population size, genetic diversity and social structure of the asian elephant in Cat Tien National Park and its adjoining areas, Vietnam based on molecular genetic analyses. Conserv Genet 8:1471–1478
Vidya TNC, Sukumar R, Melnick DJ (2009) Range-wide mtDNA phylogeography yields insights into the origins of asian elephants. Proc R Soc Lond B 276:893–902
Villesen P (2007) FaBox: an online toolbox for fasta sequences. Mol Ecol Notes 7:965–968
Wiley EO (1978) The evolutionary species concept reconsidered. Syst Zool 27:17–26
Williams C, Tiwari SK, Goswami VR, de Silva S, Kumar A, Baskaran N, Yoganand K, Menon V (2020) Elephas maximus. IUCN Red List Threat Species 2020:e.T7140A45818198
Zhang L, Dong L, Lin L, Feng L, Yan F, Wang L, Guo X, Luo A (2015) Asian elephants in China: estimating population size and evaluating habitat suitability. PLoS ONE 10(5):e0124834
Acknowledgements
We are grateful to the Lao Ministry of Agriculture and Forestry’s Department of Forestry, the Government of Lao PDR’s Provincial and District Agriculture and Forestry Offices (PAFO and DAFO) and the Nam Theun 2 Power Company Ltd. (NTPC) for their assistance with the survey of the Nakai Plateau area. This work would not have been possible without the wildlife survey guides, technicians, and administrative support from the Nam Theun 2 Watershed Management and Protection Authority (WMPA).
Funding
This research was funded by the American Philosophical Society Lewis and Clark Field Grant and the TransWorld Airline Scholarship fund. KB was funded by a National Science Foundation Graduate Research Fellowship. This is KBS contribution number 2355.
Author information
Authors and Affiliations
Contributions
KB designed the study and carried out the labwork, initial data analyses and writing of the first draft of the manuscript. JG and LLS conducted data analyses, data interpretation and contributed substantially to the writing of the final draft of the manuscript. LSE administered the project and contributed to the writing of the final draft of the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Budd, K., Gunn, J.C., Sullivan, L.L. et al. Identification of conservation priority units in the Asian elephant, Elephas maximus. Conserv Genet 24, 827–837 (2023). https://doi.org/10.1007/s10592-023-01542-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01542-1