Abstract
The biological traits of different species usually predispose them to dissimilar levels of extinction risk. Accordingly, we analysed genetic variation and population genetic structure of two endangered forest monkeys to determine how they were influenced by habitat change. The Tana River red colobus and mangabey are of similar body size, are sympatric and endemic to forests in Kenya. The mangabey is frugivorous, terrestrial and exhibits female philopatry but the colobus is folivorous, arboreal and both sexes exhibit natal dispersal. We predicted that both species would have low genetic variation due to high genetic drift, and possible inbreeding, because their populations are relatively small and subdivided in forest fragments. Nevertheless, we expected the mangabeys to have a higher genetic variation because they have a larger population, and their higher vagility would enhance gene flow. Thus, we expected a stronger population genetic structure in the colobus because lower vagility would constrain dispersal and increase genetic differentiation. Finally, we expected both species to exhibit isolation by distance due to greater gene flow among populations in close geographic proximity. We genotyped 146 colobus and 76 mangabeys with nine microsatellite markers and found that both species had relatively high heterozygosities but low allelic richness. Furthermore, allelic richness, heterozygosity and genetic differentiation were all higher in the mangabey and only the mangabey exhibited isolation by distance. Taken together, our results suggest that species with divergent behavioral ecologies may exhibit differential vulnerabilities to genetic erosion due to habitat change.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The biological traits of different species often predispose them to dissimilar levels of extinction risk. Thus beetle species that are rare and are habitat specialists are particularly vulnerable to extinction in fragmented habitats (Davies et al. 2004), vagile arboreal geckos thrive in fragmented habitats (Hoehn et al. 2007) and arboreal monkeys that subsist on low-fruit diets are highly vulnerable to agricultural expansion (Isaac and Cowlishaw 2004).
Even so, it is not always clear how, or why, particular biological traits may predispose species to different extinction threats (Davies et al. 2004; Henle et al. 2004; Isaac and Cowlishaw 2004). This is an issue of primary conservation importance because differential vulnerability to extinction threats must determine which species ultimately go extinct. Accordingly, we sought to investigate the effects of habitat loss and fragmentation, habitat change hereafter, on the genetic variation and population genetic structure of two endangered forest monkeys with divergent biological traits from eastern Kenya.
Habitat change is the most serious threat to biodiversity because its effects are diverse and widespread (Primack 2010). In particular, the breaking apart of habitat often reduces population sizes of resident species and expands the matrix which may also obstruct dispersal. The obstruction of dispersal is a serious problem because it undermines metapopulation persistence and gene flow (Clobert et al. 2001), which may decrease genetic variation through drift and inbreeding within subpopulations (Frankham et al. 2010). The inability to disperse is a ruinous prospect for any species because loss of genetic variation compromises the ability of populations to adapt to changes in the environment by undermining population persistence, evolutionary potential and individual fitness (Frankham et al. 2010; Keller and Waller 2002).
We focused on monkeys because primate biology and the variety of their responses to extinction threats are well documented relative to other taxonomic groups (Isaac and Cowlishaw 2004; Cowlishaw and Dunbar 2000). Indeed, non-human primates are a flagship order of mammals in tropical forests (sensu Simberloff 1998) because most are endemic to these forests where their species richness is positively associated with forest extent (Reed and Fleagle 1995) and their biomass often exceeds 30 % of all mammals (Fa and Purvis 1997). Even so, tropical forests everywhere are undergoing increasing habitat change due to burgeoning human populations and expanding per capita resource consumption (Laurance 2007; Mayaux et al. 2005). Consequently, studies show that even protected areas within tropical forests are becoming increasingly isolated as surrounding habitats are degraded and destroyed by development (DeFries et al. 2005). Thus, primates are disproportionately threatened by tropical forest habitat change (Mace and Balmford 2000; Mittermeier et al. 2007).
Tropical forest found in eastern Kenya is the only known habitat of two endangered monkeys, the Tana River red colobus (Procolobus rufomitratus, Peters 1879) and mangabey (Cercocebus galeritus, Peters 1879). The forest is found in fragments of various sizes with a combined total area of 26-km2, but the monkeys are found in only some of the fragments (Butynski and Mwangi 1994). The two monkey species are of similar body size, but are dissimilar in several behavioral ecology traits. The colobus are folivorous, arboreal and both males and females disperse on attaining sexual maturity (Marsh 1979; Mbora and Meikle 2004). Thus due to its arboreality, the Tana red colobus is much more dependent on a relatively continuous forest canopy than the largely terrestrial mangabey, and therefore it has lower vagility in the fragmented forest landscape. In contrast, the mangabey is a frugivore that spends at least 80 % of their time budget on the ground, only males disperse on attaining sexual maturity, and females are strongly philopatric (Kinnaird 1992; Wieczkowski and Butynski 2013). Both monkey species are endangered because the colobus population is estimated at less than 1,000 individuals and the mangabey population at about 2,000 (Mittermeier et al. 2007; Mbora unpublished data).
We predicted that both species would have low genetic variation due to high genetic drift, and possible inbreeding, because their populations are relatively small and subdivided in forest fragments. We further hypothesized that habitat change would influence the genetic variation, and population genetic structure, of the two monkey species differently given their dissimilarities in vagility, population size and patterns of natal dispersal. First, we expected that the mangabeys would have higher genetic variation than the colobus because their greater vagility enhances gene flow among forest fragments, and their population size is at least twice as large. Secondly, we expected a stronger population genetic structure in the colobus because lower vagility would constrain dispersal and enhance genetic differentiation in this species. While it is true that female philopatry could promote significant genetic differentiation in the mangabey, the tendency for new groups to form by fissioning of existing groups (Dittus 1988) should attenuate genetic structuring in this species. Third, we expected that subpopulations that were in close geographic proximity would exhibit greater gene flow among each other and therefore we expected to find isolation by distance (IBD) in both species (Wright 1943).
Materials and methods
The study area
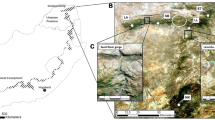
The study area comprised approximately 26 km2 of gallery forest occurring in scattered patches of various sizes on both sides of the Tana River in eastern Kenya (see Fig. 1 in Mbora and McPeek 2010). This part of Kenya is arid as it receives a mean annual rainfall of just 400 mm (Hughes 1990). As such, the forests are maintained by groundwater and annual flooding, which limits their lateral extent to about a kilometer on either side of the river (Hughes 1990). The forests are embedded in a matrix of small-scale farmland, riparian grassland and shrubs and comprise the entire global distribution range of the Tana River red colobus (Procolobus rufomitratus) and mangabey (Cercocebus galeritus).
We had mapped the forests as part of an earlier study using aerial photographs, and systematically selected 28 fragments for long-term monitoring of the primate populations (Mbora and McPeek 2010; Mbora unpublished data). We selected study fragments so as to capture the range of habitat conditions within the floodplain and to ensure that comparable forest area was studied east and west of the Tana River, and inside and outside the Tana River Primate National Reserve (TRPNR).
We surveyed each study forest entirely as a team of three observers to determine the number of resident groups of colobus and mangabeys using the sweep sampling method (Whitesides et al. 1988). Subsequently, we then identified subsets of social groups for detailed assessments of size and age-sex compositions through time as detailed in Mbora and McPeek (2010). The fecal samples used for the data reported here were collected in the field during the months of July, August and September in 2005, 2006, 2007 and 2009. The samples were collected from 37 groups of colobus in 23 forest fragments, and 27 groups of mangabey in 17 forest fragments;—please also see below under data analyses for details of how subpopulations were designated.
Collection of fecal samples and DNA extraction
We collected fecal samples as 3 observers—DNMM and two field assistants—by following the study groups, on separate days for each species, from 0600 to 1130 h, and then from 1500 h until nightfall. Upon observing an animal defecating, we collected a sample of the feces using a sterile collecting stick while wearing latex gloves. We tried to sample as many individuals from each study group and forest fragment as possible for each species.
Colobus feces are usually deposited in distinct pellets so we simply collected 1–3 pellets depending on size. The mangabey, however, does not usually produce its feces in distinct pellets so we gleaned a sample from the outermost part of the fecal deposit. In each case, the sample was placed into a tube containing 30 ml of 100 % ethanol and labeled with a permanent marker to indicate the date, species identity, and coded to identify the troop and forest. The ethanol and sample were then mixed by inversion without shaking in order to maintain the bolus and avoid losing target cells with the ethanol supernatant in the next step. After 36 h, we carefully poured off the ethanol with the tube loosely capped, and transferred the remaining solid material into a new tube, labeled as above, and containing silica for drying and storage (Nsubuga et al. 2004). The samples were then stored cool and dry in a tent in the field, and at −80 °C upon arrival in the laboratory until DNA was extracted. We extracted DNA from the fecal samples using the QIAamp DNA Stool kit (Qiagen, catalog # 51504) but instituted several modifications to the manufacturer’s protocol.
It is well established that the outermost layer of a fecal sample yields the least degraded DNA and also contains the lowest concentration of PCR inhibitors (Wehausen et al. 2004; Fernando et al. 2003). Therefore, we started our extraction procedure by carefully scraping off 15–60 mg from the outermost layer of the bolus into a 2 ml micro centrifuge tube. We used a sterile scalpel blade to scrape each sample and changed gloves between samples. We then followed the extraction procedures as outlined in the Qiagen kit but also adopted some recommendations by Wehausen et al. (2004). Specifically, we added about 2 ml of ASL buffer, vortexed the mixture thoroughly to homogenize and then heated the homogenate for 10 min at 56 °C. Additionally, we heated the elution buffer AE to 70 °C, allowed it to incubate in the spin columns for 3 min and then centrifuged the eluate through the spin columns twice to maximize DNA collection.
We were very careful to avoid cross contamination among samples of our two species, and contamination of our samples by other concentrated DNA sources. First, we worked on samples from our two study species sequentially rather than concurrently (i.e. we worked on the colobus and then mangabey samples). Second, we extracted the DNA in an area of the lab that was different and a distance away from the area where we set up the PCR reactions. Third, all the laboratory procedures reported here were performed in a section of the laboratory dedicated to the analyses of DNA from the two primates and this laboratory did not work with any other species of primates or vertebrates.
DNA amplification and genotyping
Numerous studies have demonstrated that the low quality and quantity of DNA recovered from feces can cause genotyping errors (Taberlet et al. 1999). In particular, only one allele of a heterozygous individual may be detected (Gerloff et al. 1995; Taberlet et al. 1996) and PCR amplification artifacts may be misinterpreted as true alleles (Taberlet et al. 1996). Therefore, we used well-tested procedures to minimize, or possibly eliminate, any genotyping errors (Beja-Pereira et al. 2009). First, immediately following DNA extractions, we screened the samples to ensure that they contained amplifiable DNA using a real-time quantitative PCR procedure, and genotyped a sample only if it contained at least 100 pg of amplifiable DNA (Taberlet et al. 1996; Morin et al. 2001). Whenever feasible, extractions were repeated for samples that did not contain amplifiable DNA. Second, we used same nine microsatellite markers to genotype individuals of both species in multiplex PCRs (Chamberlain et al. 1988). The nine markers were a subset of 15 originally optimized and used for the study of primate (Cercopithecidae) population genetics by Bonhomme et al. (2005, 2008). Six of these markers were tetranucleotides (D1S548, D3S1768, D4S243, D7S2204, D8S1106, and D10S1432) and the other three were dinucleotides (DQcar, MIB, MOGc; Bonhomme et al. 2005). We used these particular nine markers because they showed the highest heterozygosity, based on preliminary assessments, for our species and were therefore the most informative, and using these highly informative markers generated the same information with fewer loci, helping us to conserve template DNA and to minimize the risk of allelic dropout (Taberlet et al. 1996). We optimized multiplexed combinations of the nine markers to accommodate dye labels and product sizes as appropriate, and then ran the reactions using the QIAGEN Multiplex PCR Kit (catalog #: 206143) in a MJ Research PTC-200 Peltier thermal cycler.
A sample from each individual was amplified in four replicates, i.e. the multiple tubes approach (Taberlet et al. 1996). Each reaction contained 1.5 µL of the eluate from the extractions as template, 7.5 µL of 2X Qiagen multiplex PCR master mix, 1.5 µL of 2 µM each primer, 1.2 µL BSA and 3.3 µL of RNase-free water for a final volume of 15 µL. The thermal profile was as recommended in the PCR kit but our reactions required an annealing temperature of 57 degrees Celsius for 3 min and 35 cycles. Every 96 PCRs plate included two negative controls without template DNA, and two positive controls with high quality template DNA obtained from tissues samples. Template DNA from colobus tissue was donated by colleagues who work with closely related species of colobus in Uganda. Template DNA for the mangabey was extracted from a tissue sample acquired, and preserved in ethanol, when an individual from one of the study groups was killed by a bird of prey in August 2005 (Mbora and McPeek 2010).
Amplified products were subsequently genotyped with an ABI Prism™ 3100 genetic analyzer (Applied Biosystems). GeneMarker (SoftGenetics) and PeakScanner (Applied Biosystems) were used to score individual genotypes based on the ROX 500 size standard run with each individual with careful manual checking. The final, consensus, genotype comprised the alleles that were observed in at least three of the four replicate reactions for each individual.
Data analyses
We considered all individuals of each of the study species, within the 26 km2 of gallery forest, to be a single but fragmented population. Thus, we treated all individuals genotyped from the same forest fragment—or contiguous fragments in some cases—as comprising a subpopulation because this is how they are currently managed (Mbora, personal observations). This resulted in eight subpopulations for the colobus and seven for the mangabey;—the two species were sympatric in 6 subpopulations, two subpopulations contained only colobus and one only mangabey.
We tested for deviations from Hardy–Weinberg equilibrium (HWE) in each subpopulation of each species for each locus separately, and across loci and subpopulation using GenePop ver. 4.1.3 (Rousset 2008) and found that some of the subpopulations deviated significantly from HWE. Therefore, we used the program MICROCHECKER (van Oosterhout et al. 2004) to determine if the deviations from HWE were due to genotyping errors, allelic drop out, short allele dominance or stutter products (Miller and Waits 2003; Wattier et al. 1998; Shinde et al. 2003), or the presence of null alleles which cause false homozygotes (Shaw et al. 1999). We found that our data did not contain any genotyping errors due to allelic drop out, short allele dominance or stutter products. However, probable microsatellite null alleles were detected in two of the loci (MOGc & D7S2204) for the colobus, and three of the loci (D10S1432, D4S243 & MOGc) for the mangabey. Therefore, we used the program FREENA (Chapuis and Estoup 2007) to estimate the null allele frequencies per locus across all populations. We found that the frequency of null alleles per locus across all populations was relatively low and nearly identical in both species (colobus, mean = 0.02; SD = 0.05; mangabey, mean = 0.02, SD = 0.07). Thus, the frequency of null alleles did not differ between the two species (t = −1.0, df = 223, P = 0.30).
We evaluated the basic patterns of genetic variation between the two species as follows. For each subpopulation of each species, we calculated allelic richness, A n , as the number of alleles corrected for sample size using FSTAT 2.9.3 (Goudet 2001). We then computed the observed (H o ) and expected heterozygosity (H e ), and the inbreeding coefficient (FIS) per locus in each subpopulation of each species using GenePop ver. 4.1.3 (Rousset 2008). We then used two-sample t-tests to compare allelic richness, the inbreeding coefficient, gene diversity and heterozygosity between the two species.
We followed two approaches to investigate the population genetic differentiation of the two species. First, we tested for genic differentiation across all loci among the subpopulations of each species using GenePop ver 4.1.3 (Rousset 2008). We tested the null hypothesis that alleles were drawn from the same distribution in all subpopulations (Raymond and Rousset 1995; Goudet et al. 1996). Second, we investigated the population genetic differentiation by calculating estimates of FST as global values for each species, and as pairwise values between subpopulations, across all loci for each species. The FST estimates were calculated using the software program FREENA because our dataset harbored null alleles (Chapuis and Estoup 2007; Weir 1996). Within FREENA, we implemented the ENA method to correct for the positive bias caused by the presence of null alleles to provide accurate estimation of FST and assessed the significance of mean FST by constructing 95 % confidence intervals. We then evaluated the mean FST values between the two species using a t-test.
The interpretation of genetic differentiation values between species is often questionable because of their dependence on the levels of genetic variation. Therefore, we computed the standardized measure of genetic differentiation G’ST (Hedrick 2005) which is based on Nei’s metric (Nei 1972), as subpopulation pairwise values for each species, as global values for each species and then we used a two sample t-test to compare the mean genetic differentiation between the species.
We further evaluated if dispersal and gene flow were limited in each of the two species in two ways. First, we tested for isolation by distance by assessing the correlation between the genetic and geographic distances matrices for each species using a mantel test (Mantel 1967), and a linear regression of genetic distance against geographic distance between all pairs of populations for each species. Genetic distances were computed as FST/(1 − FST) − FST calculated as above using FREENA—and geographic distances were measured as the natural logarithm of linear centroid-to-centroid distances between forest fragments (Mbora and McPeek 2010).
Second, we performed genetic spatial autocorrelation analyses using GENALEX 6.5 (Peakall and Smouse 2012). GENALEX 6.5 uses a multivariate analysis to simultaneously assess the spatial signal generated by multiple genetic loci (Smouse and Peakall 1999; Smouse et al. 2008). Unlike the isolation by distance analysis which seeks to describe genetic structure across an entire study site, spatial autocorrelation explores the patterns of individual genotypes at a smaller geographic scale. The procedure uses pairwise geographic and pairwise squared genetic distance matrices to generate an autocorrelation coefficient, r, related to Moran’s-I (Sokal and Oden 1978), which measures genetic similarity between individuals whose geographic separation falls within specified distance classes. A significant positive autocorrelation means that individuals within a given distance class are more genetically similar than expected by chance and the results are typically presented in a correlogram (Peakall and Smouse 2012). We performed analyses for each species separately within GENALEX using uneven distance classes. We expected that samples within 2,000 m of each other should exhibit significant positive spatial autocorrelation because this is a typical dispersal distance for both colobus (Marsh 1979, 1981) and mangabey (Homewood 1978).
We used Bayesian clustering with program Structure 2.3.3 (Pritchard et al. 2000; Falush et al. 2003) to evaluate the support for our empirical subpopulation designations. We had Structure cluster our samples into k = 1–10 groups for each species, running it 5 times for each k. All analyses were run for 500,000 steps following a burn-in of 50,000 under a LOCPRIOR model with admixture (Hubisz et al. 2009). All other parameters were left at program defaults, and we evaluated support for different values of k by comparing the mean log-likelihood for each model and by calculating ΔK (Evanno et al. 2005).
Finally, we estimated the contemporary effective population size (Ne) for each species to provide us with a context for understanding whether the levels of genetic variation and patterns population genetic differentiation detected were consistent with their biological traits and population sizes. We used the bias corrected single-sample method based on linkage disequilibrium to estimate Ne (Hill 1981; Waples 2006; Waples and Do 2010) as implemented in NeEstimator V2.01 (Do et al. 2014). We assumed a random mating model, and estimated Ne sizes using a minimum alleles frequency cutoff (Pcrit) of 0.02, which provides an optimal balance between precision and bias across samples (Do et al. 2014). Additionally, 95 % Confidence Intervals were generated using the jackknife approach on loci.
Results
Genetic variation
We were to reliable genotype genotype 146 colobus from eight subpopulations and 76 mangabeys from seven subpopulations drawn from forest fragments across the distribution range of the two species. These numbers of individuals represented 14 and 4 % of the estimated current census populations of the colobus and mangabey respectively.
The study subpopulations were largely in Hardy–Weinberg equilibrium because when we evaluated the alternative hypothesis of heterozygote deficit, we failed to reject the null hypothesis of random union of gametes in 63 of 72 tests in the colobus (86 %) and 51 of 63 cases in the mangabey (81 %). Similarly, when we evaluated the alternative hypothesis of heterozygote excess, we failed to reject the null hypothesis in 61 of 72 cases in the colobus (85 %) and 60 of the 63 tests in the mangabey (95 %).
As we predicted, allelic richness was significantly higher in the mangabey than in the colobus (t = −2.4, DF = 128, P = 0.02; Fig. 1a; Table 1). Consequently, gene diversity was significantly higher in the mangabey than in the colobus (t = −3.2, DF = 114, P = 0.00; Fig. 1b; Table 1), as was the expected heterozygosity (t = −2.8, df = 116, P = 0.00; Fig. 1b; Table 1). Among the colobus subpopulations, the inbreeding coefficient, FIS, ranged from −0.60 to 0.77 (mean = −0.02, CI −0.08 to 0.04) and among mangabey subpopulations it ranged from −0.39 to 0.71 (mean = 0.05, CI −0.02 to 0.13). Therefore, the mean FIS did not deviate significantly from zero for either species, and there was no difference in FIS between the two monkeys (t = −1.6, P = 0.12; Table 1).
There were no differences in allelic richness, gene diversity or heterozygosity east and west of the river, inside or outside TRPNR for either of the species.
Population genetic differentiation
Genetic differentiation for each population pair (exact G test) across all loci was statistically significant in 16 of 28 comparisons in the colobus (57 %) and 15 of 23 comparisons in the mangabey (65 %). The subpopulation pairwise FST values ranged from 0.0 to 0.27 in the colobus and 0.0 to 0.26 in the mangabey, and global estimates of FST revealed a slightly higher level of differentiation among the subpopulations of the mangabey compared to the colobus (mangabey: FST = 0.05; 95 % CI 0.03–0.07; colobus: FST = 0.03; 95 % CI 0.02–0.04). Subpopulation pairwise standardized genetic differentiation, G’ST, ranged from 0.0 to 0.80 in the colobus and 0.0 to 0.95 in the mangabey. And, global estimates of the standardized values of G’ST indicated that differentiation was higher in the mangabey (G’ST = 0.23, 95 % CI 0.18–0.28) than in the colobus (G’ST = 0.13, 95 % CI 0.11–0.15).
We found a positive and statistically significant association between the matrices of pairwise genetic and geographic distances for the mangabey (r = 0.35, P = 0.0) but not for the colobus (r = −0.01, P = 0.5; Mantel 1967). Furthermore, we found a positive linear association between genetic distance (FST/1 − FST) and the natural logarithm of geographic distance between subpopulations for the mangabey (R 2 = 0.16, P = 0.0; Fig. 2b) but not among the colobus (R 2 = 0.0, P = 0.32; Fig. 2a). In both species, samples within 1,500 m of each other exhibited significant positive spatial autocorrelation (Fig. 3). The Bayesian clustering did not support our empirical subpopulation designations because the clustering produced designations that seemed ambiguous and arbitrary.
The estimated effective population size of the mangabey (Ne = 48, CI 32–81) was higher than that of the colobus (Ne = 19, CI 14–25). It should be noted, though, that these estimates of Ne really reflect the effective number of breeders, Nb, that produced this sample (Waples and Do 2010).
Discussion
Genetic variation and differentiation, colobus vs. mangabey
As we expected the two most common measures of genetic variation, multilocus expected heterozygosity (gene diversity, H e ) and allelic richness per locus (A n ) were higher in the mangabey than in the colobus (Fig. 1; Table 1). However, contrary to our expectations, it was the mangabey populations that exhibited higher genetic differentiation as indicated by the standardized measure of G’ST and FST and a significant isolation by distance (Fig. 2).
Given that the two monkeys are of similar body size, have largely congruent distribution within the Tana River forests, and were sympatric in six of the nine subpopulations that we analyzed, differences in body size or distribution range cannot be the reason for the differences in genetic variation (Nevo et al. 1984; Wooten and Smith 1985). So, why does the mangabey exhibit higher genetic diversity than the colobus? We believe that the higher genetic variation in the mangabey stems, in part, from a higher population size (Frankham 1996) because the current census population of the mangabey is estimated to be twice as large as that of the colobus (Wieczkowski and Butynski 2013; Mbora unpublished data). And, our estimates of the contemporary effective population size also showed that the mangabey Ne is more than twice as large as that of the colobus. Paradoxically though, it is the frugivorous mangabey that ought to have a lower population size than the folivorous colobus.
Unlike folivorous monkeys that depend, for the most part, on an abundant and widely distributed food resource in foliage, frugivorous monkeys rely on fruit and similar food types that are patchily distributed over time and space, and are often scarce. Indeed, frugivores generally live in larger social groups than folivores. As such, frugivores require more food resources, use large home ranges and should therefore be highly vulnerable to the loss of food patches and increased edge effects when forest is degraded (Johns and Skorupa 1987; Woodroffe and Ginsberg 1998; Jones et al. 2001). However, highly terrestrial frugivorous primates, like the Tana mangabey, are also known to exhibit considerable ecological flexibility (Wieczkowski and Butynski 2013; Kinnaird 1992; Homewood 1978; Fimbel 1994). And when food availability decreases, the mangabey increases the time it spends foraging, the total distance it moves, the area over which it forages per day and the diversity of items that it consumes (Homewood 1978). Therefore, it is plausible that faced with the degradation of the forest habitat, the mangabey adjusts to the changes by moving greater distances to forage more widely to satisfy its food requirements, and is therefore able to maintain a comparatively larger population size.
In contrast to the mangabey, no ecological flexibility is evident among the Tana colobus (Marsh 1981). In this canopy dwelling monkey, the population abundance, range use and probability of occupying a forest fragment are all strongly associated with the distribution and the abundance of its main food tree species (Mbora and Meikle 2004; Marsh 1981). Therefore, the colobus population abundance is particularly vulnerable to diminution by the removal of important food tree species due to forest habitat change.
So why does the mangabey exhibit greater genetic differentiation, and isolation by distance, in contrast to the colobus? We believe that the greater genetic differentiation in the mangabey stems from its pattern of natal dispersal, while the IBD stems from the dynamics of social group formation over time. The Tana mangabey, exhibits the male biased pattern of natal dispersal typical of most old-world (cercopithecine) monkeys (Pusey and Packer 1987). This, in combination with the fact that social groups in this species are composed mostly of females (Wieczkowski and Butynski 2013; Kinnaird 1992; Homewood 1978), implies that there should be close genetic relatedness within groups but high genetic differentiation among groups. Furthermore, new groups in cercopithecine monkeys are typically formed by the fissioning of existing groups along matrilines when environmental conditions are stressful (for example, food shortage in Macaca sinica: Dittus 1988). And following group fissioning, daughter groups are characterized by a higher average level of within-group relatedness than the parent group (Melnick and Kidd 1983; Whitlock and McCauley 1990). Thus, because cercopithecine monkey social groups are generally characterized by low within group genetic diversity, group fissioning followed by colonization of new areas should lead to a pattern of isolation by distance (Melnick and Hoelzer 1996; Wright 1943).
In contrast to the mangabey, the Tana River red colobus is one of a handful of old-world primate species in which both males and females repeatedly transfer among social groups (Pusey and Packer 1987). This pattern of natal dispersal should generally homogenize genetic relatedness, and reduce differentiation among social groups, just as we found in this species. Thus, in the Tana River red colobus, habitat change may not be constraining dispersal as much as we expected, or the effects of habitat change on dispersal are not yet detectable at the level of population genetics.
Genetic variation in endangered endemic species
Endemic species are expected to exhibit low genetic variation because of their restricted ranges (Nevo et al. 1984), and endangered species should have low genetic variation due to diminished populations (Frankham 1995). Accordingly, endemic endangered species, such as we studied, should exhibit disproportionately low genetic variation. However, while our study species exhibited low levels of allelic richness with an average of less than two alleles per locus, they also had relatively high average heterozygosity (Table 1). Now, as is well established, a population that has passed through a bottleneck in the effective number of breeders should have less genetic diversity than the amount it contained before experiencing the bottleneck (Nei et al. 1975; Leberg 1992). However, because rare alleles are more readily affected by drift than more common ones are, population bottlenecks typically decrease allelic richness much more than they do heterozygosity (Nei et al. 1975). As such, the low allelic richness we found, coupled with the low effective population sizes, is evidence that habitat change has taken a toll on the genetic variation of these species. Nonetheless, the high levels of heterozygosity (Table 1) also suggest that the current low census populations of these species are probably a recent occurrence (Mbora and McPeek 2010) because population bottlenecks of short duration generally have little effect on heterozygosity (Allendorf 1986; Nei et al. 1975).
Heterozygosity is often used to compare genetic diversity among species (Primack 2010; Evans and Sheldon 2008; Garner et al. 2005) and therefore we can compare the heterozygosity of our study species to that of other mammal groups. The colobus had a mean heterozygosity of 0.70 (±0.11), the mangabey 0.76 (±0.14), and taken together the two monkeys had a mean heterozygosity of 0.73 (±0.13). These mean heterozygosities compare quite favorably with the mean heterozygosities documented across all placental mammals at 0.68 (±0.01) and across all primates as a group at about 0.72 (±0.06; Garner et al. 2005). Indeed, our two species had a much higher mean heterozygosity than that found in demographically challenged mammal populations across the globe which have a mean heterozygosity of 0.50 (±0.03) (Garner et al. 2005).
We are well aware that the direct comparisons of genetic variation among species are often obfuscated by factors such as ascertainment biases, as well as demographic and genetic stochasticity. However, we believe that these comparisons are useful because they provide important baseline information for conservation purposes (Primack 2010; Evans and Sheldon 2008; Garner et al. 2005). Indeed, average heterozygosity provides a good measure of the capability of a population to respond to selection immediately following a bottleneck (Petit et al. 1998; Allendorf 1986). Therefore, our findings suggest that these two, and other species like them, should not be discounted from conservation action because of their current small populations because they have the potential to recover, genetically. However, our findings also suggest that given the low allelic richness, immediate concerted conservation efforts are needed to save these species and others like them because the number of alleles remaining is important for populations’ long-term responses to selection, and therefore survival of a species (Allendorf 1986).
In conclusion, our results suggest that the Tana colobus and mangabey exhibit differential vulnerabilities to genetic erosion by habitat change, possibly due to divergent behavioral ecology. On the one hand, the terrestrial mangabey probably uses its lomocotor and dietary versatility to adjust to changes in the forest habitat and maintain a larger population in which there is greater gene flow. On the other hand, the population of arboreal colobus is compromised by the loss of food canopy tree species. These findings are consistent with studies focused on other species and showing that biological traits, rather than stochastic processes, determine persistence or extinction of species facing different threats (Henle et al. 2004; Turner 1996). Indeed, case studies show that habitat loss and fragmentation often represent dissimilar levels of extinction risk to different species in beetles (Davies et al. 2004), non-human primates (Isaac and Cowlishaw 2004), birds (Owens and Bennett 2000), geckos (Hoehn et al. 2007) and many other species (Henle et al. 2004). In this context, our study gets at a fundamental mechanism by which habitat loss and fragmentation may elevate extinction risk for some species and not others because reduced genetic variation undermines individual fitness, population persistence, and ultimately the capacity of any species to adapt and evolve in response to environmental change.
References
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Beja-Pereira A, Oliveira R, Schwartz MK, Luikart G (2009) Advancing ecological understanding through technological transformation in noninvasive genetics. Mol Ecol Res 9:1279–1301
Bonhomme M, Blancher A, Crouau-Roy B (2005) Multiplexed microsatellites for rapid identification and characterization of individuals and populations of cercopithecidae. Am J Primatol 67:385–391
Bonhomme M, Blancher A, Cuarteros S, Chikhi L, Crouau-Roy B (2008) Origin and number of founders in an introduced insular primate: estimation from nuclear genetic data. Mol Ecol 17:1009–1019
Butynski TM, Mwangi G (1994) Conservation status and distribution of the Tana River red colobus and crested mangabey for Zoo Atlanta, Kenya Wildlife Service, National Museums of Kenya, Institute of Primate Research, and East African Wildlife Society, p 68
Chamberlain JS, Gibbs RA, Rainer JE, Nguyen PN, Thomas C (1988) Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 16:11141–11156
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Clobert J, Danchin E, Dhondt AA (2001) Dispersal. Oxford University Press, Oxford
Cowlishaw G, Dunbar R (2000) Primate conservation biology. University of Chicago Press, Chicago, Illinois
Davies KF, Margules CR, Lawrence JF (2004) A synergistic effect puts rare, specialized species at greater risk of extinction. Ecology 85:265–271
DeFries R, Hansen A, Newton AC, Hansen MC (2005) Increasing isolation of protected areas in tropical forests over the past twenty years. Ecol Appl 15:19–26
Dittus WPJ (1988) Group fission among wild toque macaques as a consequence of female resource competition and environmental stress. Anim Behav 36:1626–1645
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator V2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Evans SR, Sheldon BC (2008) Interspecific patterns of genetic diversity in birds: correlations with extinction risk. Conserv Biol 22:1016–1025
Fa JE, Purvis A (1997) Body size, diet and population density in Afrotropical forest mammals: a comparison with Neotropical species. J Anim Ecol 66:98–112
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fernando P, Vidya TNC, Rajapakse C, Dangolla A, Melnick DJ (2003) Reliable noninvasive genotyping: fantasy or reality. J Hered 94:115–123
Fimbel C (1994) Ecological correlates of species success in modified habitats may be disturbance specific and site specific—the primates of Tiwai Island. Conserv Biol 8:106–113
Frankham R (1995) Conservation genetics. Annu Rev Genet 29:305–327
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Frankham R, Ballou JD, Briscoe DA (2010) Introduction to conservation genetics, 2nd edn. Cambridge University Press, Cambridge, UK
Garner A, Rachlow JL, Hicks JF (2005) Patterns of genetic diversity and its loss in mammalian populations. Conserv Biol 19:1215–1221
Gerloff U, Schlotterer C, Rassmann K, Rambold I, Hohmann G, Fruth B, Tautz D (1995) Amplification of hypervariable simple sequence repeats (microsatellites) from excremental DNA of wild living Bonobos (Pan paniscus). Mol Ecol 4:515–518
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www2.unil.ch/popgen/softwares/fstat.htm. Institute of Ecology, Lausanne
Goudet J, Raymond M, Demeeüs T, Rousset F (1996) Testing genetic differentiation in diploid populations. Genetics 144:1933–1940
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Henle K, Davies K, Kleyer M, Margules C, Settele J (2004) Predictors of species sensitivity to fragmentation. Biodivers Conserv 13:207–251
Hill WG (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res 38:209–216
Hoehn M, Sarre SD, Henle K (2007) The tales of two geckos: does dispersal prevent extinction in recently fragmented populations? Mol Ecol 16:3299–3312
Homewood KM (1978) Feeding strategy of the Tana mangabey (Cercocebus galeritus galeritus) (Mammalia: Primates). J Zool Lond 186:375–391
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Hughes FMR (1990) The influence of flooding regimes on forest distribution and composition in the Tana River floodplain, Kenya. J Appl Ecol 27:475–491
Isaac NJB, Cowlishaw G (2004) How species respond to multiple extinction threats. Proc R Soc Lond B 271:1471–2954
Johns AD, Skorupa JP (1987) Responses of rain-forest primates to habitat disturbance—a review. Int J Primatol 8:157–191
Jones KE, Barlow KE, Vaughan N, Rodriguez-Duran A, Gannon MR (2001) Short-term impacts of extreme environmental disturbance on the bats of Puerto Rico. Anim Conserv 4:59–66
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Kinnaird MF (1992) Variable resource defense by the Tana River crested mangabey. Behav Ecol Sociobiol 31:115–122
Laurance WF (2007) Have we overstated the tropical biodiversity crisis? Trends Ecol Evol 22:65–70
Leberg PL (1992) Effects of population bottlenecks on genetic diversity as measured by allozyme electrophoresis. Evolution 46:477–494
Mace GM, Balmford A (2000) Patterns and processes in contemporary mammalian extinction. In: Entwhistle A, Dunstone N (eds) Priorities for the conservation of mammalian diversity: has the panda had its day?. Cambridge University Press, Cambridge, pp 27–52
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Can Res 27:209–220
Marsh CW (1979) Female transference and mate choice among Tana River red colobus. Nature 281:568–569
Marsh CW (1981) Ranging behavior and its relation to diet selection in Tana River red colobus (Colobus badius ruformitratus). J Zool 195:473–492
Mayaux P, Holmgren P, Achard F, Eva H, Stibig HJ, Branthomme A (2005) Tropical forest cover change in the 1990 s and options for future monitoring. Philos Trans R Soc Lon B 360:373–384
Mbora DNM, Meikle DB (2004) Forest fragmentation and the distribution, abundance and conservation of the Tana River Red Colobus (Procolobus rufomitratus). Biol Conserv 118:67–77
Mbora DNM, McPeek MA (2010) Endangered species in small habitat patches can possess high genetic diversity: the case of the Tana River red colobus and mangabey. Conserv Genet 11:1725–1735
Melnick DJ, Hoelzer GA (1996) The population genetic consequences of macaque social organization and behaviour. In: Fa JE, Lindburg DG (eds) Evolution and ecology of macaque societies. Cambridge University Press, Cambridge, pp 413–443
Melnick DJ, Kidd KK (1983) The genetic consequences of social group fission in a wild population of rhesus monkeys (Macaca mulatta). Behav Ecol Sociobiol 12:229–236
Miller CR, Waits LP (2003) The history of effective population size and genetic diversity in the Yellowstone grizzly (Ursus arctos): implications for conservation. Proc Nat Acad Sci USA 100:4334–4339
Mittermeier RA, Ratsimbazafy J, Rylands AB, Williamson L, Oates JF, Mbora DNM, Ganzhorn JU, Rodriguez-Luna E, Palacios E, Heymann EW et al (2007) Primates in peril: the world’s 25 most endangered primates, 2006-2008. Primate Conserv 22:1–40
Morin PA, Chambers KE, Boesh C, Vigilant L (2001) Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Mol Ecol 10:1835–1844
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nevo E, Bieles A, Ben-Shlomo R (1984) The evolutionary significance of genetic diversity: ecological, demographic and life history correlates. In: Mani GS (ed) Evolutionary dynamics of genetic diversity. Springer, Berlin, pp 13–213
Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L (2004) Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol Ecol 13:2089–2094
Owens IPF, Bennett PM (2000) Ecological basis of extinction risk in birds: habitat loss versus human persecution and introduced predators. Proc Natl Acad Sci USA 97:12144–12148
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Petit R, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12(844):855
Primack RB (2010) Essentials of conservation biology, 5th edn. Sinauer Associates, Inc
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pusey AE, Packer C (1987) Dispersal and philopatry. In: Smuts BB, Cheny DL, Seyfarth RM, Wrangham RW, Struhsaker TT (eds) Primate societies. University of Chicago, Chicago, pp 250–266
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Reed KE, Fleagle J (1995) Geographic and climate control of primate diversity. Proc Nat Acad Sci USA 92:7874–7876
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Shaw PW, Pierce GJ, Boyle PR (1999) Subtle population structuring within a highly vagile marine invertebrate, the veined squid Loligo forbesi, demonstrated with microsatellite DNA markers. Mol Ecol 8:407–417
Shinde D, Lai YL, Sun FZ, Arnheim N (2003) Taq DNA polymeraseslippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)(n) and (A/T)(n) microsatellites. Nucleic Acid Res 31:974–980
Simberloff D (1998) Flagships, umbrellas, and keystones: is single-species management passé in the landscape era? Biol Conserv 83:247–257
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82:561–573
Smouse PE, Peakall R, Gonzales E (2008) A heterogeneity test for fine-scale genetic structure. Mol Ecol 17:3389–3400
Sokal RR, Oden NL (1978) Spatial autocorrelation in biology. 1. Methodology. Biol J Linn Soc 10:199–228
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits PL, Bouvet J (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24:3189–3194
Taberlet P, Waits L, Luikart G (1999) Non-invasive genetic sampling: look before you leap. Trends Ecol Evol 14:323–327
Turner IM (1996) Species loss in fragments of tropical rain forest: a review of the evidence. J Appl Ecol 33:200–209
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262
Wattier R, Engel CR, Saumitou-Laprade P, Valero M (1998) Short allele dominance as a source of heterozygote deficiency at microsatellite loci: experimental evidence at the dinucleotide locus Gv1CT in Gracilaria gracilis (Rhodophyta). Mol Ecol 7:1569–1573
Wehausen JD, Ramey RR, Epps CW (2004) Experiments in DNA extraction and PCR amplification from bighorn sheep feces: the importance of DNA extraction method. J Hered 95:503–509
Weir BS (1996) Genetic data analysis II. Sinauer Associates, Sunderland, Massachusetts
Whitesides GH, Oates JF, Green SM, Kluberdanz RP (1988) Estimating primate densities from transects in a West African rainforest: a comparison of techniques. J Anim Ecol 57:345–367
Whitlock MC, McCauley DE (1990) Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44:1717–1724
Wieczkowski J, Butynski TM (2013) Cercocebus galeritus. In: Kingdon J, Happold D, Butynski TM (eds) The mammals of Africa. Bloomsbury Publishing, London
Woodroffe R, Ginsberg JR (1998) Edge effects and the extinction of populations inside protected areas. Science 280:2126–2128
Wooten MC, Smith MH (1985) Large mammals are genetically less variable? Evolution 39:210–212
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
We are grateful to our dedicated field assistants; Abio Gafo, Michael Morowa, John Kokani, Sylvia Isaya, Mary Galana, Hiribae Galana, Galana Galole, Bakari Kawa, and Mohamed Kawa. We are also indebted to three student assistants who helped with the research in the laboratory, Janel diBiccari at Dartmouth College, and Amanda Edwards and Melissa Lam at Whittier College. We further thank Drs. T. L. Goldberg and N. Ting for providing tissue samples and genomic DNA from Procolobus tephrosceles and Procolobus badius, and the anonymous reviewers for constructive feedback that greatly improved this paper. This work was funded by: The Rufford Foundation, Margot Marsh Biodiversity Foundation, National Geographic Society, and Dartmouth College. Permission to conduct research was granted by the government of Kenya via permit number: MOEST13/001/35C417/2 to DNMM.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mbora, D.N.M., McPeek, M.A. How monkeys see a forest: genetic variation and population genetic structure of two forest primates. Conserv Genet 16, 559–569 (2015). https://doi.org/10.1007/s10592-014-0680-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0680-2